

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149127.2 + phase: 0
(458 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
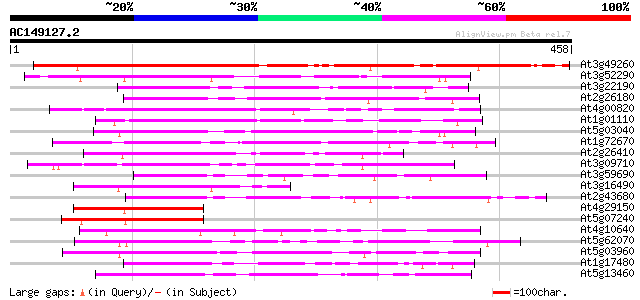
Score E
Sequences producing significant alignments: (bits) Value
At3g49260 SF16 -like protein 360 e-100
At3g52290 unknown protein (At3g52290) 175 4e-44
At3g22190 unknown protein 165 4e-41
At2g26180 putative SF16 protein {Helianthus annuus} 160 2e-39
At4g00820 unknown protein 145 5e-35
At1g01110 unknown protein 139 2e-33
At5g03040 unknown protein 137 9e-33
At1g72670 unknown protein 134 1e-31
At2g26410 putative SF16 protein {Helianthus annuus} 132 4e-31
At3g09710 putative SF16 protein 131 7e-31
At3g59690 unknown protein 127 1e-29
At3g16490 putative calmodulin-binding protein 120 1e-27
At2g43680 SF16 protein {Helianthus annuus} like protein 118 6e-27
At4g29150 unknown protein (At4g29150) 115 7e-26
At5g07240 unknown protein 114 9e-26
At4g10640 putative protein 114 9e-26
At5g62070 unknown protein 113 2e-25
At5g03960 unknown protein 110 1e-24
At1g17480 hypothetical protein 110 2e-24
At5g13460 unknown protein 109 4e-24
>At3g49260 SF16 -like protein
Length = 471
Score = 360 bits (925), Expect = e-100
Identities = 225/453 (49%), Positives = 284/453 (62%), Gaps = 42/453 (9%)
Query: 20 EHEVAEEVSFEHFPAESSPDDVTNEGSTTSTPVRD--DRNHAIAVAEATAAAASAAVVAA 77
+H+ E VSFEHFPAESSP+ + ST STP + DR HA+AVA ATAAAA AAV AA
Sbjct: 42 QHDTQEVVSFEHFPAESSPEISHDVESTASTPATNVGDRKHAMAVAIATAAAAEAAVAAA 101
Query: 78 QAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQA 137
QAAA+VVRLAGY R +E+ AA IQSHYRGYLARRALRALKGLVRLQALVRG++VRKQA
Sbjct: 102 QAAAKVVRLAGYNRQTEEDSAAVLIQSHYRGYLARRALRALKGLVRLQALVRGNHVRKQA 161
Query: 138 QMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSV 197
QMTM+CMQALVRVQ RVRARR+Q++H+ +K EE+E+ + + + R
Sbjct: 162 QMTMKCMQALVRVQGRVRARRLQVAHDRFKKQFEEEEKRSGMEKPNKGFANLKTEREKPK 221
Query: 198 SSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDV 257
+ N S+ + + + + E MKRERALAYA+ YQ+Q Q H NS + +
Sbjct: 222 KLHEVNR-----TSLYQTQGKEKERSEGMMKRERALAYAYTYQRQMQ----HTNSE-EGI 271
Query: 258 DMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVP-------RESSYMTLPSTTSTTT- 309
+ + N ++ QW WNWL+ WMSSQPY R P Y P+ +TTT
Sbjct: 272 GLSS---NGPDRNQWAWNWLDHWMSSQPYTGRQTGPGPGPGQYNPPPYPPFPTAAATTTS 328
Query: 310 DNMSEKTVEMDIMA-TPSRGNFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPGR-PSYMAP 367
D++SEKTVEMD+ T +GN +GL+ +E+ D ++ + H++ SP PSYMAP
Sbjct: 329 DDVSEKTVEMDVTTPTSLKGNI----IGLIDREYIDLG-SYRQGHKQRKSPTHIPSYMAP 383
Query: 368 TQSAKAKVRAEGPF--KQRAPYGPNWNSSIKGGSIIGSGCDSSSSGGG-TSAYKFPRSPG 424
T SAKAKVR +GP Q + P WNSS K GS+ GSGCDSSSSGG T+ Y PRSP
Sbjct: 384 TASAKAKVRDQGPTVKLQGTSFMPYWNSSTKNGSVNGSGCDSSSSGGAITTGYPGPRSPN 443
Query: 425 PKVNGVRSESRRTVGGSQDYTEDWAVPLGAHGW 457
PK + +R R+ V SQ T G GW
Sbjct: 444 PK-SDIR---RKPVSPSQSPT-----GFGKRGW 467
>At3g52290 unknown protein (At3g52290)
Length = 430
Score = 175 bits (444), Expect = 4e-44
Identities = 139/414 (33%), Positives = 197/414 (47%), Gaps = 98/414 (23%)
Query: 13 KENKEKLEHEVAEEVSFEHFPAESSPDDVTNEGSTTST-PVRDDR---------NHAIAV 62
K+ KE+ H+ + + +S DVTN G+ S V+D + HA +V
Sbjct: 18 KQKKEQKPHKSKK------WFGKSKKLDVTNSGAAYSPRTVKDAKLKEIEEQQSRHAYSV 71
Query: 63 AEATAAAASAAVVAAQAAARVVRLAGYGRH---NKEERAATFIQSHYRGYLARRALRALK 119
A ATAAAA AAV AAQAAA VVRL+ R + EE AA IQ+ +RGY+ARRALRAL+
Sbjct: 72 AIATAAAAEAAVAAAQAAAEVVRLSALSRFPGKSMEEIAAIKIQTAFRGYMARRALRALR 131
Query: 120 GLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSH--ECLEKAMEEDEEEE 177
GLVRL++LV+G VR+QA T++ MQ L RVQ ++R RR++LS + L + +++ ++
Sbjct: 132 GLVRLKSLVQGKCVRRQATSTLQSMQTLARVQYQIRERRLRLSEDKQALTRQLQQKHNKD 191
Query: 178 FVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAF 237
F + E W++ S +K + N L K A M+RE+ALAYAF
Sbjct: 192 FDKTGE--------------------NWNDSTLSREKVEANMLNKQVATMRREKALAYAF 231
Query: 238 NYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVPRESS 297
+ H+N+ + MG+ D WGW+WLERWM+++P N H +
Sbjct: 232 S----------HQNTWKNSTKMGSQTFMDPNNPHWGWSWLERWMAARP-NENHSL----- 275
Query: 298 YMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTF-------- 349
T + S ++V M+ N+ P G SSP
Sbjct: 276 ------TPDNAEKDSSARSVASRAMSEMIPRGKNLSPRGKTPNSRRGSSPRVRQVPSEDS 329
Query: 350 ------------NRQHQ---------------RPPSPGRPSYMAPTQSAKAKVR 376
NR+H S P YMAPTQ+AKA+ R
Sbjct: 330 NSIVSFQSEQPCNRRHSTCGSIPSTRDDESFTSSFSQSVPGYMAPTQAAKARAR 383
>At3g22190 unknown protein
Length = 400
Score = 165 bits (418), Expect = 4e-41
Identities = 114/289 (39%), Positives = 153/289 (52%), Gaps = 40/289 (13%)
Query: 89 YGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALV 148
Y ++E RAAT IQ+ YRG+LARRALRALKGLVRLQALVRGH VRKQA +T+RCMQALV
Sbjct: 81 YDEQSRENRAATRIQTAYRGFLARRALRALKGLVRLQALVRGHAVRKQAAVTLRCMQALV 140
Query: 149 RVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNK 208
RVQARVRARRV+L+ E +E + ++ ++Q + + GW +
Sbjct: 141 RVQARVRARRVRLALE-----LESETSQQTLQQQ---------LADEARVREIEEGWCDS 186
Query: 209 CQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDE 268
SV++ + L++ EAA KRERA+AYA +Q Q + L +S + P +
Sbjct: 187 IGSVEQIQAKLLKRQEAAAKRERAMAYALTHQWQAGTRLLSAHSG--------FQP---D 235
Query: 269 KVQWGWNWLERWMSSQPYNVRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRG 328
K WGWNWLERWM+ +P+ R + L ++N+ KT + P+
Sbjct: 236 KNNWGWNWLERWMAVRPWENRFLDSNLRDDAKLGENGMEQSENV-PKTQIKSVSKMPNTS 294
Query: 329 NFNMGPMGLM---AQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAK 374
N G M Q DSS SPG S + AK+K
Sbjct: 295 NLVSGVSSQMTGPCQSDGDSS-----------SPGISSSIPVVSKAKSK 332
>At2g26180 putative SF16 protein {Helianthus annuus}
Length = 416
Score = 160 bits (404), Expect = 2e-39
Identities = 110/313 (35%), Positives = 157/313 (50%), Gaps = 46/313 (14%)
Query: 94 KEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQAR 153
+EE AA IQ+ +RG+LARRALRALKG+VRLQALVRG VRKQA +T+RCMQALVRVQAR
Sbjct: 82 REEWAAIRIQTAFRGFLARRALRALKGIVRLQALVRGRQVRKQAAVTLRCMQALVRVQAR 141
Query: 154 VRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVK 213
VRARRV+++ +E ++ + +H T +S + GW ++ +V
Sbjct: 142 VRARRVRMT-------VEGQAVQKLLDEHRT---------KSDLLKEVEEGWCDRKGTVD 185
Query: 214 KAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWG 273
K ++ E A KRERALAYA +Q + + +N + + +K WG
Sbjct: 186 DIKSKLQQRQEGAFKRERALAYALAQKQWRSTTSSNLKTNS---SISYLKSQEFDKNSWG 242
Query: 274 WNWLERWMSSQPYNVRHM-------VPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPS 326
W+WLERWM+++P+ R M P + L S + + V + A P
Sbjct: 243 WSWLERWMAARPWETRLMDTVDTAATPPPLPHKHLKSPETADVVQVRRNNVTTRVSAKPP 302
Query: 327 RGNFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPG----------------RPSYMAPTQS 370
+ P EF++SS + + P G +PSYM+ T+S
Sbjct: 303 PHMLSSSP----GYEFNESSGSSSICTSTTPVSGKTGLVSDNSSSQAKKHKPSYMSLTES 358
Query: 371 AKAKVRAEGPFKQ 383
KAK R +Q
Sbjct: 359 TKAKRRTNRGLRQ 371
>At4g00820 unknown protein
Length = 534
Score = 145 bits (366), Expect = 5e-35
Identities = 117/356 (32%), Positives = 182/356 (50%), Gaps = 41/356 (11%)
Query: 33 PAESSPDDVTNEGSTTSTPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRH 92
PA+ S + T T + V + R A + AT +AAS + + L
Sbjct: 71 PAQKSTETTTIINPTVLSSVTEQRYDA-STPPATVSAASETHPPS-TTKELPNLTRRTYT 128
Query: 93 NKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQA 152
+E+ AA IQ+ +RGYLARRALRALKGLV+LQALVRGHNVRKQA+MT+RCMQALVRVQ+
Sbjct: 129 AREDYAAVVIQTGFRGYLARRALRALKGLVKLQALVRGHNVRKQAKMTLRCMQALVRVQS 188
Query: 153 RVRARRVQLSHECLEKAMEEDEEEEFVRQH-ETITKPMSPMRRSSVSSNNNNGWDNKCQS 211
RV +R +LSH+ K+ D + ++ + I+ S R S + + WD++ +
Sbjct: 189 RVLDQRKRLSHDGSRKSAFSDTQSVLESRYLQEISDRRSMSREGSSIAED---WDDRPHT 245
Query: 212 VKKAKENDLRKHEAAMKRE--RALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEK 269
+++ K ++ + A++RE +++ AF++Q ++ + G+Y D+ +
Sbjct: 246 IEEVKAMLQQRRDNALRRESNNSISQAFSHQVRRTR--------------GSYSTGDEYE 291
Query: 270 VQWGWNWLERWMSSQPYNVRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDI-MATPSRG 328
+ WL+RWM+S+P++ R S+ +P KTVE+D +RG
Sbjct: 292 EERP-KWLDRWMASKPWD-----KRASTDQRVPPV---------YKTVEIDTSQPYLTRG 336
Query: 329 NFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQR 384
N G +Q S T + Q S PS P +S ++R+ P QR
Sbjct: 337 NSRTGASPSRSQRPSSPSRTSHHYQQHNFSSATPS---PAKSRPIQIRSASPRIQR 389
>At1g01110 unknown protein
Length = 527
Score = 139 bits (351), Expect = 2e-33
Identities = 110/331 (33%), Positives = 165/331 (49%), Gaps = 61/331 (18%)
Query: 71 SAAVVAAQAAARVV--RLAGYGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALV 128
SA V A +A++ + R Y R N AA IQ+ +RGYLARRALRALKGLV+LQALV
Sbjct: 97 SAVVPIATSASKTLAPRRIYYARENY---AAVVIQTSFRGYLARRALRALKGLVKLQALV 153
Query: 129 RGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKP 188
RGHNVRKQA+MT+RCMQALVRVQ+RV +R +LSH+ K+ D F ++
Sbjct: 154 RGHNVRKQAKMTLRCMQALVRVQSRVLDQRKRLSHDGSRKSAFSDSHAVFESRYLQDLSD 213
Query: 189 MSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERA-LAYAFNYQQQQQKQH 247
M R S+ + WD++ ++ K R+ + A++ ++ L+ AF+ Q+ +
Sbjct: 214 RQSMSREGSSAAED--WDDRPHTIDAVKVMLQRRRDTALRHDKTNLSQAFS--QKMWRTV 269
Query: 248 LHRNSNG-DDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVPRESSYMTLPSTTS 306
++++ G +V++ P WL+RWM+++P++ R S+ +
Sbjct: 270 GNQSTEGHHEVELEEERP----------KWLDRWMATRPWDKR------------ASSRA 307
Query: 307 TTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSY-- 364
+ +S KTVE+D SR G QRP SP R S+
Sbjct: 308 SVDQRVSVKTVEIDTSQPYSRTGAGSPSRG-----------------QRPSSPSRTSHHY 350
Query: 365 ---------MAPTQSAKAKVRAEGPFKQRAP 386
+P +S +R+ P QR P
Sbjct: 351 QSRNNFSATPSPAKSRPILIRSASPRCQRDP 381
>At5g03040 unknown protein
Length = 446
Score = 137 bits (346), Expect = 9e-33
Identities = 109/332 (32%), Positives = 157/332 (46%), Gaps = 44/332 (13%)
Query: 69 AASAAVVAAQAAARVVRLAG---YGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQ 125
A VV + +A VVR A + + EE AA IQ+ +RGYLARRALRA++GLVRL+
Sbjct: 85 ATDVPVVPSSSAPGVVRRATPTRFAGKSNEEAAAILIQTIFRGYLARRALRAMRGLVRLK 144
Query: 126 ALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETI 185
L+ G V++QA T++CMQ L RVQ+++RARR+++S EE RQ + +
Sbjct: 145 LLMEGSVVKRQAANTLKCMQTLSRVQSQIRARRIRMS------------EENQARQKQLL 192
Query: 186 TKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQK 245
K + N + W++ QS +K + N L K+EA M+RERALAY++++QQ +
Sbjct: 193 QKHAKEL----AGLKNGDNWNDSIQSKEKVEANLLSKYEATMRRERALAYSYSHQQNWKN 248
Query: 246 QHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVPRESSYMTLPSTT 305
N D T+ P ++ N S + R S T P+T
Sbjct: 249 NSKSGNPMFMDPSNPTWVPRKNKSNSNNDNAASVKGSINRNEAAKSLTRNGS--TQPNTP 306
Query: 306 STTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTF------NRQH------ 353
S+ K TPSR N DS T NR+H
Sbjct: 307 SSARGTPRNKN-SFFSPPTPSRLN-----QSSRKSNDDDSKSTISVLSERNRRHSIAGSS 360
Query: 354 -----QRPPSPGRPSYMAPTQSAKAKVRAEGP 380
SP PSYM PT+SA+A+++ + P
Sbjct: 361 VRDDESLAGSPALPSYMVPTKSARARLKPQSP 392
>At1g72670 unknown protein
Length = 414
Score = 134 bits (337), Expect = 1e-31
Identities = 118/384 (30%), Positives = 172/384 (44%), Gaps = 67/384 (17%)
Query: 36 SSPDDVTNEGSTTSTPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRHNKE 95
SS + G + TP + + ++ AA +A + A +V K
Sbjct: 42 SSSKGFKSRGGSYGTPSLGSDPPSFSADDSFTAAVAAVIRAPPKDFFLV---------KR 92
Query: 96 ERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVR 155
E AAT IQ+ +R +LAR+ALRALK +VR+QA+ RG VRKQA +T+RCMQALVRVQARVR
Sbjct: 93 EWAATRIQAAFRAFLARQALRALKAVVRIQAIFRGRQVRKQADVTLRCMQALVRVQARVR 152
Query: 156 ARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKA 215
A C + E E KP S ++ + GW + S+ +
Sbjct: 153 A-------HCNRGPSDGQELE----------KP-SDQQKDDPAKQAEKGWCDSPGSINEV 194
Query: 216 KENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWN 275
+ + E A+KRERA+ YA +Q + + G N K GWN
Sbjct: 195 RTKLQMRQEGAIKRERAMVYALTHQPRTCPSPAKASKQG-----SVKKNNGSCKSSPGWN 249
Query: 276 WLERWMSSQPYNVRHMV-PRESSYMTLPSTTSTT---TDNMSEKTVEMDIMATPSRGNFN 331
WL+RW++ +P+ R M P SS S +S + T + + + ++A P
Sbjct: 250 WLDRWVADRPWEGRLMEGPTNSSENARKSESSVSEHDTVQVRKNNLTTRVLARPP----- 304
Query: 332 MGPMGLMAQEFHDSSPTFNRQHQRP-PSPG----------RPSYMAPTQSAKAKVRAEG- 379
PM A SS + Q P P G +PSYM+ TQS KAK R G
Sbjct: 305 --PMSSSATSSESSSTS-----QSPVPFSGSFLEEGGYYRKPSYMSLTQSIKAKQRRSGS 357
Query: 380 -------PFKQRAPYGPNWNSSIK 396
PF+++ N + +++
Sbjct: 358 SSSCSKTPFEKKQSMSYNGDVNVR 381
>At2g26410 putative SF16 protein {Helianthus annuus}
Length = 516
Score = 132 bits (332), Expect = 4e-31
Identities = 92/274 (33%), Positives = 149/274 (53%), Gaps = 38/274 (13%)
Query: 61 AVAEATAAAASAAVVAAQAAARVVRLAGYG----RHNKEERAATFIQSHYRGYLARRALR 116
A+A A+A AA AAVVAA AAA V+RL +KEE AA IQ+ YR Y ARR LR
Sbjct: 98 ALALASAVAAEAAVVAAHAAAEVIRLTTPSTPQIEESKEETAAIKIQNAYRCYTARRTLR 157
Query: 117 ALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEE 176
AL+G+ RL++L++G V++Q + MQ L R+Q +++ RR +LS E + ++
Sbjct: 158 ALRGMARLKSLLQGKYVKRQMNAMLSSMQTLTRLQTQIQERRNRLSAENKTRHRLIQQKG 217
Query: 177 EFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYA 236
H+ + V++ N +D+ +S ++ + + EA+++RERALAYA
Sbjct: 218 HQKENHQNL-----------VTAGN---FDSSNKSKEQIVARSVNRKEASVRRERALAYA 263
Query: 237 FNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPY---------N 287
+++QQ RNS+ + D WGW+WLERWM+S+P+ +
Sbjct: 264 YSHQQ------TWRNSS----KLPHQTLMDTNTTDWGWSWLERWMASRPWDAESIDDQVS 313
Query: 288 VRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDI 321
V+ + RE+S + P+ S T + S+ +++ +
Sbjct: 314 VKSSLKRENSIKSSPA-RSKTQKSASQSSIQWPV 346
>At3g09710 putative SF16 protein
Length = 454
Score = 131 bits (330), Expect = 7e-31
Identities = 107/377 (28%), Positives = 174/377 (45%), Gaps = 58/377 (15%)
Query: 15 NKEKLEHEVAE-EVSFEHFPA------ESSP------DDVTNEGSTTSTPVRDDRNHAIA 61
+ +KL+HE E + S +P SSP D+V E P D A
Sbjct: 18 DSKKLKHESVECQDSVISYPVLIATSRSSSPQFEVRVDEVNYEQKKNLYPPSSDSVTA-T 76
Query: 62 VAEATAAAASAAVVAAQAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRALRALKGL 121
VA + ++ + A V R AG +KEE AA IQS +RG+LARR + ++G
Sbjct: 77 VAHVLVDSPPSSPESVHQAIVVNRFAG---KSKEEAAAILIQSTFRGHLARRESQVMRGQ 133
Query: 122 VRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQ 181
RL+ L+ G V++QA +T++CMQ L RVQ+++R+RR+++S E + ++ +++
Sbjct: 134 ERLKLLMEGSVVQRQAAITLKCMQTLSRVQSQIRSRRIRMSEE------NQARHKQLLQK 187
Query: 182 HETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQ 241
H K + ++ N W+ QS ++ + L K+EA M+RERALAYAF +QQ
Sbjct: 188 H---AKELGGLK-------NGGNWNYSNQSKEQVEAGMLHKYEATMRRERALAYAFTHQQ 237
Query: 242 QQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPY--------------- 286
+ + + + M +P WGW+WLERWM+ +P+
Sbjct: 238 NLKS----FSKTANPMFMDPSNPT------WGWSWLERWMAGRPWESSEKEQNTTNNDNS 287
Query: 287 NVRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSS 346
+V++ R S +++ N S K +T +R P+ +
Sbjct: 288 SVKNSTNRNSQGGETAKSSNRNKLNSSTKPNTPSASSTATRNPRKKRPIPSSIKSKSSDD 347
Query: 347 PTFNRQHQRPPSPGRPS 363
+ + R PS RPS
Sbjct: 348 EAKSSERNRRPSIARPS 364
>At3g59690 unknown protein
Length = 517
Score = 127 bits (320), Expect = 1e-29
Identities = 103/308 (33%), Positives = 150/308 (48%), Gaps = 62/308 (20%)
Query: 102 IQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQL 161
IQ+ +RGY+ARR+ RALKGLVRLQ +VRGH+V++Q M+ MQ LVRVQ +V++RR+Q+
Sbjct: 175 IQAAFRGYMARRSFRALKGLVRLQGVVRGHSVKRQTMNAMKYMQLLVRVQTQVQSRRIQM 234
Query: 162 SHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLR 221
LE D+++ VSS ++ WD+ SV +E D+R
Sbjct: 235 ----LENRARNDKDD-----------------TKLVSSRMSDDWDD---SVLTKEEKDVR 270
Query: 222 KH---EAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLE 278
H +A +KRER++AYA+++Q L +NS D+ T WNW++
Sbjct: 271 LHRKIDAMIKRERSMAYAYSHQ-------LWKNSPKSAQDIRT------SGFPLWWNWVD 317
Query: 279 RWMS-SQPYNVRHMVPRES--------SYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGN 329
R + +QP+ + P S ++ L ++ T+T N S+ T TPSR
Sbjct: 318 RQKNQNQPFRLTPTRPSLSPQPQSSNQNHFRLNNSFDTSTPNSSKST-----FVTPSRPI 372
Query: 330 FNMGPMGLMAQEF--------HDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPF 381
P + DS + P PSYMAPT SAKAK+RA
Sbjct: 373 HTPQPYSSSVSRYSRGGGRATQDSPFKDDDSLTSCPPFSAPSYMAPTVSAKAKLRANSNP 432
Query: 382 KQRAPYGP 389
K+R P
Sbjct: 433 KERMDRTP 440
>At3g16490 putative calmodulin-binding protein
Length = 389
Score = 120 bits (302), Expect = 1e-27
Identities = 83/188 (44%), Positives = 110/188 (58%), Gaps = 23/188 (12%)
Query: 53 RDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLA------GYGRHNKEERAATFIQSHY 106
++ HAIAVA ATAAAA AAV AAQAA VVRL GY + E AA IQS +
Sbjct: 58 KEQNKHAIAVAAATAAAADAAVAAAQAAVAVVRLTSNGRSGGYSGNAMERWAAVKIQSVF 117
Query: 107 RGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARRVQLSH--- 163
+GYLAR+ALRALKGLV+LQALVRG+ VRK+A T+ MQAL+R Q VR++R+ ++
Sbjct: 118 KGYLARKALRALKGLVKLQALVRGYLVRKRAAETLHSMQALIRAQTSVRSQRINRNNMFH 177
Query: 164 --ECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDLR 221
LE+ + E R ++ K S++NNN +D ++ K E D
Sbjct: 178 PRHSLERLDDSRSEIHSKRISISVEKQ---------SNHNNNAYD---ETSPKIVEIDTY 225
Query: 222 KHEAAMKR 229
K ++ KR
Sbjct: 226 KTKSRSKR 233
>At2g43680 SF16 protein {Helianthus annuus} like protein
Length = 668
Score = 118 bits (296), Expect = 6e-27
Identities = 107/379 (28%), Positives = 167/379 (43%), Gaps = 93/379 (24%)
Query: 95 EERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARV 154
+ +AT IQ +RGY+AR++ RALKGLVRLQ +VRG++V++Q M+ MQ +VRVQ+++
Sbjct: 321 QHASATKIQGAFRGYMARKSFRALKGLVRLQGVVRGYSVKRQTINAMKYMQQVVRVQSQI 380
Query: 155 RARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKK 214
++RR+++ LE + +++E + + S N+ WD+ + ++
Sbjct: 381 QSRRIKM----LENQAQVEKDE----------------AKWAASEAGNDNWDDSVLTKEE 420
Query: 215 AKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGW 274
RK +A +KRER++AYA++ + L +NS D ++ QW W
Sbjct: 421 RDSRSQRKTDAIIKRERSMAYAYS-------RKLWKNSPKSTQDNRSF-------PQW-W 465
Query: 275 NWLERW--------MSSQPYNVRHMVP-----------RESSYMTLPSTTSTTTDNMSEK 315
NW++R SQP + P + ++ L + T+T S
Sbjct: 466 NWVDRQNPLASPAPSYSQPQRDFRLTPSRLCPSPLSQSSKQHHIRLDNHFDTSTPRSSRS 525
Query: 316 TVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKV 375
T TPSR + G F P PSYMAPT SAKAKV
Sbjct: 526 TFH-----TPSR-PIHTGTSRYSRGRLRGQDSPFKDDDSLTSCPPFPSYMAPTVSAKAKV 579
Query: 376 RAEGPFKQRAPYGP----------------NWNSSIKGGSIIGSGCDSSSSGGGTSAYKF 419
R K+R P WN GS++ S +S+++
Sbjct: 580 RPNSNPKERVMGTPVSEKRRMSYPPTQDTFRWNK----GSLVMS---------NSSSHRG 626
Query: 420 PRSPGPKVNGVRSESRRTV 438
P SPG GV E +T+
Sbjct: 627 PGSPG----GVVLEKHKTL 641
>At4g29150 unknown protein (At4g29150)
Length = 383
Score = 115 bits (287), Expect = 7e-26
Identities = 66/113 (58%), Positives = 82/113 (72%), Gaps = 7/113 (6%)
Query: 53 RDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRH-------NKEERAATFIQSH 105
++ R HAIAVA ATAAAA AAV AA+AAA VVRL G G+ ++E RAA IQ
Sbjct: 65 KERRTHAIAVAAATAAAADAAVAAAKAAAAVVRLQGQGKSGPLGGGKSREHRAAMQIQCA 124
Query: 106 YRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRARR 158
+RGYLAR+ALRAL+G+V++QALVRG VR QA T+R M+ALVR Q V+ +R
Sbjct: 125 FRGYLARKALRALRGVVKIQALVRGFLVRNQAAATLRSMEALVRAQKTVKIQR 177
>At5g07240 unknown protein
Length = 401
Score = 114 bits (286), Expect = 9e-26
Identities = 73/135 (54%), Positives = 88/135 (65%), Gaps = 19/135 (14%)
Query: 43 NEGSTTSTPVRDDRN------HAIAVAEATAAAASAAVVAAQAAARVVRLAGYGRHN--- 93
N+ S+ S+ R D + HAIAVA ATAA A AA+ AA+AAA VVRL GR++
Sbjct: 34 NDSSSHSSKRRGDEDVLNGDKHAIAVAAATAAVAEAALAAARAAAEVVRLTNGGRNSSVK 93
Query: 94 ----------KEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRC 143
+E +AA IQS +RGYLARRALRALK LV+LQALV+GH VRKQ +R
Sbjct: 94 QISRSNRRWSQEYKAAMKIQSAFRGYLARRALRALKALVKLQALVKGHIVRKQTADMLRR 153
Query: 144 MQALVRVQARVRARR 158
MQ LVR+QAR RA R
Sbjct: 154 MQTLVRLQARARASR 168
>At4g10640 putative protein
Length = 407
Score = 114 bits (286), Expect = 9e-26
Identities = 100/341 (29%), Positives = 158/341 (46%), Gaps = 64/341 (18%)
Query: 58 HAIAVAEATAAAASAAVVAA--QAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRAL 115
H + EA +V++ + +V+L + AA IQ+ +RGYL+RRAL
Sbjct: 60 HTVRTVEAEEKEKPPVIVSSVEEGVTEIVKLTATPGFIRRHWAAIIIQTAFRGYLSRRAL 119
Query: 116 RALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARV------RARRVQLSHECLEKA 169
RALKG+V+LQALVRG+NVR QA++T+RC++ALVRVQ +V + RV LS
Sbjct: 120 RALKGIVKLQALVRGNNVRNQAKLTLRCIKALVRVQDQVLNHHQQQRSRVLLSPPSRNYN 179
Query: 170 MEEDEEEEFVRQH---ETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAKENDL---RKH 223
+E F + +T T R S+S + N+C + ++E +L +K
Sbjct: 180 IEARRNSMFAESNGFWDTKTYLQDIRSRRSLSRDM-----NRCNNEFYSEETELILQKKL 234
Query: 224 EAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSS 283
E A+KRE+A A A + Q + + S GDD ++ E+ Q WL+RWM++
Sbjct: 235 EIAIKREKAQALALSNQIRSRSS--RNQSAGDDREL-------LERTQ----WLDRWMAT 281
Query: 284 QPYNVRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFH 343
+ ++ T T + N+ + ++ + T H
Sbjct: 282 KQWD----------------DTITNSTNVRDPIKTLEAVTT----------------HHH 309
Query: 344 DSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQR 384
S R P+YM+ T+SAKAK R + ++R
Sbjct: 310 QRSYPATPPSCRASRSVMPNYMSATESAKAKARTQSTPRRR 350
>At5g62070 unknown protein
Length = 403
Score = 113 bits (282), Expect = 2e-25
Identities = 119/390 (30%), Positives = 164/390 (41%), Gaps = 93/390 (23%)
Query: 54 DDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAG-----------------YGRHNK-- 94
D HAIAVA ATAA A AA+ AA AAA VVRL GR N+
Sbjct: 54 DADKHAIAVAAATAAVAEAALTAAHAAAEVVRLTSGNGGRNVGGGGNSSVFQIGRSNRRW 113
Query: 95 --EERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQA 152
E AA IQS +RGYLARRALRALK LV+LQALVRGH VRKQ +R MQ LVR+Q+
Sbjct: 114 AQENIAAMKIQSAFRGYLARRALRALKALVKLQALVRGHIVRKQTADMLRRMQTLVRLQS 173
Query: 153 RVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSV 212
+ RAR + SH + T + P SS++ +C S
Sbjct: 174 QARARASRSSHSS--------------ASFHSSTALLFPS-----SSSSPRSLHTRCVSN 214
Query: 213 KKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDV-DMGTYHPNDDEKVQ 271
+ D H KR ++Q ++ + NGD + ++ T+ P+ K
Sbjct: 215 AEVSSLD---HRGGSKR-------LDWQAEE-------SENGDKILEVDTWKPHYHPKP- 256
Query: 272 WGWNWLERWMSSQPYNVRHMVPRESSYMTL-PSTTSTTTDNMSEKTVEMDIMATPSRGNF 330
+ S+ N PR+ L P +T + S + SR +
Sbjct: 257 ---------LRSERNNES---PRKRQQSLLGPRSTENSPQVGSSGSRRRTPFTPTSRSEY 304
Query: 331 NMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQRAPYG-- 388
+ G +H P+YMA T+S KAKVR++ KQR
Sbjct: 305 SWGCNNYYYSGYH------------------PNYMANTESYKAKVRSQSAPKQRVEVSNE 346
Query: 389 -PNWNSSIKGGSIIGSGCDSSSSGGGTSAY 417
+ S++G + + S G++ Y
Sbjct: 347 TSGYKRSVQGQYYYYTAVEEESLDVGSAGY 376
>At5g03960 unknown protein
Length = 403
Score = 110 bits (276), Expect = 1e-24
Identities = 104/350 (29%), Positives = 157/350 (44%), Gaps = 49/350 (14%)
Query: 44 EGSTTSTPVRDDRNHAIAVAEATAAAASAAVVAAQAAARVVRLAG-------YGRHNKEE 96
E T + +D R HA+ VA ATAAAA AAV AA+AAA VVR+AG + +
Sbjct: 48 ETRTLNEATQDQRKHAMNVAIATAAAAEAAVAAAKAAAEVVRMAGNAFTSQHFVKKLAPN 107
Query: 97 RAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQARVRA 156
AA IQS +R LAR+ALRALK LVRLQA+VRG VR++ ++ + + +
Sbjct: 108 VAAIKIQSAFRASLARKALRALKALVRLQAIVRGRAVRRKVSALLKSSHSNKASTSNIIQ 167
Query: 157 RRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVKKAK 216
R+ + H K+ E +EE V H S NGWD+ + + K
Sbjct: 168 RQTERKHWSNTKS--EIKEELQVSNHSLCN-----------SKVKCNGWDSSALTKEDIK 214
Query: 217 ENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWGWNW 276
LRK E +KR+R L Y+ + ++++ L + D+ M +
Sbjct: 215 AIWLRKQEGVIKRDRMLKYSRSQRERRSPHMLVESLYAKDMGMRSCR------------- 261
Query: 277 LERWMSSQPYNV--RHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMGP 334
LE W S+ ++P E M +P+ + + D + R +F+
Sbjct: 262 LEHWGESKSAKSINSFLIPSE---MLVPTKVKLRSLQRQDSGDGQDSPFSFPRRSFSR-- 316
Query: 335 MGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQSAKAKVRAEGPFKQR 384
+ Q + F R S G YM+ T+SA+ K+R+ +QR
Sbjct: 317 ---LEQSILEDESWFQR------SNGFQPYMSVTESAREKMRSLSTPRQR 357
>At1g17480 hypothetical protein
Length = 371
Score = 110 bits (274), Expect = 2e-24
Identities = 98/299 (32%), Positives = 135/299 (44%), Gaps = 64/299 (21%)
Query: 94 KEERAATFIQSHYRGYLARRALRALKGLVRLQALVRGHNVRKQAQMTMRCMQALVRVQAR 153
K E A+T IQ+ +R +LAR+A RALK +VR+QA+ RG VRKQA +T+RCMQALVRVQ+R
Sbjct: 92 KREWASTRIQAAFRAFLARQAFRALKAVVRIQAIFRGRQVRKQAAVTLRCMQALVRVQSR 151
Query: 154 VRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMSPMRRSSVSSNNNNGWDNKCQSVK 213
VRA R S E ++ V+Q E GW +S+K
Sbjct: 152 VRAHRRAPSDSL--------ELKDPVKQTE-------------------KGWCGSPRSIK 184
Query: 214 KAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHRNSNGDDVDMGTYHPNDDEKVQWG 273
+ K K E A+KRERA+ YA +Q R T+H K G
Sbjct: 185 EVKTKLQMKQEGAIKRERAMVYALTHQS--------RTCPSPSGRAITHH--GLRKSSPG 234
Query: 274 WNWLERWMSSQPYNVRHMVPRESSYMTLPSTTSTTTDNMSEKTVEMDIMATPSRGNFNMG 333
WNW + ESS ++ T + +N+S V +A P +
Sbjct: 235 WNWYD------DVGTFSRKSSESSVLSEYETVTVRKNNLSSTRV----LARPP---LLLP 281
Query: 334 PM--GLMAQEFHDSSPTFNRQHQRPPSPG-----------RPSYMAPTQSAKAKVRAEG 379
P+ G+ HD + T + Q P + +PSYM+ TQS +AK R G
Sbjct: 282 PVSSGMSYDSLHDETST-SSTSQSPVAFSSSVLDGGGYYRKPSYMSLTQSTQAKQRQSG 339
>At5g13460 unknown protein
Length = 443
Score = 109 bits (272), Expect = 4e-24
Identities = 82/307 (26%), Positives = 141/307 (45%), Gaps = 48/307 (15%)
Query: 71 SAAVVAAQAAARVVRLAGYGRHNKEERAATFIQSHYRGYLARRALRALKGLVRLQALVRG 130
S + + A VV+ + +E AAT IQ+ +RG+LAR+ALRALKG+V+LQA +RG
Sbjct: 86 SKGSTSPETADLVVQYQMFLNRQEEVLAATRIQTAFRGHLARKALRALKGIVKLQAYIRG 145
Query: 131 HNVRKQAQMTMRCMQALVRVQARVRARRVQLSHECLEKAMEEDEEEEFVRQHETITKPMS 190
VR+QA T++C+Q++V +Q++V +R Q + + D EE + +
Sbjct: 146 RAVRRQAMTTLKCLQSVVNIQSQVCGKRTQ-----IPGGVHRDYEESNIFNDNIL----- 195
Query: 191 PMRRSSVSSNNNNGWDNKCQSVKKAKENDLRKHEAAMKRERALAYAFNYQQQQQKQHLHR 250
V +N WD+ + ++ + + K EA+++RER YA +++ +
Sbjct: 196 -----KVDTNGQKRWDDSLLTKEEKEAVVMSKKEASLRRERIKEYAVTHRKSAESYQKRS 250
Query: 251 NSNGDDVDMGTYHPNDDEKVQWGWNWLERWMSSQPYNVRHMVPRESSYMTLPSTTSTTTD 310
N+ +W + WL+ W+ +Q + + + S T P
Sbjct: 251 NT------------------KWKY-WLDEWVDTQLTKSKELEDLDFSSKTKP-------- 283
Query: 311 NMSEKTVEMDIMATPSRGNFNMGPMGLMAQEFHDSSPTFNRQHQRPPSPGRPSYMAPTQS 370
++T+ + TP N P L+ S + Q + P+YM T+S
Sbjct: 284 --KDETLNEKQLKTPR----NSSPRRLVNNHRRQVSIGEDEQSPAAVTITTPTYMVATES 337
Query: 371 AKAKVRA 377
AKAK R+
Sbjct: 338 AKAKSRS 344
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.127 0.376
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,484,010
Number of Sequences: 26719
Number of extensions: 460071
Number of successful extensions: 2429
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 2235
Number of HSP's gapped (non-prelim): 163
length of query: 458
length of database: 11,318,596
effective HSP length: 103
effective length of query: 355
effective length of database: 8,566,539
effective search space: 3041121345
effective search space used: 3041121345
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149127.2


