

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.3 + phase: 0
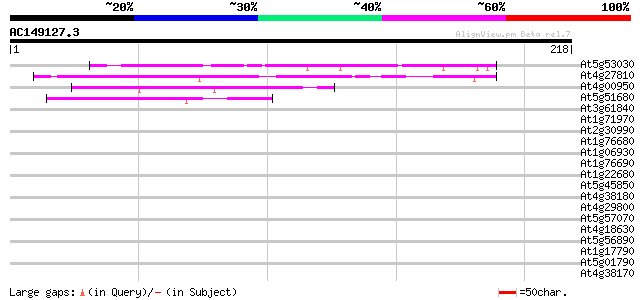
(218 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g53030 unknown protein 87 5e-18
At4g27810 hypothetical protein 72 2e-13
At4g00950 unknown protein 58 3e-09
At5g51680 unknown protein 41 4e-04
At3g61840 unknown protein 39 0.002
At1g71970 unknown protein 37 0.008
At2g30990 hypothetical protein 36 0.014
At1g76680 12-oxophytodienoate reductase (OPR1) 33 0.089
At1g06930 hypothetical protein 33 0.12
At1g76690 12-oxophytodienoate reductase (OPR2) 32 0.20
At1g22680 hypothetical protein 32 0.20
At5g45850 putative protein 32 0.26
At4g38180 unknown protein (At4g38180) 32 0.26
At4g29800 unknown protein 32 0.26
At5g57070 putative protein 32 0.34
At4g18630 unknown protein 31 0.44
At5g56890 unknown protein 31 0.58
At1g17790 unknown protein (At1g17790) 31 0.58
At5g01790 putative protein 30 0.98
At4g38170 hypothetical protein 30 0.98
>At5g53030 unknown protein
Length = 245
Score = 87.4 bits (215), Expect = 5e-18
Identities = 71/193 (36%), Positives = 94/193 (47%), Gaps = 46/193 (23%)
Query: 32 TRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELREREVNNVVRALELPPRLL 91
T PG ++ PP S+PF WEEAPGKPR L ++ VVR+LELPPRL+
Sbjct: 33 TTPGLAT-----PPVNIAGSVPFLWEEAPGKPRRVKKPARLNQK---GVVRSLELPPRLV 84
Query: 92 SLERESTGNIYAPSPTTVLEGPY--------VGRAVSFTTSYRD-----NNNKDSVNFGS 138
L EST + PSPTTVL+GPY + R+ + R ++ + GS
Sbjct: 85 -LPGEST-TVNEPSPTTVLDGPYDLRRRSLSLPRSAAVIRKLRGVPAPAPEKEERLVGGS 142
Query: 139 SRWGGLKKNNRIDREGSFDFSSWSVEGGD------------------KVKITRVKRRGSF 180
SRWG + EG FDFS + +G D KVK+ R+ ++GSF
Sbjct: 143 SRWGSFGNCKEVS-EGIFDFSRFRDDGYDCRRDWAGGGGVGNFAGDAKVKLYRIIKKGSF 201
Query: 181 ---SHGT-SHFWV 189
SH T S FW+
Sbjct: 202 FNLSHTTKSDFWL 214
>At4g27810 hypothetical protein
Length = 182
Score = 72.0 bits (175), Expect = 2e-13
Identities = 67/189 (35%), Positives = 81/189 (42%), Gaps = 32/189 (16%)
Query: 10 DHDDELRSNYDTPKKLSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTR 69
+ DD+ R + KL L +I + PP S+PF WEEAPGKPR
Sbjct: 4 EFDDQER--FCATPKLPLFSIPFNRACDTPGLATPPVNIAGSVPFLWEEAPGKPRVSDEN 61
Query: 70 PEL------REREVNNVVRALELPPRLLSLERESTGNIYAPSPTTVLEGPYVGRAVSFTT 123
L RE VVR LELPPRL + PSPTTVL+GPY S +
Sbjct: 62 KPLASKQNEREGGGGGVVRCLELPPRLFFPADDE------PSPTTVLDGPYDVPRRSLSV 115
Query: 124 SYRDNNNKDSVNFGSSRWGGLKKNNRIDREGSFDFSSWSVEGGDKVKITRVKRRGS---F 180
R + F SR N+R G GG VKI+RV+R+GS
Sbjct: 116 IRRSERASEG-RFEFSR----STNSRCCDGG----------GGTTVKISRVRRKGSLLNL 160
Query: 181 SHGTSHFWV 189
SH S F V
Sbjct: 161 SHSKSQFLV 169
>At4g00950 unknown protein
Length = 291
Score = 58.2 bits (139), Expect = 3e-09
Identities = 38/112 (33%), Positives = 53/112 (46%), Gaps = 15/112 (13%)
Query: 25 LSLPTILTRPGFSSEAPTPPPRTTV-ISIPFKWEEAPGKPRSCHTRPELREREVN----- 78
+ LP + T+P S + + P +++ S+PF WEE PGKP+ T
Sbjct: 16 MKLPVLPTKPNTHSHSMSSPIHSSISASVPFSWEEEPGKPKQHSTSSSSSSSSSPLTSYS 75
Query: 79 ----NVVRALELPPRLLSLERESTGNIYAPSPTTVLEGPYVGRAVSFTTSYR 126
++LELPPRL LE++ SP TV +GPY S TTS R
Sbjct: 76 SSPFETHKSLELPPRLHLLEKDGGSVTKLHSPITVFDGPY-----SMTTSKR 122
>At5g51680 unknown protein
Length = 343
Score = 41.2 bits (95), Expect = 4e-04
Identities = 27/91 (29%), Positives = 43/91 (46%), Gaps = 12/91 (13%)
Query: 15 LRSNYDTPKKLSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCH---TRPE 71
L ++ + P +L P + T P P P V S+PF WEE PG+P C + PE
Sbjct: 77 LPASSNDPPQLPHPPLETATPTPLPPPVPVPVKQVTSVPFDWEETPGQPYPCFVDTSPPE 136
Query: 72 LREREVNNVVRALELPPRLLSLERESTGNIY 102
L ++ LPP + + E++ +I+
Sbjct: 137 LLDQ---------PLPPPPMYGDVETSSDIF 158
Score = 37.7 bits (86), Expect = 0.005
Identities = 21/46 (45%), Positives = 25/46 (53%), Gaps = 6/46 (13%)
Query: 22 PKKLSLPTILTRPGFSSEAPTPPPRTT----VISIPFKWEEAPGKP 63
PKK P++ T S P PPP V S+PF+WEE PGKP
Sbjct: 33 PKKNWQPSLATF--VPSPPPLPPPIPVPVKLVTSVPFRWEETPGKP 76
Score = 28.1 bits (61), Expect = 3.7
Identities = 13/26 (50%), Positives = 16/26 (61%), Gaps = 3/26 (11%)
Query: 42 TPPPRTTVI---SIPFKWEEAPGKPR 64
T PPR + S+PF WEE PG P+
Sbjct: 9 TKPPRKQLRQPPSVPFIWEERPGFPK 34
>At3g61840 unknown protein
Length = 211
Score = 38.9 bits (89), Expect = 0.002
Identities = 43/155 (27%), Positives = 57/155 (36%), Gaps = 42/155 (27%)
Query: 25 LSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELREREVNNVVRAL 84
LS P + T+ S +P + SIPF WEE PGKP+ P + + L
Sbjct: 78 LSSP-VTTKDVSLSLMSSPDDSLSRASIPFSWEEEPGKPKHHVRHP--------SYSKCL 128
Query: 85 ELPPRLLSLERESTGNIYAPSPTTVLEGPYVGRAVSFTTSYRDNNNKDSVNFGSSRWGGL 144
+LPPR+L + + P T G Y+ G RW
Sbjct: 129 DLPPRMLFPDE------FTKMPRTANHGRYLA--------------------GWKRWFRW 162
Query: 145 KKNNRIDR----EGSFDFSSWSVEGGDKVKITRVK 175
KK D GS+ + S E D K TR K
Sbjct: 163 KKERDGDAYVAGSGSYIYPS---EDEDDTKTTRTK 194
>At1g71970 unknown protein
Length = 225
Score = 37.0 bits (84), Expect = 0.008
Identities = 20/56 (35%), Positives = 30/56 (52%), Gaps = 5/56 (8%)
Query: 36 FSSEAPTPPPRTTV--ISIPFKWEEAPGKPRS-CHTRPELREREVNNVVRALELPP 88
F ++P P T + + +PF WE+ PGKP+ H R E +N++ L LPP
Sbjct: 49 FPGDSPLNSPATPLRLLGVPFSWEQLPGKPKDYSHRLNNRRNNESSNLL--LPLPP 102
>At2g30990 hypothetical protein
Length = 581
Score = 36.2 bits (82), Expect = 0.014
Identities = 37/137 (27%), Positives = 63/137 (45%), Gaps = 16/137 (11%)
Query: 34 PGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELREREVNNVVRALELPP---RL 90
P + S+ + P R ++PF+WE PGKP+ +P L+ + V +LPP R+
Sbjct: 43 PVYKSDIKSGPVRNPG-TVPFQWEHKPGKPKD-ERKPVLQSFVEPHFVP--KLPPGRERV 98
Query: 91 LSLER--ESTGNIYAPSPTTVLEGPYVGRAVSFTTSYRDNNNKDSVNFGSSRWGGLKKNN 148
+ L R ESTG + + + V A S ++ Y D+++ + L +
Sbjct: 99 VELGRKPESTGADHQTKTVSSSDKYLVEDAKSNSSRYDDDDDDSDGTY-------LDATD 151
Query: 149 RIDREGSFDFSSWSVEG 165
+ R SF F+ +V G
Sbjct: 152 TLSRTESFFFNCSAVSG 168
>At1g76680 12-oxophytodienoate reductase (OPR1)
Length = 372
Score = 33.5 bits (75), Expect = 0.089
Identities = 24/87 (27%), Positives = 38/87 (43%), Gaps = 2/87 (2%)
Query: 63 PRSCHTRPELREREVNNVVRALELPPRLLSLER-ESTGNIYAPSPTTVLEGPYVGRAVSF 121
P SC +P + + N + AL PPR L +E N + + +E + G +
Sbjct: 125 PISCSDKPLMPQIRSNGIDEALFTPPRRLGIEEIPGIVNDFRLAARNAMEAGFDGVEIHG 184
Query: 122 TTSYR-DNNNKDSVNFGSSRWGGLKKN 147
Y D KD+VN + +GG +N
Sbjct: 185 ANGYLIDQFMKDTVNDRTDEYGGSLQN 211
>At1g06930 hypothetical protein
Length = 169
Score = 33.1 bits (74), Expect = 0.12
Identities = 30/116 (25%), Positives = 47/116 (39%), Gaps = 7/116 (6%)
Query: 51 SIPFKWEEAPGKPRSCHTRPEL----REREVNNVVRALELPPRLLSLERESTGNIYAPSP 106
++PFKWE PG PR R + + N V A PP S+ +P
Sbjct: 31 AVPFKWESQPGTPRRLSKRSSSSGFNSDSDFNFPVSAPLTPPPSYFYASPSSTKHVSPKK 90
Query: 107 TTVLEGPYVGRAVSFTTSYRDNNNKDSVNFGSSRWGGLKKNNRIDREGSFDFSSWS 162
T L + + S +S +++ S + SS L+ ++ +R S F S S
Sbjct: 91 TNTLFSSLLSKNRSVPSSPASSSSSSSSSVPSS---PLRTSDLSNRRRSMWFESGS 143
>At1g76690 12-oxophytodienoate reductase (OPR2)
Length = 374
Score = 32.3 bits (72), Expect = 0.20
Identities = 24/87 (27%), Positives = 37/87 (41%), Gaps = 2/87 (2%)
Query: 63 PRSCHTRPELREREVNNVVRALELPPRLLSLER-ESTGNIYAPSPTTVLEGPYVGRAVSF 121
P SC +P + + N + A PPR LS+E N + + +E + G +
Sbjct: 127 PISCTGKPIMPQMRANGIDEARFTPPRRLSIEEIPGIVNDFRLAARNAMEAGFDGVEIHG 186
Query: 122 TTSYR-DNNNKDSVNFGSSRWGGLKKN 147
Y D KD VN + +GG +N
Sbjct: 187 AHGYLIDQFMKDKVNDRTDEYGGSLQN 213
>At1g22680 hypothetical protein
Length = 185
Score = 32.3 bits (72), Expect = 0.20
Identities = 20/55 (36%), Positives = 27/55 (48%), Gaps = 11/55 (20%)
Query: 36 FSSEAPTPPPRTTVISIPFKWEEAPGKPR--SCHTRPELREREVNNVVRALELPP 88
FS +P P + IPF WE+ PGKP+ S H + E ++ L LPP
Sbjct: 42 FSGNSPATPLHP--LGIPFSWEKLPGKPKEFSYHRKNE-------SLSNLLPLPP 87
>At5g45850 putative protein
Length = 444
Score = 32.0 bits (71), Expect = 0.26
Identities = 13/36 (36%), Positives = 24/36 (66%), Gaps = 5/36 (13%)
Query: 51 SIPFKWEEAPGKPRSCHTRPELREREVNNVVRALEL 86
S+PFKWE+ PGKP+ + E +++++A+E+
Sbjct: 72 SVPFKWEQTPGKPKD-----KKNLVEESDLIKAMEM 102
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 32.0 bits (71), Expect = 0.26
Identities = 12/35 (34%), Positives = 21/35 (59%)
Query: 94 ERESTGNIYAPSPTTVLEGPYVGRAVSFTTSYRDN 128
E +S GN++ P +++ + G V+F T+YR N
Sbjct: 275 EDQSVGNVFWADPKAIMDFTHFGDTVTFDTTYRSN 309
>At4g29800 unknown protein
Length = 525
Score = 32.0 bits (71), Expect = 0.26
Identities = 52/178 (29%), Positives = 68/178 (37%), Gaps = 24/178 (13%)
Query: 19 YDTPKKLSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELREREVN 78
Y+ P+ L +P RPG S P+P T + PG P S P R ++
Sbjct: 71 YEDPRLLWIPQSPLRPGDSEAGPSPRSPLTPNGVVL-----PGTPSSSFRSPRGRICVLS 125
Query: 79 ---NVVRALELPPRLLSLE---RESTG--NIYAPSPTTVLEGPYVG---RAVSFTTSYRD 127
+R L L+ LE +E +G N V G VG A+ F T RD
Sbjct: 126 IDGGGMRGLLAGKSLIYLEQMLKEKSGDPNARIADYFDVAAGSGVGGVFAAMIFAT--RD 183
Query: 128 NNNKDSVNFGSSRWGGLKKNNRIDREGSFDFSSWSVEGGDKVKITRVKRRGSFSHGTS 185
N + W L +N EG + S S GG I RV R GS S +S
Sbjct: 184 GNR--PIFKAEDTWKFLVEN----AEGFYRSGSGSGGGGAGAAIKRVIRSGSGSGSSS 235
>At5g57070 putative protein
Length = 607
Score = 31.6 bits (70), Expect = 0.34
Identities = 19/65 (29%), Positives = 27/65 (41%), Gaps = 3/65 (4%)
Query: 14 ELRSNYDTPKKLSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELR 73
E+ PK++ + T + +P S P PP T P E P KPR H R
Sbjct: 221 EIEEEESEPKEIQIDTFVVKP---SSPPQQPPATPPPPPPPPPVEVPQKPRRTHRSVRNR 277
Query: 74 EREVN 78
+ + N
Sbjct: 278 DLQEN 282
>At4g18630 unknown protein
Length = 479
Score = 31.2 bits (69), Expect = 0.44
Identities = 22/61 (36%), Positives = 28/61 (45%), Gaps = 8/61 (13%)
Query: 4 ENNKKPDHDDELRSNYDTPKKLSLPTILTRPGFSSEAPTPPPRTTVISIPFKWEEAPGKP 63
EN K ELRS T ++ + P S + T P SIPF WE+ PGKP
Sbjct: 29 ENKKTTITRPELRSCPSTKQETP---VFVLPDQSFDHLTEPA-----SIPFMWEQIPGKP 80
Query: 64 R 64
+
Sbjct: 81 K 81
>At5g56890 unknown protein
Length = 1113
Score = 30.8 bits (68), Expect = 0.58
Identities = 23/92 (25%), Positives = 39/92 (42%), Gaps = 7/92 (7%)
Query: 22 PKKLSLPTIL-TRPGFS------SEAPTPPPRTTVISIPFKWEEAPGKPRSCHTRPELRE 74
P K S P+I + P F+ S + TPPP +T ++ P S H +
Sbjct: 294 PHKASPPSIAPSAPKFNRHSHHTSPSTTPPPDSTPSNVHHHPSSPSPPPLSSHHQHHQER 353
Query: 75 REVNNVVRALELPPRLLSLERESTGNIYAPSP 106
+++ + LPP L+S ++ + P P
Sbjct: 354 KKIADSPAPSPLPPHLISPKKSNRKGSMTPPP 385
>At1g17790 unknown protein (At1g17790)
Length = 487
Score = 30.8 bits (68), Expect = 0.58
Identities = 26/73 (35%), Positives = 31/73 (41%), Gaps = 9/73 (12%)
Query: 27 LPTILTRPGFSSEAPTPPPRTTVISIPFK-WEE-----APGKPRSCHTRPELRERE---V 77
LP I+ P SS P PPP + + WE P +P + T PE E E V
Sbjct: 266 LPAIVPSPSPSSPPPPPPPPVAAPVLENRTWEREESMTIPVEPEAVITAPEKAEEEEAPV 325
Query: 78 NNVVRALELPPRL 90
NN LE RL
Sbjct: 326 NNRDLTLEEKRRL 338
>At5g01790 putative protein
Length = 188
Score = 30.0 bits (66), Expect = 0.98
Identities = 36/129 (27%), Positives = 50/129 (37%), Gaps = 34/129 (26%)
Query: 51 SIPFKWEEAPGKPRSCHTRPELREREVNNVVRALELPPRLLSL--------ERESTGNIY 102
++PF+WE PG P+ H EL + L PP S ++ST I
Sbjct: 54 AVPFEWESHPGTPK--HPSSEL------PTLPPLTPPPSHFSFSGDQIRRRSKKSTKKIL 105
Query: 103 APSPTTV--LEGPYVGRAVSFTTS---------YRDNNNKDSVNFGSSRWGGLKKNNRID 151
A PT + L G + G+A + S D N D F + R N +
Sbjct: 106 ALIPTRLFWLSGDHNGKAKKLSASSPPSLSERVLIDENEYDLFKFQTER-------NVMR 158
Query: 152 REGSFDFSS 160
R SFD S+
Sbjct: 159 RFSSFDSSA 167
>At4g38170 hypothetical protein
Length = 531
Score = 30.0 bits (66), Expect = 0.98
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Query: 82 RALELPPRLLSLERESTGNIYAPSPTTVLEGPYVGRAVSFTTSYR 126
R LE P L ++E + GN++ PT L Y G + F T+YR
Sbjct: 14 RQLENPGFLYAIE-DDCGNVFWADPTCRLNYTYFGDTLVFDTTYR 57
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,799,431
Number of Sequences: 26719
Number of extensions: 271048
Number of successful extensions: 901
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 864
Number of HSP's gapped (non-prelim): 51
length of query: 218
length of database: 11,318,596
effective HSP length: 95
effective length of query: 123
effective length of database: 8,780,291
effective search space: 1079975793
effective search space used: 1079975793
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC149127.3


