

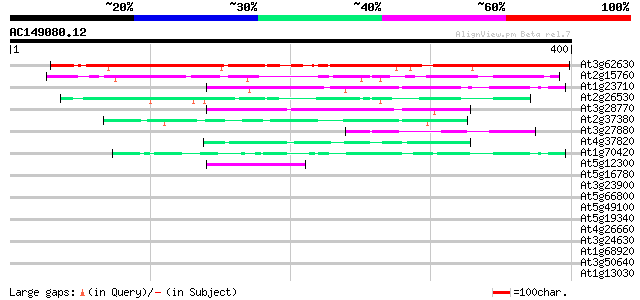
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149080.12 - phase: 0
(400 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g62630 unknown protein 330 8e-91
At2g15760 unknown protein 115 6e-26
At1g23710 unknown protein 75 8e-14
At2g26530 AR781, similar to yeast pheromone receptor 69 3e-12
At3g28770 hypothetical protein 49 5e-06
At2g37380 unknown protein 49 6e-06
At3g27880 unknown protein 47 2e-05
At4g37820 unknown protein 44 2e-04
At1g70420 unknown protein 43 4e-04
At5g12300 putative protein 42 6e-04
At5g16780 putative protein 40 0.003
At3g23900 unknown protein 40 0.003
At5g66800 unknown protein 39 0.007
At5g49100 unknown protein 38 0.009
At5g19340 putative protein 38 0.011
At4g26660 putative protein 38 0.011
At3g24630 hypothetical protein 38 0.011
At1g68920 putative bHLH transcription factor (bHLH049) 37 0.019
At3g50640 unknown protein 37 0.025
At1g13030 unknown protein 36 0.033
>At3g62630 unknown protein
Length = 380
Score = 330 bits (846), Expect = 8e-91
Identities = 200/386 (51%), Positives = 256/386 (65%), Gaps = 47/386 (12%)
Query: 30 SNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSI--TASSSYHHQTTTSSVPMS 87
S CSTP+VSAPSSPGRG PPP GYF+SAP+SP+HF + +SSS + + +S
Sbjct: 25 SACSTPFVSAPSSPGRG--PPP---GYFFSAPSSPMHFFLCSASSSSENPKKLDTSSCGD 79
Query: 88 YEFEFSARFGSTGSAASG-SMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEED 146
+EF+FS+R S+ G SMTSA+ELF NGQI+PMKLSSHL+RPQ+L+PLLDLE EEED
Sbjct: 80 FEFDFSSRLSSSSGPLGGVSMTSAEELFSNGQIKPMKLSSHLQRPQILSPLLDLENEEED 139
Query: 147 EEE-----GEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACE 201
+++ GE+ RGRDL+LR +SV R+ RS+SPLRN ++ +W + E + E
Sbjct: 140 DDDETKPNGEMK--RGRDLKLRSRSVHRKARSLSPLRNAAY-QWNQEEGEEE-------E 189
Query: 202 IEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKD 261
+ E K +C + ++ DE + E TP SASSSRSSS GR+SK+W+FLKD
Sbjct: 190 VAGEREVK------EC--IRKLQEDE-NVPSAETTPSCSASSSRSSSYGRNSKKWIFLKD 240
Query: 262 FL-RSKSEGRSNNK--FWSTISFSPT--KDKKSSSNNQNLQAQISKEKTCETQRNGSSSS 316
L RSKSEGR N K FWS ISFSP+ KDKK S+ +EK + + + S
Sbjct: 241 LLHRSKSEGRGNGKEKFWSNISFSPSNFKDKKLKSSQL-------EEKPIQETVDAAVES 293
Query: 317 SSSSSKGSWGKK--MTGKPMNGVGKRR-VPPSPHELHYKANRAQAEELRRKTFLPYRQGL 373
K KK + GKP NG+ KRR + PS HELHY NRAQAEE++++T+LPYR GL
Sbjct: 294 KKQKQKQPPAKKAPVNGKPTNGIAKRRGLQPSAHELHYTTNRAQAEEMKKRTYLPYRHGL 353
Query: 374 LGCLGFSSKGYGAMNGFARALNPVSS 399
GCLGFSSKGY A+NG AR+LNPVSS
Sbjct: 354 FGCLGFSSKGYSALNGLARSLNPVSS 379
>At2g15760 unknown protein
Length = 315
Score = 115 bits (287), Expect = 6e-26
Identities = 121/386 (31%), Positives = 170/386 (43%), Gaps = 113/386 (29%)
Query: 27 DIDSNCSTPYVSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSS----YHHQTTTS 82
+ DS S+PY++APSSP R G F+SAP SP S + SS+ + Q T
Sbjct: 14 NFDSTTSSPYITAPSSPTRFGNN-------FFSAPTSP---SPSTSSNIPFDWDDQPRTP 63
Query: 83 SVPMSYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLL-DLE 141
+ FE F +G S ++ADELF G+IRP++ P V +P LE
Sbjct: 64 KKRSAIGFEDDFEFNFSGQLEKTSFSAADELFDGGKIRPLRTPL---TPTVSSPRSRGLE 120
Query: 142 EEEEDEEEGEVVVVRGRDLRLRDKSVR---RRTRSMSPLRNNSHLEWTENEDDNNNKNNV 198
E+ D+++ RGRD S R + +RSMSPLR
Sbjct: 121 IEDSDDQKD-----RGRDRSPGSSSSRYDRKGSRSMSPLR-------------------- 155
Query: 199 ACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSA--------- 249
+ ++ VDE E ++ T + ++++S S+
Sbjct: 156 ---------------------VSDIMVDEE--EEVQSTKMVASNTSNQKSSVFLSAILFP 192
Query: 250 GRSSKRWVFLKDFL--RSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCE 307
GR+ K+W LKD L RS S+GR PTK+ + + I +K E
Sbjct: 193 GRAYKKWK-LKDLLLFRSASDGRP----------IPTKESLN-------RYDILTKKDAE 234
Query: 308 TQRNGS-SSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTF 366
RN S S S S S ++ G + S HE+HY NRA +EEL+RKTF
Sbjct: 235 EVRNSSIRSRESCESSVSRSRRRNGAVV----------SAHEMHYTENRAVSEELKRKTF 284
Query: 367 LPYRQGLLGCLGFSSKGYGAMNGFAR 392
LPY+QG LGCLGF+ A+N AR
Sbjct: 285 LPYKQGWLGCLGFNP----AVNEIAR 306
>At1g23710 unknown protein
Length = 295
Score = 74.7 bits (182), Expect = 8e-14
Identities = 74/268 (27%), Positives = 116/268 (42%), Gaps = 41/268 (15%)
Query: 141 EEEEEDEEEGEVVVVRGRDLRLRDKSVRR--RTRSMSPLRNNSHLEWTENEDDNNNKNNV 198
E++EE+EEE V G + + R + PL N L ENEDD N+ +V
Sbjct: 56 EDDEEEEEEFSFACVNGEGSPITADEAFEDGQIRPVFPLFNRDLLFEYENEDDKNDNVSV 115
Query: 199 ACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPL----------DSASSSRSSS 248
E + E+ N + E + G E+ + P S + R S+
Sbjct: 116 TDENRPRLRKLFVEDRNGNGDGEETE----GSEKEPLGPYCSWTGGTVAEASPETCRKSN 171
Query: 249 AGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCET 308
+ SK W F LRS S+GR F + S T+ + SSS++ + K+ E
Sbjct: 172 STGFSKLWRFRDLVLRSNSDGRDAFVFLNN-SNDKTRTRSSSSSSSTAAEENDKKVITEK 230
Query: 309 QRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLP 368
++ +S+SS +K KK T S HE Y NRA EE++ +++LP
Sbjct: 231 KKGKEKTSTSSETK----KKTT-----------TTKSAHEKLYMRNRAMKEEVKHRSYLP 275
Query: 369 YRQGLLGCLGFSSKGYGAMNGFARALNP 396
Y+Q +GF + +NG +R ++P
Sbjct: 276 YKQ-----VGF----FTNVNGLSRNIHP 294
>At2g26530 AR781, similar to yeast pheromone receptor
Length = 317
Score = 69.3 bits (168), Expect = 3e-12
Identities = 96/384 (25%), Positives = 142/384 (36%), Gaps = 127/384 (33%)
Query: 37 VSAPSSPGRGGGPPPISSGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSYEFEFSARF 96
++APSSP + SG F SAP SP F+ T S ++ F++
Sbjct: 6 LTAPSSPRQ-------LSGCFLSAPTSPRRFNEFYREFEEAATRNFSDRLTVPFDWEETP 58
Query: 97 GST-----------------GSAASGSMTSADELFLNGQIRPMKLSSHLE-----RPQVL 134
G+ G + A+ELF G+I+P+K +L+ +PQ+L
Sbjct: 59 GTPRKITNDDDDDIDFAFEIGGKLETTSLFAEELFDGGKIKPLKPPPYLQLDHHHQPQIL 118
Query: 135 APL-------------------------LDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRR 169
+P +D E D+ + RGR R S RR
Sbjct: 119 SPRSPRSPIAHGKNIIRKAFSPRKKPDNVDPFEVAMDKARNGLGEERGRGRR--QNSGRR 176
Query: 170 RTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMG 229
RS+SP R +++ W E E + E E V E
Sbjct: 177 VARSLSPFRVSAY-PWEEQEQEQ--------------------------EQEQRDVQEQR 209
Query: 230 LERIEITPLDSASSSRSSSAGRSSKRWVFLKDFL--RSKSEGRSNNKFWSTISFSPTKDK 287
+ P S+S+ S + SSK+W LKDFL RS SEGR+ +
Sbjct: 210 KGTLSSIPSTSSSACVSCKSS-SSKKWR-LKDFLLFRSASEGRARH-------------- 253
Query: 288 KSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPH 347
N +++ S + E +N SS SSS S H
Sbjct: 254 ----NKDSVKTFTSLFRKQEDTKNSSSRGRGSSSV----------------------SAH 287
Query: 348 ELHYKANRAQAEELRRKTFLPYRQ 371
E HY + +A+ ++L++KTFLPY Q
Sbjct: 288 EFHYMSKKAETKDLKKKTFLPYMQ 311
>At3g28770 hypothetical protein
Length = 2081
Score = 48.9 bits (115), Expect = 5e-06
Identities = 38/193 (19%), Positives = 87/193 (44%), Gaps = 12/193 (6%)
Query: 141 EEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVAC 200
EEE ++++E E + ++ + + + + H E + + + K+
Sbjct: 1064 EEETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDME-- 1121
Query: 201 EIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLK 260
++ED+ +NK E+ N+ + ++VK+ + ++ E + S ++ + +S K V K
Sbjct: 1122 KLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKK 1181
Query: 261 DFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQIS-----KEKTCETQRNGSSS 315
+ SK + + K +++KK N ++ + Q S K+K + ++N
Sbjct: 1182 EKKSSKDQQKKKEK-----EMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKD 1236
Query: 316 SSSSSSKGSWGKK 328
+++K S GKK
Sbjct: 1237 DKKNTTKQSGGKK 1249
Score = 38.1 bits (87), Expect = 0.009
Identities = 34/146 (23%), Positives = 68/146 (46%), Gaps = 5/146 (3%)
Query: 189 EDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSS 248
+D ++K+ ++D+ +++NG D +E+ N K D + EI D++ ++ +SS
Sbjct: 1744 KDSKDSKSVEINGVKDDSLKDDSKNG-DINEINNGKEDSVKDNVTEIQGNDNSLTNSTSS 1802
Query: 249 AGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSP-TKDKKSSSNNQNLQAQISKEKTCE 307
K + E + + ST + TK+ S NNQN+Q S E +
Sbjct: 1803 EPNGDKLDTNKDSMKNNTMEAQGGSNGDSTNGETEETKESNVSMNNQNMQDVGSNENSMN 1862
Query: 308 TQRNGSSS---SSSSSSKGSWGKKMT 330
Q G+ S+++ ++ + K++T
Sbjct: 1863 NQTTGTGDDIISTTTDTESNTSKEVT 1888
Score = 35.4 bits (80), Expect = 0.056
Identities = 31/209 (14%), Positives = 81/209 (37%), Gaps = 19/209 (9%)
Query: 139 DLEEEEEDEEEGEVVVVRGR-------DLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDD 191
D +E++E+EE+ E + D + + S ++ + ++ + + +NE+D
Sbjct: 1152 DKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEED 1211
Query: 192 NNNKNNVA------------CEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLD 239
+ +V + +D+ N ++G ME+ + ++ + T
Sbjct: 1212 RKKQTSVEENKKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQA 1271
Query: 240 SASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQ 299
+ S++ ++ + D E ++ + + ++ + Q A+
Sbjct: 1272 DSDESKNEILMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAE 1331
Query: 300 ISKEKTCETQRNGSSSSSSSSSKGSWGKK 328
K+K + ++N +++K S GKK
Sbjct: 1332 NKKQKETKEEKNKPKDDKKNTTKQSGGKK 1360
Score = 34.3 bits (77), Expect = 0.12
Identities = 38/199 (19%), Positives = 81/199 (40%), Gaps = 20/199 (10%)
Query: 128 LERPQVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTE 187
LE + D + ++ +EE ++ +D DKSV + + E E
Sbjct: 711 LENKESQTDSKDDKSVDDKQEEAQIYGGESKD----DKSVEAKGKKK---------ESKE 757
Query: 188 NEDDNNNKNNVACEIEDEMNNKNN----ENGNDCSEMENVKVDEMGLERIEITPLDSASS 243
N+ N+N V + E+ NK E G + V+ +++ T +
Sbjct: 758 NKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAK 817
Query: 244 SRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKE 303
RS + K KD+ +++ ++ N T + +D K ++++++ + +KE
Sbjct: 818 ERSGEDNKEDKEES--KDYQSVEAKEKNENGGVDT-NVGNKEDSKDLKDDRSVEVKANKE 874
Query: 304 KTCETQRNGSSSSSSSSSK 322
++ + +R + SS+K
Sbjct: 875 ESMKKKREEVQRNDKSSTK 893
Score = 33.9 bits (76), Expect = 0.16
Identities = 36/191 (18%), Positives = 71/191 (36%), Gaps = 18/191 (9%)
Query: 142 EEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENE----DDNNNKNN 197
+ + DE + E+++ + + R + S N E E + DD N
Sbjct: 1295 QADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQKETKEEKNKPKDDKKNTTK 1354
Query: 198 VACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWV 257
+ ++ M +++ E N + D + + DS + S S S S +
Sbjct: 1355 QSGGKKESMESESKEAENQQKSQATTQADSDESKNEILMQADSQADSHSDSQADSDE--- 1411
Query: 258 FLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSS 317
SK+E +T + +D+K Q A+ K+K + ++N
Sbjct: 1412 -------SKNEILMQADSQATTQRNNEEDRKK----QTSVAENKKQKETKEEKNKPKDDK 1460
Query: 318 SSSSKGSWGKK 328
++++ S GKK
Sbjct: 1461 KNTTEQSGGKK 1471
Score = 31.2 bits (69), Expect = 1.1
Identities = 41/224 (18%), Positives = 88/224 (38%), Gaps = 8/224 (3%)
Query: 144 EEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNS-HLEWTENEDDNNNKNNVACEI 202
E EE +G D + K V + + + H+ + N+++ +K + E+
Sbjct: 624 ETSLEEKREQTQKGHDNSINSKIVDNKGGNADSNKEKEVHVGDSTNDNNMESKEDTKSEV 683
Query: 203 EDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDF 262
E + N+ ++E G + E D M +++E + S S + + ++ +
Sbjct: 684 EVKKNDGSSEKGEEGKENNK---DSMEDKKLENKESQTDSKDDKSVDDKQEEAQIYGGES 740
Query: 263 LRSKS-EGRSNNKFWSTISFSPTKDKKSSSNNQNLQA-QISKEKTCETQRNGSSSSSSSS 320
KS E + K + T + + + +N+Q + EK + ++ S + S
Sbjct: 741 KDDKSVEAKGKKKESKENKKTKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAKSVE 800
Query: 321 SKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRK 364
+K + KK++ K R E ++ Q+ E + K
Sbjct: 801 TKDN--KKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEK 842
Score = 28.1 bits (61), Expect = 8.9
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 3/55 (5%)
Query: 287 KKSSSNNQNLQAQISKEKTCETQR--NGSSSSSSSSSKGSWGK-KMTGKPMNGVG 338
K ++ + + +IS E ++ N SSSSSSSSS S K+ G+ + G+G
Sbjct: 2024 KIATESEKQFSLEISTENGYHMKKSYNSSSSSSSSSSSSSRSDLKLNGEHLKGMG 2078
>At2g37380 unknown protein
Length = 321
Score = 48.5 bits (114), Expect = 6e-06
Identities = 71/269 (26%), Positives = 103/269 (37%), Gaps = 51/269 (18%)
Query: 68 SITASSSYHHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTS--ADELFLNGQIRPMKLS 125
S T+SSS+ TSS P S EFEF + + ASG T+ ADELF GQ+ P+ L
Sbjct: 34 SSTSSSSFFSFPVTSSPPQSREFEFQM---CSSAVASGESTTSPADELFYKGQLLPLHLP 90
Query: 126 SHLERPQVLAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEW 185
L+ Q L L E + S R +SP R +S
Sbjct: 91 PRLKMVQKL-----LLASSSSTAATETPI-----------SPRAAADVLSPRRFSS---- 130
Query: 186 TENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSR 245
E D N ++ E++ + + N GN S+ + +S ++
Sbjct: 131 CEIGQDENCFFEISTELKRFIESNENHLGNSWSKK-----------------IKHSSITQ 173
Query: 246 SSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNL--------Q 297
A R+ + +F K SE K + S K+ +S N L
Sbjct: 174 KLKASRAYIKALFSKQACSDSSEINPRFKIEPS-KVSRKKNPFVNSENPLLIHRRSFSGV 232
Query: 298 AQISKEKTCETQRNGSSSSSSSSSKGSWG 326
Q + C T + SSS+SS SS S+G
Sbjct: 233 IQRHSQAKCSTSSSSSSSASSLSSSFSFG 261
>At3g27880 unknown protein
Length = 242
Score = 47.0 bits (110), Expect = 2e-05
Identities = 43/137 (31%), Positives = 59/137 (42%), Gaps = 46/137 (33%)
Query: 240 SASSSRSSSAGRSSKRWVFLKDFL-RSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQA 298
S S+ SS++ S+KRW L+DFL RSKS+G+ + KF +
Sbjct: 136 SKSTGSSSTSTWSTKRWR-LRDFLKRSKSDGKQSLKFLNP-------------------- 174
Query: 299 QISKEKTCETQRNGSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQA 358
T R+ SS S K S V S HE Y N+A
Sbjct: 175 ---------TNRDDDDESSKSKKKVSVS---------------VTVSAHEKFYLRNKAIK 210
Query: 359 EELRRKTFLPYRQGLLG 375
EE +RK++LPY+Q L+G
Sbjct: 211 EEDKRKSYLPYKQDLVG 227
>At4g37820 unknown protein
Length = 532
Score = 43.5 bits (101), Expect = 2e-04
Identities = 48/191 (25%), Positives = 71/191 (37%), Gaps = 27/191 (14%)
Query: 139 DLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNS-HLEWTENEDDNNNKNN 197
++E+EE + EG V D D+ V R + NN E T NE+ N + NN
Sbjct: 82 EVEDEEGSKNEGGGDV--STDKENGDEIVEREEEEKAVEENNEKEAEGTGNEEGNEDSNN 139
Query: 198 VACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWV 257
E E +E GN+ S E +++ G D ASS S V
Sbjct: 140 G----ESEKVVDESEGGNEISNEEAREINYKG---------DDASSEVMHGTEEKSNEKV 186
Query: 258 FLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSS 317
+ EG S + +S + +S N+ L+ + KE + T NGS
Sbjct: 187 --------EVEGESKSNSTENVS---VHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGE 235
Query: 318 SSSSKGSWGKK 328
+K K
Sbjct: 236 QQETKSELDSK 246
Score = 35.8 bits (81), Expect = 0.043
Identities = 39/181 (21%), Positives = 72/181 (39%), Gaps = 13/181 (7%)
Query: 146 DEEEGEVVVVRGRDLRLRDKSVRRRTRSMSP----LRNNSHLEWTENEDDNNNKNNVACE 201
+E+ E V V G ++V P L + E + N +N + + E
Sbjct: 179 EEKSNEKVEVEGESKSNSTENVSVHEDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQE 238
Query: 202 IEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKD 261
+ E+++K E G S E + + E T +S+ S S S+G+S+
Sbjct: 239 TKSELDSKTGEKGFSDSNGELPETNLSTSNATETT--ESSGSDESGSSGKST-------G 289
Query: 262 FLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSS 321
+ ++K+E K S+ S K+ + + + SKE+ E ++ SSS
Sbjct: 290 YQQTKNEEDEKEKVQSSEEESKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGK 349
Query: 322 K 322
+
Sbjct: 350 E 350
Score = 35.0 bits (79), Expect = 0.073
Identities = 51/241 (21%), Positives = 83/241 (34%), Gaps = 35/241 (14%)
Query: 142 EEEEDEEEG--EVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVA 199
E+ DE EG E+ R++ + + ++N +E E E +N+ NV+
Sbjct: 143 EKVVDESEGGNEISNEEAREINYKGDDASSEVMHGTEEKSNEKVE-VEGESKSNSTENVS 201
Query: 200 CEIEDEMNNKNN---------------ENGNDCSEMENVKVDEMGLERIEITPLDSASSS 244
EDE KN ENG+D E + K + LDS +
Sbjct: 202 VH-EDESGPKNEVLEGSVIKEVSLNTTENGSDDGEQQETKSE-----------LDSKTGE 249
Query: 245 R--SSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISK 302
+ S S G + + + + S+ S S + K + +Q+ +
Sbjct: 250 KGFSDSNGELPETNLSTSNATETTESSGSDESGSSGKSTGYQQTKNEEDEKEKVQSSEEE 309
Query: 303 EKTCETQRN---GSSSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAE 359
K E+ +N SSS S + KK G GK P + + E
Sbjct: 310 SKVKESGKNEKDASSSQDESKEEKPERKKKEESSSQGEGKEEEPEKREKEDSSSQEESKE 369
Query: 360 E 360
E
Sbjct: 370 E 370
Score = 31.2 bits (69), Expect = 1.1
Identities = 39/173 (22%), Positives = 74/173 (42%), Gaps = 31/173 (17%)
Query: 186 TENEDDNNNKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSR 245
T+NE+D K V E+ ++ +N D S + DE E+ E + +SS
Sbjct: 293 TKNEEDEKEK--VQSSEEESKVKESGKNEKDASSSQ----DESKEEKPERKKKEESSSQG 346
Query: 246 SSSAGRSSKRWVFLKDFLRSKSEGR-----------SNNKFWSTISFSPTKDKKSSSNNQ 294
KR K+ S+ E + S+++ + I + K+K+ SS+ +
Sbjct: 347 EGKEEEPEKRE---KEDSSSQEESKEEEPENKEKEASSSQEENEIKETEIKEKEESSSQE 403
Query: 295 NLQAQISKEKTCETQRNGSSSS-----------SSSSSKGSWGKKMTGKPMNG 336
+ + +++K+ E+QR +++S SS++ KG K K +G
Sbjct: 404 GNENKETEKKSSESQRKENTNSEKKIEQVESTDSSNTQKGDEQKTDESKRESG 456
>At1g70420 unknown protein
Length = 272
Score = 42.7 bits (99), Expect = 4e-04
Identities = 79/323 (24%), Positives = 120/323 (36%), Gaps = 99/323 (30%)
Query: 74 SYHHQTTTSSVPMSYEFEFSARFGSTGSAASGSMTSADELFLNGQIRPMKLSSHLERPQV 133
S+ +Q T E +FS F S A S +ADE F +GQIRP+
Sbjct: 48 SWWNQNGTDKDDEGREEDFS--FASVN--ADNSPITADEAFEDGQIRPVY---------- 93
Query: 134 LAPLLDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNN 193
PL + +D EE K++R SPL+ +E T E++
Sbjct: 94 --PLFNRNIFFDDPEE---------------KTLR------SPLKK-LFVESTTTEEEEE 129
Query: 194 NKNNVACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSS 253
+ V G CS N V++ S + R S++ S
Sbjct: 130 ESDTV---------------GPYCS-WTNRTVEQA-----------SPETCRKSNSTGFS 162
Query: 254 KRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGS 313
K W F LRS S+G+ F S S S+S+ + A++S ++ G
Sbjct: 163 KLWRFRDLVLRSNSDGKDAFVFLSNGS-------SSTSSTSSTAARLSG--VVKSSEKGK 213
Query: 314 SSSSSSSSKGSWGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQAEELRRKTFLPYRQGL 373
+ + K K S HE Y NRA EE +R+++LPY+
Sbjct: 214 EKTKTDKKKDKMRTK----------------SAHEKLYMRNRAMREEGKRRSYLPYKH-- 255
Query: 374 LGCLGFSSKGYGAMNGFARALNP 396
+GF + +NG R ++P
Sbjct: 256 ---VGF----FTNVNGLTRNVHP 271
>At5g12300 putative protein
Length = 374
Score = 42.0 bits (97), Expect = 6e-04
Identities = 22/72 (30%), Positives = 43/72 (59%), Gaps = 1/72 (1%)
Query: 141 EEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDN-NNKNNVA 199
EEEEE+ + E ++ + + +S+++ T S++ ++ L +E+D+ NN NN A
Sbjct: 283 EEEEEENKMNEETTMQKQIAEMYMRSMQQFTESLAKMKLPMDLHNKPHEEDHSNNNNNTA 342
Query: 200 CEIEDEMNNKNN 211
+I+++ NN NN
Sbjct: 343 TQIQNQNNNANN 354
>At5g16780 putative protein
Length = 820
Score = 39.7 bits (91), Expect = 0.003
Identities = 53/209 (25%), Positives = 98/209 (46%), Gaps = 20/209 (9%)
Query: 141 EEEEED-----EEEGEVVVVRGRDL-RLRDKSVRRRTRSMSPLRNNSHLEWTENEDDN-N 193
+E+E D +E+ E + RGRD R +DKS R N H + ENE DN
Sbjct: 60 KEKERDRKRSRDEDTEKEISRGRDKEREKDKSRDRVKEKDKEKERNRHKD-RENERDNEK 118
Query: 194 NKNNVACEIEDEMNNKNNENGNDCSEMEN--VKVDEMGLERIEITPLDSASSSRSSSAGR 251
K+ +++ + K++E+ ++ + D GL +D+ASS + +SA
Sbjct: 119 EKDKDRARVKERASKKSHEDDDETHKAAERYEHSDNRGLNE-GGDNVDAASSGKEASALD 177
Query: 252 SSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRN 311
R + +++ + K+E S+ W ++ S ++K ++ Q Q Q+S + E Q N
Sbjct: 178 LQNRILKMREERKKKAEDASDALSW--VARSRKIEEKRNAEKQRAQ-QLS--RIFEEQDN 232
Query: 312 GSSSSSSSSSKGSWGKKMTG-KPMNGVGK 339
+ + G G+ ++G K ++G+ K
Sbjct: 233 ---LNQGENEDGEDGEHLSGVKVLHGLEK 258
>At3g23900 unknown protein
Length = 989
Score = 39.7 bits (91), Expect = 0.003
Identities = 56/255 (21%), Positives = 104/255 (39%), Gaps = 26/255 (10%)
Query: 164 DKSVRRRTRSMSPLRNNSHLEWTENEDD---NNNKNNVACEIEDEMNNKNNENGND---- 216
+ V+RRTRS S +S ++ D+ ++ K + + ED ++ +D
Sbjct: 648 ENKVKRRTRSRSRSVEDSADIKDKSRDEELKHHKKRSRSRSREDRSKTRDTSRNSDEAKQ 707
Query: 217 ----CSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSN 272
S +++ D E +++ + +S S +S +D+ + GRS
Sbjct: 708 KHRQRSRSRSLENDNGSHENVDVAQDNDLNSRHSKRRSKSLD-----EDYDMKERRGRSR 762
Query: 273 NKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQR--NGSSSSSSSSSKGSWGKKMT 330
++ T + S K+K N + + S+ K+ E +R N + S SK G++
Sbjct: 763 SRSLETKNRSSRKNKLDEDRNTGSRRRRSRSKSVEGKRSYNKETRSRDKKSKRRSGRRSR 822
Query: 331 GKPMNGVGKRRVPPSP------HELHYKANRAQAEELRRKTFLPYRQGLLGCLGFSSKGY 384
G R + SP H + +R+++ E ++ + R L SS G
Sbjct: 823 SPSSEGKQGRDIRSSPGYSDEKKSRHKRHSRSRSIE-KKNSSRDKRSKRHERLRSSSPGR 881
Query: 385 GAMNGFARALNPVSS 399
G R+L+PVSS
Sbjct: 882 DKRRG-DRSLSPVSS 895
>At5g66800 unknown protein
Length = 183
Score = 38.5 bits (88), Expect = 0.007
Identities = 38/111 (34%), Positives = 50/111 (44%), Gaps = 23/111 (20%)
Query: 46 GGGPPPISSGYFYS---APASPLHFSITASSSYHHQTTTSSVPMSYEFEFSARFGSTGSA 102
GGG P+S +S P T SS Q +SS S FEFS
Sbjct: 11 GGGGTPMSPRISFSNDFVEIRPETTKTTRSSPLSKQEGSSS-SFSDNFEFSV-------- 61
Query: 103 ASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEE---EEDEEEG 150
++ +M ADELF G++ P K ++ ++R L EE EEDEEEG
Sbjct: 62 SNYTMMPADELFSKGKLLPFKETNQVQR--------TLREELLVEEDEEEG 104
>At5g49100 unknown protein
Length = 396
Score = 38.1 bits (87), Expect = 0.009
Identities = 35/100 (35%), Positives = 45/100 (45%), Gaps = 10/100 (10%)
Query: 240 SASSSRSSSAG----RSSKRWVFLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQN 295
+ASSS S++A RS + S R N FWS +K SS N
Sbjct: 109 TASSSSSTTANIVYKRSQSTRTTKTTYGDSDLSPRKRNGFWSFFHLYSSKQHGSSKKVGN 168
Query: 296 LQAQISKEKT----CETQRNGSSSSSSSSSKGSWGKKMTG 331
IS+ +T ET GSSSSSS+SS S K++ G
Sbjct: 169 FHQPISQTETKTELAETTTVGSSSSSSASS--SMSKRVVG 206
Score = 33.5 bits (75), Expect = 0.21
Identities = 36/121 (29%), Positives = 50/121 (40%), Gaps = 12/121 (9%)
Query: 208 NKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKS 267
+ +N NG D V V+E G IE+TP + SRS S G S+ F DF +
Sbjct: 210 SSSNRNGID------VIVEEDGSPNIEVTPSER-KVSRSRSVGCGSRS--FSGDFFERIT 260
Query: 268 EGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNG---SSSSSSSSSKGS 324
G + S + K + + N + + C G +SSSSSSS S
Sbjct: 261 NGFGDCTLRRVESQREGNNNKGNKVSSNPSNGVREMVRCGGIFGGFMIMTSSSSSSSSSS 320
Query: 325 W 325
W
Sbjct: 321 W 321
>At5g19340 putative protein
Length = 255
Score = 37.7 bits (86), Expect = 0.011
Identities = 40/156 (25%), Positives = 65/156 (41%), Gaps = 33/156 (21%)
Query: 104 SGSMTSADELFLNGQIRPMKLSSHLERPQ--VLAPLLDLEEEEED----EEEGEVVVVRG 157
+ +M SADELF G++ P H E+ + L P +++++EEED EEG V
Sbjct: 60 NATMLSADELFSEGKLLPFWQVKHSEKLKNVTLKPKVEVQQEEEDHKVVNEEGFV----- 114
Query: 158 RDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDC 217
+N E N ++NNN N + ++D+ + + +
Sbjct: 115 ---------------------HNKEQE-NNNNNNNNNNNRGSWFLDDDPSPRPPKCTVLW 152
Query: 218 SEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSS 253
E+ +K T + S S S SSS+ SS
Sbjct: 153 KELLRLKKQRTTTTTTASTRVSSLSPSSSSSSTSSS 188
>At4g26660 putative protein
Length = 806
Score = 37.7 bits (86), Expect = 0.011
Identities = 47/196 (23%), Positives = 76/196 (37%), Gaps = 14/196 (7%)
Query: 144 EEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNNVAC--E 201
+ DE EG+ V+V G D+ +K + + S + ++ D + + C +
Sbjct: 187 KRDESEGKEVIVDGEDVLELNKHIEKLHGSYDSVHSSFASASCYEADSMSGDDEDVCSED 246
Query: 202 IEDEMNNKNNENG---NDCSEMENVKVDEMGL-----ERIEITPLDSASSSRSSSAGRSS 253
+E M+ + + ND S+++NV+ D G ++ L+ S S
Sbjct: 247 LEKPMHGNHKDVDFVDNDPSQLDNVEFDTTGSGISIRSQLPSCVLEEPIFSESPKFKNVQ 306
Query: 254 KRWVFLKDFL---RSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQR 310
K F R+ SE SN SP KK S +L A + +
Sbjct: 307 KSVAASTKFSANPRNVSES-SNIGDMKVNQISPCMSKKVSGPTDSLAASLQRGLQIIDYH 365
Query: 311 NGSSSSSSSSSKGSWG 326
GSS S SSS S+G
Sbjct: 366 QGSSLSKSSSVSFSFG 381
>At3g24630 hypothetical protein
Length = 724
Score = 37.7 bits (86), Expect = 0.011
Identities = 68/334 (20%), Positives = 128/334 (37%), Gaps = 36/334 (10%)
Query: 53 SSGYFYSAPASPLHFSITASSSYHHQTTTSSVPMSYEFEFSARFG---STGSAASGSMTS 109
SSG S+ +S ++ S+ + +S VP + G ST S+
Sbjct: 216 SSGAVSSSTSSSQSSMVSGSTK---SSASSDVPQRAPSLIARLMGLDVSTQEPRKSSVNH 272
Query: 110 ADE---LFLNGQIRPMKLSSHLERPQVL-------APLLDLEEEEEDEEEGEVVVVRGRD 159
D+ L L+ + + + E P+++ A L L EE E +V++R
Sbjct: 273 IDKPDILKLSSERQEKVKKNSKESPEIVRCNSTREAALQSLPEETPSENPSTIVLIRPMR 332
Query: 160 LRLRDKSVRRRTRSMSP-LRNNSHLEWTENEDDNNNKNNVACEIEDEMNNKNNENGNDCS 218
+ + ++ P ++ H D+ + ++ M K+ E
Sbjct: 333 VVKPEPGSKQPVVPKKPRMQGEVHPRMINQRKDHQANGSNKMKLPLRMTKKDKEPKEMVP 392
Query: 219 EMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWST 278
++E + + L ++P ++ +R ++K+ V KD + +EG+ ++
Sbjct: 393 KVEENEGKVIKL----MSPSNAKVLTRDRKPLETNKKLVVKKDDI---AEGKDRHRALKP 445
Query: 279 ISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGSW------GKKMTGK 332
S +P K S++++ +S+ K+ + R SSSSS S K S KK +
Sbjct: 446 PS-NPVSQKISNNSSD-----VSRNKSRRSSRLSSSSSSGSREKKSGEASRPNAKKKLRQ 499
Query: 333 PMNGVGKRRVPPSPHELHYKANRAQAEELRRKTF 366
N +G S E H N+ EE F
Sbjct: 500 QDNDLGSENNSCSSQETHGSLNQLSTEETTSSEF 533
>At1g68920 putative bHLH transcription factor (bHLH049)
Length = 486
Score = 37.0 bits (84), Expect = 0.019
Identities = 24/94 (25%), Positives = 39/94 (40%), Gaps = 1/94 (1%)
Query: 265 SKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGS 324
++S G + K T S + + + N++ Q+ S++ E NG + S S
Sbjct: 218 TQSSGGNGQKGRETSSNTKKRKRNGQKNSEAAQSHRSQQSEEEPDNNGDEKRNDEQSPNS 277
Query: 325 WGKKMTGKPMNGVGKRRVPPSPHELHYKANRAQA 358
GKK G + PP +H +A R QA
Sbjct: 278 PGKKSNSGKQQG-KQSSDPPKDGYIHVRARRGQA 310
>At3g50640 unknown protein
Length = 166
Score = 36.6 bits (83), Expect = 0.025
Identities = 23/53 (43%), Positives = 28/53 (52%), Gaps = 8/53 (15%)
Query: 101 SAASGSMTSADELFLNGQIRPMKLSSHLERPQVLAPLLDLEEEEEDEEEGEVV 153
S SM ADE+FL G+I P K +SH+ R L EE EEEG +V
Sbjct: 53 SVTDYSMIPADEIFLKGKILPFKETSHVHR--------TLGEELLTEEEGSMV 97
>At1g13030 unknown protein
Length = 608
Score = 36.2 bits (82), Expect = 0.033
Identities = 37/137 (27%), Positives = 53/137 (38%), Gaps = 6/137 (4%)
Query: 203 EDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPL-DSASSSRSSSAGRSSKRWVFLKD 261
E + K E+ + C + + E G IE + L D S SSS + +
Sbjct: 466 EFPIEKKTEEDDDFCMQPDTSLYKEDGSLEIEFSALLDVRSVKTSSSDSAEVAKSALPEP 525
Query: 262 FLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSS 321
+K S NK T P K+ S + L +S +K +Q N + SSS
Sbjct: 526 DQSAKKPKLSANKELQT----PAKENGEVSPWEELSEALSAKKAALSQANNGWNKKGSSS 581
Query: 322 KGSWG-KKMTGKPMNGV 337
GSW K + G M V
Sbjct: 582 GGSWSYKALRGSAMGPV 598
Score = 31.6 bits (70), Expect = 0.81
Identities = 45/186 (24%), Positives = 70/186 (37%), Gaps = 35/186 (18%)
Query: 178 RNNSHLEWTENEDDNNNKNNVACEIED-------EMNNKNNENGNDCSEMENV-KVDEMG 229
+ S LE + D N N A E+E+ EM N E + E+ + DE+
Sbjct: 92 KKESLLEIVGEDSDENVYN--AIEVEERPQIRPGEMLLANEEFQKETGGYESESEEDELE 149
Query: 230 LERIEITPLDSASSSR-SSSAGRSSKRWVFLKDFLRSKSEGRSNNKFWSTI--------- 279
E E P AS R +SS +S+KR D + R N S +
Sbjct: 150 EEAEEFVPEKKASKKRKTSSKNQSTKRKKCKLDTTEESPDERENTAVVSNVVKKKKKKKS 209
Query: 280 --------------SFSP-TKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSSSSSSKGS 324
S P TK K+SS ++ + + + ET++ S S+ +K
Sbjct: 210 LDVQSANNDEQNNDSTKPMTKSKRSSQQEESKEHNDLCQLSAETKKTPSRSARRKKAKRQ 269
Query: 325 WGKKMT 330
W ++ T
Sbjct: 270 WLREKT 275
Score = 30.0 bits (66), Expect = 2.3
Identities = 33/181 (18%), Positives = 65/181 (35%), Gaps = 40/181 (22%)
Query: 138 LDLEEEEEDEEEGEVVVVRGRDLRLRDKSVRRRTRSMSPLRNNSHLEWTENEDDNNNKNN 197
LD EE DE E VV + + KS+ ++ N++ NN+
Sbjct: 181 LDTTEESPDERENTAVVSNVVKKKKKKKSL--------------DVQSANNDEQNNDSTK 226
Query: 198 VACEIEDEMNNKNNENGNDCSEMENVKVDEMGLERIEITPLDSASSSRSSSAGRSSKRWV 257
+ + + ++ ND ++ S+ + RS++R
Sbjct: 227 PMTKSKRSSQQEESKEHNDLCQL---------------------SAETKKTPSRSARRKK 265
Query: 258 FLKDFLRSKSEGRSNNKFWSTISFSPTKDKKSSSNNQNLQAQISKEKTCETQRNGSSSSS 317
+ +LR K++ + + +P++ + + Q +KEK CET N +
Sbjct: 266 AKRQWLREKTKLEKEELLQTQLVVAPSQKPVIT-----IDHQATKEKHCETLENQQAEEV 320
Query: 318 S 318
S
Sbjct: 321 S 321
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.307 0.125 0.352
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,358,090
Number of Sequences: 26719
Number of extensions: 520121
Number of successful extensions: 4094
Number of sequences better than 10.0: 199
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 159
Number of HSP's that attempted gapping in prelim test: 3498
Number of HSP's gapped (non-prelim): 489
length of query: 400
length of database: 11,318,596
effective HSP length: 101
effective length of query: 299
effective length of database: 8,619,977
effective search space: 2577373123
effective search space used: 2577373123
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC149080.12


