

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149080.4 + phase: 0
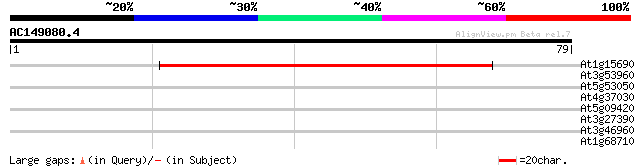
(79 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g15690 Unknown protein 69 4e-13
At3g53960 transporter like protein 32 0.074
At5g53050 unknown protein 27 2.4
At4g37030 hypothetical protein 26 3.1
At5g09420 putative subunit of TOC complex 25 5.3
At3g27390 unknown protein 25 5.3
At3g46960 putative helicase 25 9.0
At1g68710 ATPase, putative 25 9.0
>At1g15690 Unknown protein
Length = 770
Score = 68.9 bits (167), Expect = 4e-13
Identities = 36/47 (76%), Positives = 39/47 (82%)
Query: 22 LSMKITTYVNARTTLEARKGKRKSSAVAFRSGAVKGFLLAANGLLLL 68
L MKI TY NARTTLEARKG K+ VAFRSGAV GFLLAA+GLL+L
Sbjct: 163 LGMKIATYANARTTLEARKGVGKAFIVAFRSGAVMGFLLAASGLLVL 209
>At3g53960 transporter like protein
Length = 620
Score = 31.6 bits (70), Expect = 0.074
Identities = 22/78 (28%), Positives = 39/78 (49%), Gaps = 4/78 (5%)
Query: 2 FRVSGCCLLIITLKQTSDIKLSMKITTYVNARTTLEARKGKRKSSAVAFRSGAVKGFLLA 61
F +S ++ +T D+K+++K T Y + TTL G + A R G V L
Sbjct: 59 FGISTNLVVYLTTILHQDLKMAVKNTNYWSGVTTLMPLLGGFVADAYLGRYGTV----LL 114
Query: 62 ANGLLLLSLIPISITWTV 79
A + L+ LI ++++W +
Sbjct: 115 ATTIYLMGLILLTLSWFI 132
>At5g53050 unknown protein
Length = 396
Score = 26.6 bits (57), Expect = 2.4
Identities = 18/50 (36%), Positives = 26/50 (52%), Gaps = 2/50 (4%)
Query: 22 LSMKITTYVNARTTL--EARKGKRKSSAVAFRSGAVKGFLLAANGLLLLS 69
L+M+ Y R TL + +GK +S F + GFLL GLL+L+
Sbjct: 326 LTMETPGYFKRRLTLIRSSTEGKNVASPAHFIAEKFHGFLLFLFGLLVLA 375
>At4g37030 hypothetical protein
Length = 519
Score = 26.2 bits (56), Expect = 3.1
Identities = 14/40 (35%), Positives = 20/40 (50%), Gaps = 9/40 (22%)
Query: 49 AFRSGAVKGF---------LLAANGLLLLSLIPISITWTV 79
AF GA+KG L+ N ++L L P +TWT+
Sbjct: 24 AFFLGAIKGLIVGPIAGLTLIVGNVGVILCLFPAHVTWTI 63
>At5g09420 putative subunit of TOC complex
Length = 616
Score = 25.4 bits (54), Expect = 5.3
Identities = 10/26 (38%), Positives = 16/26 (61%)
Query: 4 VSGCCLLIITLKQTSDIKLSMKITTY 29
+SGCC + I L + D +S+ + TY
Sbjct: 422 MSGCCQVTIPLGEHGDRPISVSLLTY 447
>At3g27390 unknown protein
Length = 599
Score = 25.4 bits (54), Expect = 5.3
Identities = 12/35 (34%), Positives = 18/35 (51%), Gaps = 9/35 (25%)
Query: 53 GAVKGFLLA---------ANGLLLLSLIPISITWT 78
G +KG +L N ++LSL+P+ I WT
Sbjct: 28 GFIKGIVLCPLVCLVVTIGNSAVILSLLPVHIVWT 62
>At3g46960 putative helicase
Length = 1347
Score = 24.6 bits (52), Expect = 9.0
Identities = 11/27 (40%), Positives = 16/27 (58%)
Query: 49 AFRSGAVKGFLLAANGLLLLSLIPISI 75
AFR A +LL N L +SL+P+ +
Sbjct: 619 AFRRSAASNWLLLINKLSKMSLLPVVV 645
>At1g68710 ATPase, putative
Length = 1200
Score = 24.6 bits (52), Expect = 9.0
Identities = 13/41 (31%), Positives = 19/41 (45%)
Query: 12 ITLKQTSDIKLSMKITTYVNARTTLEARKGKRKSSAVAFRS 52
+T IK S+ T Y T +E G+RK + F+S
Sbjct: 435 LTCNSMEFIKCSVAGTAYGRGVTEVEMAMGRRKGGPLVFQS 475
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,315,537
Number of Sequences: 26719
Number of extensions: 33958
Number of successful extensions: 124
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 120
Number of HSP's gapped (non-prelim): 9
length of query: 79
length of database: 11,318,596
effective HSP length: 55
effective length of query: 24
effective length of database: 9,849,051
effective search space: 236377224
effective search space used: 236377224
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 52 (24.6 bits)
Medicago: description of AC149080.4


