

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.6 - phase: 0
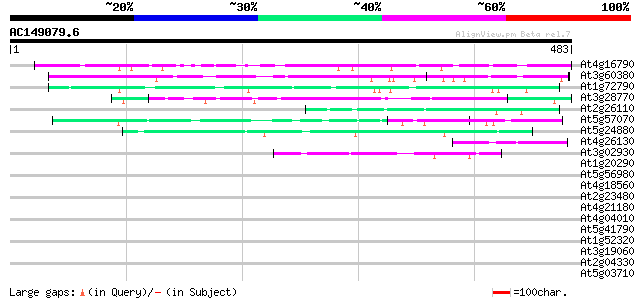
(483 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g16790 glycoprotein homolog 206 3e-53
At3g60380 putative protein 130 1e-30
At1g72790 unknown protein 62 7e-10
At3g28770 hypothetical protein 55 7e-08
At2g26110 unknown protein 50 4e-06
At5g57070 putative protein 49 8e-06
At5g24880 glutamic acid-rich protein 44 2e-04
At4g26130 uncharacterized protein 42 8e-04
At3g02930 unknown protein 42 8e-04
At1g20290 hypothetical protein 41 0.001
At5g56980 unknown protein 41 0.002
At4g18560 pherophorin - like protein 41 0.002
At2g23480 hypothetical protein 41 0.002
At4g21180 unknown protein 40 0.002
At4g04010 hypothetical protein 40 0.002
At5g41790 myosin heavy chain-like protein 40 0.003
At1g52320 bZIP protein, putative 40 0.003
At3g19060 hypothetical protein 40 0.004
At2g04330 hypothetical protein 40 0.004
At5g03710 putative protein 39 0.005
>At4g16790 glycoprotein homolog
Length = 473
Score = 206 bits (523), Expect = 3e-53
Identities = 183/516 (35%), Positives = 249/516 (47%), Gaps = 112/516 (21%)
Query: 22 QNQQEPNKFHYNFAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAIS 81
+ Q P KF+ F +KA I+ + ++P+ SQ PE +Q TR ELLHL+FVGIA+S
Sbjct: 16 KEDQNPRKFYSRFIFKALILTVLCAVVPVFLSQTPELANQ---TRLLELLHLVFVGIAVS 72
Query: 82 YGLFSRRNQEPD----KDNNNNTKFD----NAQSLVSRFLQVSSFFEDEVDHQNQSE--- 130
YGLFSRRN + N+++ K D N+ S V + L+VSS F V H+++SE
Sbjct: 73 YGLFSRRNYDGGGGGGTSNSDHNKADHSNNNSHSYVPKILEVSSVFN--VGHESESEPSD 130
Query: 131 --SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSL 188
S D K QTW N+ Y K P+V+ + F D SS N EKPLLLPVRSL
Sbjct: 131 DSSGDQRKFQTWKNK---YHMK-----IPEVE----TRFVDRVSSENR-EKPLLLPVRSL 177
Query: 189 K-SRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENA-V 246
SR+SD S D +S + +VR+ E LK ++N+ V
Sbjct: 178 NYSRVSDS-------SGD----------NSGRWEKVRSKREL-------LKTLGDDNSDV 213
Query: 247 LPSPIPWRSRSASARMMEPKQEAIEKALKASMVME----------PKQEANENASKP--- 293
LPSPIPWRSRS+S+ K+ ++K +E P ++ S P
Sbjct: 214 LPSPIPWRSRSSSSSSSSSKEVESLPSVKNLTTVESQPLIKNLTPPSSFSSPRKSNPIPN 273
Query: 294 --------------------SMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY--PPPP 331
+ +SS K P + E + K F + PPPP
Sbjct: 274 LASEFHPSPPPPPPPPPPLPAFYNSSSRKDHPGIYRVERRESSVHKTKFAGGEFHPPPPP 333
Query: 332 PPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEK---EDEDEEEKEYSTSSMQEQ 388
PPPPP +YKS K R N R S + ++ +K D E KE T E+
Sbjct: 334 PPPPPVEYYKSPPTKFRLS--NERRKSSEQKMKRNAPKKVWWSDPIVESKEQDT----EK 387
Query: 389 TDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDV 448
D+ + + + E+E + E+ E+ V+ EE S ++ G DV
Sbjct: 388 NDQRSNLGSKAVEESENGEQRRGEN--EIHDEVEKKIVEEEGVSEINNG--------SDV 437
Query: 449 DKKADEFIAKFREQIRLQRIESIKRST-RVARNSSR 483
DKKADEFIAKFREQIRLQRIESIKRST +++ NSSR
Sbjct: 438 DKKADEFIAKFREQIRLQRIESIKRSTNKISANSSR 473
>At3g60380 putative protein
Length = 743
Score = 130 bits (328), Expect = 1e-30
Identities = 133/507 (26%), Positives = 215/507 (42%), Gaps = 100/507 (19%)
Query: 34 FAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAISYGLFSRRNQEPD 93
F K+ + +F + LPL PSQAP+F+ + +LT+ WEL+HLLFVGIA++YGLFSRRN E
Sbjct: 32 FFCKSVLFALFLLALPLFPSQAPDFVGETVLTKFWELIHLLFVGIAVAYGLFSRRNVESA 91
Query: 94 KDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDH---------------------------- 125
D +++ S VSR QVSS F++E D
Sbjct: 92 VDLRMTRVDESSLSYVSRIFQVSSVFDEEFDDNSCEFVDVRSDESVSARASVVGKSESFV 151
Query: 126 ------QNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEK 179
+ SE + N+++ W+ +Q+++ K V+VA L V +
Sbjct: 152 VESGELEESSEFGETNEVRAWN--SQYFQGKSKVVVARPAYGLDGHVV----------HQ 199
Query: 180 PLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQ 239
PL LP+R L+S L D+ + D AE E L+ D
Sbjct: 200 PLGLPIRRLRSSLRDNAALQDKSFADSCDGAVN--------------AEAESLLADNF-- 243
Query: 240 KEEENAVLPSPIPWRSRSASARMMEPKQEAIEK-ALKASMVMEPKQEANENASKPSMVKT 298
+E A SP+PW++R + + + ++ ++ + ++S+ S
Sbjct: 244 FDEVLAAPASPVPWQARPEMMGIGDNYPSNFQPISVDETLKSISSRSTGSSSSQTSYASQ 303
Query: 299 SSIKFTPSESVA-----KNSEDLIKKKSFYYK------SYPP----PPPPPPPTMFYKST 343
+ +F+PS SV+ N E+L+K+KS S PP P PP P + T
Sbjct: 304 NQNRFSPSRSVSAESLNSNVEELVKEKSRQSSSRSSSPSLPPSPSLSPSPPSPELVPNDT 363
Query: 344 FLKS--RYGEKNGRNMSMGKSIEEGTNEKED-----EDEEEKEYSTSSMQEQTDKERFIE 396
+S + R S + +G+ +ED E+E E E K+
Sbjct: 364 RRRSPELVTDDTPRRASHSRHYSDGSLLEEDVRRGFENELEGSKVRGRKAEFFSKKERGS 423
Query: 397 KRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCS-SSLSGGIDTVSDEGPDVDKKADEF 455
K + L AE + ++ KS ++ P S SS GG D + D+ +K++
Sbjct: 424 KSLNLAAESS-----------RRGNKSRRSYPPESISSPVGGADDSTTRRQDLQQKSNCH 472
Query: 456 IAKFREQIRLQRIESIKRSTRVARNSS 482
+ E IR + +E+ + RV + S
Sbjct: 473 L--LEENIR-KGVEADHNNLRVKKGRS 496
Score = 48.1 bits (113), Expect = 1e-05
Identities = 45/135 (33%), Positives = 64/135 (47%), Gaps = 19/135 (14%)
Query: 360 GKSIEEGTNEKEDEDEEEKEY-----STSSMQEQTD----KERFIEKRVILEAEETDSDE 410
GK ++ + ED E + E S QE+ +E+ E R E EE +E
Sbjct: 612 GKDVKTDGDSSEDRAEAKVESRGRTKSRRPRQEELSIVLHQEKSSETRAKSEPEEVAMEE 671
Query: 411 DEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIES 470
+ E + V + EE S S S + +VD+KA EFIAKFREQIRLQ++ S
Sbjct: 672 PQ--AEQQPEVTFEEEEEAAWESQSNA----SHDHNEVDRKAGEFIAKFREQIRLQKLIS 725
Query: 471 IKR----STRVARNS 481
++ T + RNS
Sbjct: 726 GEQPRGGGTGIFRNS 740
>At1g72790 unknown protein
Length = 561
Score = 62.0 bits (149), Expect = 7e-10
Identities = 103/518 (19%), Positives = 190/518 (35%), Gaps = 102/518 (19%)
Query: 34 FAYKATIVLIFFVILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAISYGLFSRRNQEPD 93
FA A I+++ F+I P S + +L+ ++W+ L+ + V A+ G SR +
Sbjct: 42 FAGTAAILIVVFIIPPFF-SSVSQIFRPHLVRKSWDYLNFVLVLFAVLCGFLSRNTNNDE 100
Query: 94 ----KDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRN 149
K+ + KF + S++ R +VS+ + ++ +Q + R
Sbjct: 101 SNHHKEEDIRNKFSTSPSIIDRRSRVSN-------SGTTPRYWNDDRGGGGGDQTVYKRF 153
Query: 150 KPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSV----- 204
+ V+ DE+ F ++ R + R D D + +QS
Sbjct: 154 SRLRSVSSYPDLRLREYEADERWRFYDD-------TRVSQCRYEDVDPIYPNQSYRNWHE 206
Query: 205 -------------DGLSKTTTK-RFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSP 250
DG + +K R + +V A E V ++LK +PSP
Sbjct: 207 EGKPPPEDVDQTEDGDNGEGSKVRNGGSETEKVEVVATAEAEVVEELKVPSAP-PYIPSP 265
Query: 251 IPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVA 310
P R A+ + K + + + E K+E ++ + + + + + S
Sbjct: 266 PPSPPRPPPAKQAKRKTNRVYQDVSPQ---EEKKERDDFVATTTPIPPPATVYQKSNKQE 322
Query: 311 KN----SEDLI-----KKKSFYYKS---------------YPPPPPPPPPTMFYKSTFLK 346
K ++D + KKK +S Y PPPPPPPP F++ F
Sbjct: 323 KKKGGATKDFLIALRRKKKKQRQQSIDGLDLLFGSDPPLVYSPPPPPPPPPPFFQGLF-- 380
Query: 347 SRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEET 406
K G++ + E E STS +++ + R + + +
Sbjct: 381 ---SSKKGKSKKNNSNPPPPPPPPPPERRYESRASTSKLRKAPVESRTSKPNPPAKVTQY 437
Query: 407 DSDEDEDV-----------------------GELKQ-----SVKSVKTEEPCSSSLSGGI 438
E G+ + S+ S + ++P + +G
Sbjct: 438 VGTGSESPLMPIPPPPPPPPFKMPAWKFVKRGDYVRMASDISISSDEPDDPDVAQSAGSK 497
Query: 439 DTVSD---EGPDVDKKADEFIAKFREQIRLQRIESIKR 473
+ PDVD KAD+FIA+FR ++L+++ S+KR
Sbjct: 498 EAAGSMFCPSPDVDTKADDFIARFRAGLKLEKMNSVKR 535
>At3g28770 hypothetical protein
Length = 2081
Score = 55.5 bits (132), Expect = 7e-08
Identities = 79/403 (19%), Positives = 156/403 (38%), Gaps = 31/403 (7%)
Query: 88 RNQEPDKDN--NNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQ 145
+ +E DK NN K + + S E+ D++ + ESED + +
Sbjct: 957 KKKEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYE 1016
Query: 146 HYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVD 205
++K + ++ Q+ E++ S +++K R LK++ +++ + +S +
Sbjct: 1017 EKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKE-KEESRDLKAKKKEEETKEKKESEN 1075
Query: 206 GLSK---TTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARM 262
SK + + S + + E + E K ++KEE+ + S
Sbjct: 1076 HKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDK 1135
Query: 263 MEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSF 322
E K+ K +K + K+E E + + + S K + K+S+D KKK
Sbjct: 1136 NEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEK 1195
Query: 323 YYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYST 382
K + + +KN + S+EE N+K+ E ++EK
Sbjct: 1196 EMKE------------------SEEKKLKKNEEDRKKQTSVEE--NKKQKETKKEKNKPK 1235
Query: 383 SSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVS 442
+ T + K+ +E+E ++ E++ + S +++ D+ S
Sbjct: 1236 DDKKNTTKQSG--GKKESMESESKEA-ENQQKSQATTQADSDESKNEILMQADSQADSHS 1292
Query: 443 DEGPDVDKKADEFIAKFREQIRLQR--IESIKRSTRVARNSSR 483
D D D+ +E + + Q QR E K+ T VA N +
Sbjct: 1293 DSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAENKKQ 1335
Score = 43.9 bits (102), Expect = 2e-04
Identities = 68/313 (21%), Positives = 131/313 (41%), Gaps = 44/313 (14%)
Query: 120 EDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVF---EDEQSSFNE 176
+DE N+ E++D I T S Q + K + ++ +NS E+++ +
Sbjct: 918 KDEKKEGNKEENKDT--INTSSKQKGKDKKK-------KKKESKNSNMKKKEEDKKEYVN 968
Query: 177 NEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDK 236
NE L + K ++ ++ ++ D K + S +S ++ R E+E E K
Sbjct: 969 NE--LKKQEDNKKETTKSENSKLKEENKDNKEK----KESEDSASKNREKKEYE---EKK 1019
Query: 237 LKQKEE-ENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSM 295
K KEE + S R S K++ + LKA E +E E+ + S
Sbjct: 1020 SKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSK 1079
Query: 296 VKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGR 355
K + ++S+ K + KKK + +S KSR E++ +
Sbjct: 1080 KKEDKKEHEDNKSMKKEEDKKEKKK--HEES-------------------KSRKKEEDKK 1118
Query: 356 NMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVG 415
+M + + +KED++E++K +++++DK+ E E +E +S + +
Sbjct: 1119 DMEKLED-QNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETKEIESSKSQKNE 1177
Query: 416 ELKQSVKSVKTEE 428
K+ KS K ++
Sbjct: 1178 VDKKEKKSSKDQQ 1190
Score = 36.2 bits (82), Expect = 0.041
Identities = 64/387 (16%), Positives = 147/387 (37%), Gaps = 38/387 (9%)
Query: 88 RNQEPDKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHY 147
+ Q+ K N K D + + S + + +NQ +S+ + + ++N+
Sbjct: 1222 KKQKETKKEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESKNE-- 1279
Query: 148 RNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKS--RLSDDDCVVQSQSVD 205
+++ Q +S D Q+ +E++ +L+ S + R +++D Q+ +
Sbjct: 1280 -----ILMQADSQADSHS---DSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVAE 1331
Query: 206 GLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQ--KEEENAVLPSPIPWRSRSASARMM 263
+ TK + + +N + G ++ ++ KE EN + A
Sbjct: 1332 NKKQKETKEEKNKPKDDKKNTTKQSGGKKESMESESKEAENQQKSQA------TTQADSD 1385
Query: 264 EPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFY 323
E K E + +A + Q ++ + +++ S + +N+E+ KK++
Sbjct: 1386 ESKNEILMQADSQADSHSDSQADSDESKNEILMQADS-----QATTQRNNEEDRKKQTSV 1440
Query: 324 YKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTS 383
++ K K + +KN S GK + KE E++++ + +T
Sbjct: 1441 AENKKQKET--------KEEKNKPKDDKKNTTEQSGGKKESMESESKEAENQQKSQATTQ 1492
Query: 384 SMQEQTDKERFIEKRVILEAE-ETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGI---- 438
+++ E ++ + + D DE E+ S + S I
Sbjct: 1493 GESDESKNEILMQADSQADTHANSQGDSDESKNEILMQADSQADSQTDSDESKNEILMQA 1552
Query: 439 DTVSDEGPDVDKKADEFIAKFREQIRL 465
D+ +D D D+ +E + + Q ++
Sbjct: 1553 DSQADSQTDSDESKNEILMQADSQAKI 1579
Score = 30.0 bits (66), Expect = 3.0
Identities = 60/287 (20%), Positives = 121/287 (41%), Gaps = 31/287 (10%)
Query: 193 SDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEF------EGLVEDKLKQKEEENAV 246
S DD V+++ SK K + + NRVRN E E +K ++KE ++A
Sbjct: 740 SKDDKSVEAKGKKKESKENKK--TKTNENRVRNKEENVQGNKKESEKVEKGEKKESKDAK 797
Query: 247 LPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPS 306
+ S++ E K+ + E E K+E+ + S + K + +
Sbjct: 798 SVETKDNKKLSSTENRDEAKERSGEDN------KEDKEESKDYQSVEAKEKNENGGVDTN 851
Query: 307 ESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSM------G 360
++S+DL +S K+ ++ ++ NM + G
Sbjct: 852 VGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQKGSG 911
Query: 361 KSIEEGTNEKEDEDEEEKE--YSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDV-GEL 417
+S++ +EK++ ++EE + +TSS Q+ DK++ ++ ++ + D+ E V EL
Sbjct: 912 ESVKYKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEYVNNEL 971
Query: 418 KQSVKSVKTEEPCSSSLSGGIDTVSDEGPD--VDKKADEFIAKFREQ 462
K+ + K +S + +E D K++++ +K RE+
Sbjct: 972 KKQEDNKKETTKSENS------KLKEENKDNKEKKESEDSASKNREK 1012
Score = 30.0 bits (66), Expect = 3.0
Identities = 75/423 (17%), Positives = 162/423 (37%), Gaps = 53/423 (12%)
Query: 68 WELLHLLFVGIAISYGLFSRRNQEPDKDNNNNTKFDNA---QSLVSRFLQVSSFFEDEVD 124
WE V I + + S+ + E + +++T+ +N ++V + S +++ D
Sbjct: 169 WEQTITRIVKIVVE--VKSKSSSEASSEESSSTEHNNVTTGSNMVETNGENSESTQEKGD 226
Query: 125 HQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLP 184
S D++ N+ + + V+ + ++ E E + +
Sbjct: 227 GVEGSNGGDVSMENLQGNKVEDLKEGNNVVENGETKENNGENVESNNEKEVEGQGESIGD 286
Query: 185 VRSLKSRLSDDDCV--VQSQSVDGLSKTTT--KRFSSNSFNRVRNYAEFEGL-------- 232
K+ S +D V++ DG S T + +N + + N E EG
Sbjct: 287 SAIEKNLESKEDVKSEVEAAKNDGSSMTENLGEAQGNNGVSTIDNEKEVEGQGESIEDSD 346
Query: 233 VEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEK-ALKASMVMEPKQEANENAS 291
+E L+ KE+ + + + +++A + M +EA + + M + + + ++
Sbjct: 347 IEKNLESKEDVKSEVEA-----AKNAGSSMTGKLEEAQRNNGVSTNETMNSENKGSGEST 401
Query: 292 KPSMVKTSSI----KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKS 347
MV ++ K E +N+ + +K ++ K+ K L++
Sbjct: 402 NDKMVNATTNDEDHKKENKEETHENNGESVKGENLENKAGNEES--------MKGENLEN 453
Query: 348 RYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETD 407
+ G + + S+E TN E EE++E S S + +KE + V ++ E
Sbjct: 454 KVGNEE---LKGNASVEAKTNN-ESSKEEKREESQRSNEVYMNKETTKGENVNIQGESIG 509
Query: 408 SDEDEDVGELKQSVK----------SVKTEEPCSSSLSGGIDT----VSDEGPDVDKKAD 453
++ E K+ VK + E + ++ G+ T + + G D KK D
Sbjct: 510 DSTKDNSLENKEDVKPKVDANESDGNSTKERHQEAQVNNGVSTEDKNLDNIGADEQKKND 569
Query: 454 EFI 456
+ +
Sbjct: 570 KSV 572
Score = 28.5 bits (62), Expect = 8.6
Identities = 23/80 (28%), Positives = 39/80 (48%), Gaps = 6/80 (7%)
Query: 351 EKNGRNMSMGKSIEEGTNEK-EDEDEEEKEYSTSSMQEQT--DKERFIEKRVILEAEETD 407
+KN + G+ +E + ED+ E KE T S +++ DK+ E+ I E D
Sbjct: 686 KKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDKQ---EEAQIYGGESKD 742
Query: 408 SDEDEDVGELKQSVKSVKTE 427
E G+ K+S ++ KT+
Sbjct: 743 DKSVEAKGKKKESKENKKTK 762
>At2g26110 unknown protein
Length = 309
Score = 49.7 bits (117), Expect = 4e-06
Identities = 57/239 (23%), Positives = 96/239 (39%), Gaps = 28/239 (11%)
Query: 255 SRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSE 314
S S S++ +P Q I+++ SM+ K + + P S ++F PS ++
Sbjct: 68 SSSFSSKSNDPNQTQIQRS--PSMIHRLKSINFSSFTSPDK---SHLEFPPSTPEDNSNN 122
Query: 315 DLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDED 374
+ + S + P P + +F Y + N+ + S T E + E
Sbjct: 123 NFHQPASI--EQNQPFLSRSPSVLHRIKSFNLYNYISQEPTNI-IEASPPSVTVETKQEQ 179
Query: 375 EEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGEL-----KQSVKSVKTEEP 429
+E+E +E+ E K + T SD + G K+ KS T+ P
Sbjct: 180 VQEQEVKEEQEEEEQSLEEVYSKLNLNHVARTKSDTEPAAGIRPPKLPKKMKKSASTKSP 239
Query: 430 CSSSLSGGID---------------TVSDEGPDVDKKADEFIAKFREQIRLQRIESIKR 473
S I TV + +VD KAD+FI +F+ Q++LQRI+SI +
Sbjct: 240 FSHFQEDEISVEARRPATVKVPRVTTVEEADEEVDAKADDFINRFKHQLKLQRIDSITK 298
>At5g57070 putative protein
Length = 607
Score = 48.5 bits (114), Expect = 8e-06
Identities = 68/369 (18%), Positives = 139/369 (37%), Gaps = 48/369 (13%)
Query: 38 ATIVLIFFV-ILPLLPSQAPEFISQNLLTRNWELLHLLFVGIAISYGLFSRRNQEP---- 92
A I L+F ++P S + + + R W+ ++++ V AI G+ +RRN +
Sbjct: 38 AAIFLVFVTFVVPTFLSVTSQILQPASVKRGWDSINVVLVVFAILCGVLARRNDDGLSSE 97
Query: 93 -----DKDNNNNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHY 147
+++ N + V ++SS V Q + D ++++ + + +
Sbjct: 98 SLHGGEEEEVGGGAVTNGEMTVGEISKISSS-SSTVSEQWFDDVYDSDRLKIYESVSSRS 156
Query: 148 RNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGL 207
+ + + N SS+ + + + + R DD + + +S D
Sbjct: 157 FSHGLPVTG-------NVPLRRSSSSYPDLRQGVFRETGDRRFRFYDDFEIDKYRSQDSS 209
Query: 208 SKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQ 267
S Y +F+ L + +++++E E P + + + P Q
Sbjct: 210 S-----------------YQQFQNLSKTEIEEEESE------PKEIQIDTFVVKPSSPPQ 246
Query: 268 EAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY 327
+ A+ P E KP S ++ + AK SE K+ +F
Sbjct: 247 QP-----PATPPPPPPPPPVEVPQKPRRTHRS-VRNRDLQENAKRSETKFKR-TFQPPPS 299
Query: 328 PPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQE 387
PPPPPPPPP + + G R + K I+ ++ +++K+ S +E
Sbjct: 300 PPPPPPPPPPQPLIAATPPRKQGTLQRRKSNAAKEIKMVFASLYNQGKKKKKLQKSKRKE 359
Query: 388 QTDKERFIE 396
+ + +E
Sbjct: 360 RIESSPMVE 368
Score = 44.7 bits (104), Expect = 1e-04
Identities = 47/165 (28%), Positives = 71/165 (42%), Gaps = 21/165 (12%)
Query: 326 SYPPPPPPPPP--TMFYKSTFLKSRYGEKNGR--NMSMGKSIEEGTNEKEDEDEEEKEYS 381
S P PPPPPPP T F T + K+GR + K+ E N + +
Sbjct: 419 SVPAPPPPPPPRYTQFDPQTPPRR---VKSGRPPRPTKPKNFNEENNGQGSPLIQITPPP 475
Query: 382 TSSMQEQTDKERFIEKRVILEAEETDSD-------EDEDVG---ELKQSVKSVKTEEPCS 431
+ +++ + S E D+G EL QS V+T+
Sbjct: 476 PPPPPFRVPPLKYVVSGDFAKIRSNQSSRCSSPEREVFDIGWGLELTQSDGGVETK---- 531
Query: 432 SSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTR 476
+++SGG PDVD KAD FIA+ R++ RL +I S+ R ++
Sbjct: 532 AAVSGGGMPGFCPSPDVDTKADNFIARLRDEWRLDKINSVNRKSK 576
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 43.9 bits (102), Expect = 2e-04
Identities = 78/366 (21%), Positives = 140/366 (37%), Gaps = 25/366 (6%)
Query: 98 NNTKFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAP 157
NN D+ Q L+ R SF + S S ++ + R++P V
Sbjct: 70 NNNSLDDNQKLLRR----RSFDRPPSSLTSPSTSASPRIQKSLNVSPSRSRDRPAVPREK 125
Query: 158 QVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSS 217
V L++S F ++ PV KS L+ +S+ +G K+ S
Sbjct: 126 PVTALRSSSFHGSRNVPKGGNTAKSPPVAPKKSGLNSSSTSSKSKK-EGSENVRIKKASD 184
Query: 218 N--SFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALK 275
+ + + E E+ LK + + V I + + ++E EK ++
Sbjct: 185 KEIALDSASMSSAQEDHQEEILKVESDHLQVSDHDIE------EPKYEKEEKEVQEKVVQ 238
Query: 276 ASMVMEPKQEANENASKPSMV--KTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPP 333
A+ +E K E++ S V +++ E + KN +D+ +K +
Sbjct: 239 ANESVEEKAESSGPTPVASPVGKDCNAVVAELEEKLIKNEDDIEEKTEEMKEQDNNQANK 298
Query: 334 PPPTMFYKSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDE---------DEEEKEYSTSS 384
K ++ EK +S+EE T EKE+E +EEEKE
Sbjct: 299 SEEEEDVKKKIDENETPEKVDTESKEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKVK 358
Query: 385 MQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDE 444
+Q +K EK + EE + ++E+ E K+ + VK ++ S+ + I + E
Sbjct: 359 EDDQKEKVEEEEKEKVKGDEEKEKVKEEESAEGKKK-EVVKGKKESPSAYNDVIASKMQE 417
Query: 445 GPDVDK 450
P +K
Sbjct: 418 NPRKNK 423
Score = 30.4 bits (67), Expect = 2.3
Identities = 28/124 (22%), Positives = 52/124 (41%), Gaps = 20/124 (16%)
Query: 363 IEEGTNEKEDEDEEEKEYSTS-SMQEQTDKERFIE---------KRVILEAEETDSDEDE 412
IEE EKE+++ +EK + S++E+ + V+ E EE ++
Sbjct: 220 IEEPKYEKEEKEVQEKVVQANESVEEKAESSGPTPVASPVGKDCNAVVAELEEKLIKNED 279
Query: 413 DVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIK 472
D+ E + +K + S+E DV KK DE + + +ES++
Sbjct: 280 DIEEKTEEMKEQDNNQA----------NKSEEEEDVKKKIDENETPEKVDTESKEVESVE 329
Query: 473 RSTR 476
+T+
Sbjct: 330 ETTQ 333
>At4g26130 uncharacterized protein
Length = 286
Score = 42.0 bits (97), Expect = 8e-04
Identities = 27/100 (27%), Positives = 54/100 (54%), Gaps = 7/100 (7%)
Query: 382 TSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTV 441
+ S + T K++ K+++ A E +E+ ++V++V+ P + + ++
Sbjct: 192 SESSKPATKKKKKATKKMMKSASERHIGREEE-----ETVEAVEKRRPETMRVER-TTSI 245
Query: 442 SDEGPD-VDKKADEFIAKFREQIRLQRIESIKRSTRVARN 480
D G + VD KA FI KF++Q++LQR++S R + +N
Sbjct: 246 GDGGEEGVDDKASNFINKFKQQLKLQRLDSFLRYREMLKN 285
>At3g02930 unknown protein
Length = 806
Score = 42.0 bits (97), Expect = 8e-04
Identities = 49/207 (23%), Positives = 87/207 (41%), Gaps = 30/207 (14%)
Query: 228 EFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEAN 287
E E + D LK+ E+E L + R A A ++ K + ++K + ++ E
Sbjct: 578 EKESQMRDCLKEVEDEVIYLQETL----REAKAETLKLKGKMLDKETEFQSIVHENDELR 633
Query: 288 ENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKS 347
S+ K + E++AK + + S K Y P
Sbjct: 634 VKQDD-SLKKIKELSELLEEALAKKHIEENGELSESEKDYDLLPKVV------------- 679
Query: 348 RYGEKNGRNMSMGKSIE----EGTNEKEDEDEEEKEYSTSSMQEQTDKERF-------IE 396
+ E+NG + KS + +G N K +ED E+KE S +++T + F IE
Sbjct: 680 EFSEENGYRSAEEKSSKVETLDGMNMKLEEDTEKKEKKERSPEDETVEVEFKMWESCQIE 739
Query: 397 KRVILEAEETDSDEDEDVGELKQSVKS 423
K+ + +E+ +E+ED+ + QS K+
Sbjct: 740 KKEVFH-KESAKEEEEDLNVVDQSQKT 765
>At1g20290 hypothetical protein
Length = 497
Score = 41.2 bits (95), Expect = 0.001
Identities = 29/148 (19%), Positives = 70/148 (46%), Gaps = 2/148 (1%)
Query: 336 PTMFY--KSTFLKSRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKER 393
P+M + + T +++ E+ + EE E+E+E+EEE+E +E+ ++E
Sbjct: 350 PSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 409
Query: 394 FIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKAD 453
E+ E EE + +E+E+ E ++ + + EE + +E + +++ +
Sbjct: 410 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 469
Query: 454 EFIAKFREQIRLQRIESIKRSTRVARNS 481
E + E+ ++ +K+ + R++
Sbjct: 470 EEEEEEEEEEEEKKYMIVKKKKKKRRST 497
>At5g56980 unknown protein
Length = 379
Score = 40.8 bits (94), Expect = 0.002
Identities = 34/128 (26%), Positives = 62/128 (47%), Gaps = 17/128 (13%)
Query: 361 KSIEEGTNEKEDEDEEEKEYSTSSMQEQ--------TDKERFIEKRVILEAEETDSDEDE 412
KSI+ + + D D ++K+ + E+ K+ +K+ L + E
Sbjct: 246 KSIKLSSFYRSDPDLDQKQNPDPVLHEEHKHVRSKSESKKPVKKKKKALTKMTKSASEKS 305
Query: 413 DVG------ELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPD-VDKKADEFIAKFREQIRL 465
G E ++V+S++ P ++ + T +G D VD KA +FI KF++Q++L
Sbjct: 306 GFGFAGSHAEAPETVESLERRRPDTTRVERS--TSFGDGEDGVDAKASDFINKFKQQLKL 363
Query: 466 QRIESIKR 473
QR++SI R
Sbjct: 364 QRLDSILR 371
>At4g18560 pherophorin - like protein
Length = 637
Score = 40.8 bits (94), Expect = 0.002
Identities = 37/157 (23%), Positives = 63/157 (39%), Gaps = 13/157 (8%)
Query: 182 LLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVEDKLKQKE 241
++ +R L S SDD + SQ GL + K SN ++ L E Q+
Sbjct: 160 IVELRKLVSSESDDHALSVSQRFQGLMDVSAK---SNLIRSLKRVGSLRNLPEPITNQEN 216
Query: 242 EENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVKTSSI 301
++ S S A +++ IE ++S E + ++ + + + +
Sbjct: 217 TNKSI--------SSSGDADGDIYRKDEIESYSRSSNSEELTESSSLSTVRSRVPRVP-- 266
Query: 302 KFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTM 338
K P S++ + KS PPPPPPPPP +
Sbjct: 267 KPPPKRSISLGDSTENRADPPPQKSIPPPPPPPPPPL 303
>At2g23480 hypothetical protein
Length = 705
Score = 40.8 bits (94), Expect = 0.002
Identities = 50/239 (20%), Positives = 101/239 (41%), Gaps = 17/239 (7%)
Query: 238 KQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVK 297
K+KE E P+P A A ++ K A + ++A E Q + + K
Sbjct: 12 KRKEIE---APAPAKTEKVKAPAEKVKEKVPAKKAKVQAPAKKEKVQAPAKKEKVQAPAK 68
Query: 298 TSSIKFTP-SESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF---LKSRYGEKN 353
+ ++ T + + A+ + + + + P P M STF LK E+
Sbjct: 69 KAKVQATAKTTATAQTTATAMATTAAPTTTAPTTAPTTESPMLDDSTFYDALKHIPAEET 128
Query: 354 GRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDED 413
NM + +E+ ++E ++EE S+ ++ +D E + + A + E +D
Sbjct: 129 QENMQTDE-VEDENEKEEGSEKEESGSSSQTLGSDSDSEETETNKEVACANPVEEAERQD 187
Query: 414 VGELKQSVKSVKTEEPCSSSLSGGID---TVSDEGPDVDKKADEFIAKFREQIRLQRIE 469
G + ++ EE SS+ ++ +V DEG D D++ ++ I + + + + I+
Sbjct: 188 DG-----LAVIEEEEERSSASDEDVNVEKSVEDEG-DEDERDEDVIVQVEKPVEERTID 240
>At4g21180 unknown protein
Length = 643
Score = 40.4 bits (93), Expect = 0.002
Identities = 19/54 (35%), Positives = 36/54 (66%), Gaps = 1/54 (1%)
Query: 360 GKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDED 413
G++ EEG E+ED++ EE++Y + +++ DK+R +K+V E+ +S DE+
Sbjct: 591 GENAEEGL-EEEDDEIEEEDYESEYSEDEEDKKRGSKKKVNKESSSEESGSDEE 643
>At4g04010 hypothetical protein
Length = 836
Score = 40.4 bits (93), Expect = 0.002
Identities = 67/343 (19%), Positives = 144/343 (41%), Gaps = 37/343 (10%)
Query: 157 PQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDD----CVVQSQSVD--GLSKT 210
P+ ++ N V + + SF + E +L + ++ RL + CV +++ G +K
Sbjct: 283 PEFFKVVNLVNDVSEVSFYDTEFQHILGLVNMGYRLKKTEWNTRCVDVCAALEEVGQNKI 342
Query: 211 TTKRFSSNSFNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQE-- 268
K S +R+ ++ED K+ E +L + ++ S E KQE
Sbjct: 343 NNKISDSEKLDRILT------ILEDLNKRVELIERILDIRMEEKNNQRSEEDEERKQEDE 396
Query: 269 AIEKALKASMVMEPKQEA-NENASKPSMVKTSSIKFTPSESVAKNSEDLIKKKSFYYKSY 327
+E+ +A +++A N+N S ++ + ++ +N++ + + S
Sbjct: 397 GLERQPEAEEEGGLERKAKNDNESFEDSIREPNTQYGSYLGDDENTQRYVGDEVAEESSK 456
Query: 328 PPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGK--------SIEEGTN---EKEDEDEE 376
P P T + F +S +K+ + +S G+ S+ N K+ +
Sbjct: 457 DKSPTPRSSTPNFNILFEESLDVQKDKKRVSRGRNENKRVKPSVHAEDNLKTRKQVPRKR 516
Query: 377 EKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSG 436
+K+ ++ + QT KE +KR I+ + ++D D D + + + P +S G
Sbjct: 517 QKQVDSADVDVQTRKEAQSKKRKIIGNDGDNADNDGDNDDFQPA--------PQRNSKRG 568
Query: 437 GIDTVSDEGP-DVDKKADEFIAKFR--EQIRLQRIESIKRSTR 476
+ ++ + P +KK + F + RL+++ K+S +
Sbjct: 569 TVPSIHTQAPFTAEKKKHPILHPFAKVDATRLEKLAEWKKSRK 611
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 40.0 bits (92), Expect = 0.003
Identities = 78/393 (19%), Positives = 164/393 (40%), Gaps = 35/393 (8%)
Query: 103 DNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNKP-MVIVAPQVQQ 161
++++ LV+ F Q + E+E +Q +E N+IQ N Q ++ + + V++
Sbjct: 76 ESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNEIQEAQNTMQELMSESGQLKESHSVKE 135
Query: 162 LQ----NSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSS 217
+ + E Q + L + S K ++SD +++ + + ++ +
Sbjct: 136 RELFSLRDIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETM 195
Query: 218 NSFNRVRN-----YAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEK 272
N + +N AE G ++D ++KE E + L R +S + E +E +E
Sbjct: 196 NKLEQTQNTIQELMAEL-GKLKDSHREKESELSSLVEVHETHQRDSSIHVKE-LEEQVES 253
Query: 273 ALKASMVMEPKQEANENASKPSMVKTSSIKFTPSESVAKNS-EDLIKKKSFYYKSYPPPP 331
+ K +V E Q N + ++ + + A+N+ ++L+ + +S+
Sbjct: 254 SKK--LVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKESHSVKD 311
Query: 332 PPPPPTMFYKSTFLK---SRYGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQ 388
T + +R E + S + I + T + +D +EE K S+ ++ E
Sbjct: 312 RDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEENKAISSKNL-EI 370
Query: 389 TDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDV 448
DK + +A+ T + +++GELK K ++E SS + V+D +
Sbjct: 371 MDK--------LEQAQNTIKELMDELGELKDRHKEKESE--LSSLVKSADQQVADMKQSL 420
Query: 449 DKKADEFIAKFREQIRLQRIESIKRSTRVARNS 481
D +E +++ QRI I + A+ +
Sbjct: 421 DNAEEE------KKMLSQRILDISNEIQEAQKT 447
Score = 30.8 bits (68), Expect = 1.7
Identities = 57/316 (18%), Positives = 122/316 (38%), Gaps = 27/316 (8%)
Query: 101 KFDNAQSLVSRFLQVSSFFEDEVDHQNQSESEDINKIQTWSNQNQHYRNKPMVIVAPQVQ 160
K + A S + ++ + ++E+D +SE +++ + N+ + V+
Sbjct: 971 KINVASSEIMALTELINNLKNELDSLQVQKSETEAELEREKQEKSELSNQITDVQKALVE 1030
Query: 161 Q-LQNSVFEDEQSSFNENEKPLLLPVRSLKSRLSDDDCVVQSQSVDGLSKTTTKRFSSNS 219
Q + E+E NE K + + + +++ + + S+ +T +
Sbjct: 1031 QEAAYNTLEEEHKQINELFKETEATLNKVTVDYKEAQRLLEERGKEVTSRDSTIGVHEET 1090
Query: 220 FNRVRNYAEFEGLVEDKLKQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMV 279
+RN E +G + L +K N + + + + +++ K+EA K +
Sbjct: 1091 MESLRNELEMKGDEIETLMEKIS-NIEVKLRLSNQKLRVTEQVLTEKEEAFRKEEAKHLE 1149
Query: 280 MEPKQEANENASKPS---MVKTSSIKFTPS-ESVAKNSEDLIKKKSFYYKSYPPPPPPPP 335
+ E N + + M+K + K + + SE L +K+ Y K
Sbjct: 1150 EQALLEKNLTMTHETYRGMIKEIADKVNITVDGFQSMSEKLTEKQGRYEK---------- 1199
Query: 336 PTMFYKSTFLKSRYG---EKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKE 392
T+ S L + E+N M K IE+ +DEE K+ +++ +KE
Sbjct: 1200 -TVMEASKILWTATNWVIERNHEKEKMNKEIEK-------KDEEIKKLGGKVREDEKEKE 1251
Query: 393 RFIEKRVILEAEETDS 408
E + L E+ ++
Sbjct: 1252 MMKETLMGLGEEKREA 1267
>At1g52320 bZIP protein, putative
Length = 798
Score = 40.0 bits (92), Expect = 0.003
Identities = 20/71 (28%), Positives = 36/71 (50%), Gaps = 7/71 (9%)
Query: 329 PPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKS--IEEGTNEKEDEDEEEKEYSTSSMQ 386
PPPPPPPP ++ + G G + G + E+G + +D+D+++ + S
Sbjct: 116 PPPPPPPPLPLQRAATMPEMNGRSGGGHAGSGLNGIEEDGALDNDDDDDDDDDDS----- 170
Query: 387 EQTDKERFIEK 397
E +++R I K
Sbjct: 171 EMENRDRLIRK 181
>At3g19060 hypothetical protein
Length = 1647
Score = 39.7 bits (91), Expect = 0.004
Identities = 75/361 (20%), Positives = 149/361 (40%), Gaps = 50/361 (13%)
Query: 131 SEDINKIQTWSNQNQHYRNKPMVIVAPQVQQLQNSVFEDEQSSFNENEKPLLLPVRSLKS 190
+ED K++ + +K + ++ V + SVF D S + + LL +L
Sbjct: 1166 TEDSKKLEKLTRDGMAISDKMLQLICENVDKA--SVFADTVQSLQIDVQELLSENLNLHD 1223
Query: 191 RLSDDDCVVQSQSVDGLSKTTTKRFSSNSFNRVRNYAEFEGLVED-----KLKQKEEENA 245
L D V++ S D + + +SNS ++ E VE LK E E+A
Sbjct: 1224 ELLRKDDVLKGLSFD---LSLLQESASNSRDKKDETKEIMVHVEALEKTLALKTFELEDA 1280
Query: 246 VLPSPIPWRSRSASARMMEPK-QEAIEKALKASMVMEPKQEANENASKPSMVKTSSIKFT 304
V + A+M+E + QE+ E + E ++ E S
Sbjct: 1281 V-----------SHAQMLEVRLQESKEITRNLEVDTEKARKCQEKLS------------A 1317
Query: 305 PSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTFLKSRYGEKNGRNMSMGKSIE 364
++ + +EDL+ +K + +M + L++ G+ N + +
Sbjct: 1318 ENKDIRAEAEDLLAEKCSLEEEMIQTKKVSE-SMEMELFNLRNALGQLNDTVAFTQRKLN 1376
Query: 365 EGTNEKEDEDEE----EKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQS 420
+ +E+++ +E ++E+ + + + R+IE + I E+ +T +DE E+ E+K
Sbjct: 1377 DAIDERDNLQDEVLNLKEEFGKMKSEAKEMEARYIEAQQIAESRKTYADEREE--EVKLL 1434
Query: 421 VKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIAKFREQIRLQRIESIKRSTRVARN 480
SV+ E + L ++ V DE E RE++ ++ + +I++ ARN
Sbjct: 1435 EGSVEELEYTINVLENKVNVVKDEA--------ERQRLQREELEME-LHTIRQQMESARN 1485
Query: 481 S 481
+
Sbjct: 1486 A 1486
>At2g04330 hypothetical protein
Length = 564
Score = 39.7 bits (91), Expect = 0.004
Identities = 60/230 (26%), Positives = 92/230 (39%), Gaps = 32/230 (13%)
Query: 238 KQKEEENAVLPSPIPWRSRSASARMMEPKQEAIEKALKASMVMEPKQEANENASKPSMVK 297
K+KE E P+P A A ++ K A + ++A E Q + A + K
Sbjct: 12 KRKEIE---APAPTKTEKVKAPAEKVKEKVPAKKAKVQAPAKKEKVQAPAKKAKVQAPAK 68
Query: 298 TSSIKFTPSESVAKNSEDLIKKKSFYYKSYPPPPPPPPPTMFYKSTF---LKSRYGEKNG 354
T++ T + ++A N+ + + P P M STF LK E+
Sbjct: 69 TTATAQTIATAMATNAAPTTAALT---TTAPTTAPTTESPMLDDSTFYDALKHIPAEETQ 125
Query: 355 RNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKR-------VILEAEETD 407
NM T+E EDE+E+E+ T + +E E VI E EE
Sbjct: 126 ENMQ--------TDEVEDENEKEEGKETETNKELACANPVEEAERQDDGLAVIEEEEERS 177
Query: 408 SDEDEDVGELKQSVKSVKTEEPCSSSLSGGIDTVSDEGPDVDKKADEFIA 457
S DEDV +++SV+ E+ + V E P ++ DE IA
Sbjct: 178 SASDEDV-NVEKSVEDEGDEDERD-------EDVIVEKPVEERTIDEDIA 219
>At5g03710 putative protein
Length = 81
Score = 39.3 bits (90), Expect = 0.005
Identities = 21/73 (28%), Positives = 40/73 (54%)
Query: 364 EEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKS 423
EE E+E+E+EEE+E +E+ ++E E+ E EE + +E+E+ E ++ +
Sbjct: 8 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEED 67
Query: 424 VKTEEPCSSSLSG 436
+ EE + + G
Sbjct: 68 REREERAFAFVPG 80
Score = 38.1 bits (87), Expect = 0.011
Identities = 19/57 (33%), Positives = 33/57 (57%)
Query: 360 GKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGE 416
G+ EE E+E+E+EEE+E +E+ ++E E+ E EE + +E+E+ E
Sbjct: 5 GEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 61
Score = 37.4 bits (85), Expect = 0.019
Identities = 19/65 (29%), Positives = 36/65 (55%)
Query: 364 EEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKS 423
EE E+E+E+EEE+E +E+ ++E E+ E EE + +E+E+ E ++ +
Sbjct: 7 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 66
Query: 424 VKTEE 428
+ E
Sbjct: 67 DRERE 71
Score = 37.4 bits (85), Expect = 0.019
Identities = 18/63 (28%), Positives = 36/63 (56%)
Query: 366 GTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSVK 425
G E+E+E+EEE+E +E+ ++E E+ E EE + +E+E+ E ++ + +
Sbjct: 5 GEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 64
Query: 426 TEE 428
E+
Sbjct: 65 EED 67
Score = 35.8 bits (81), Expect = 0.054
Identities = 18/64 (28%), Positives = 34/64 (53%)
Query: 349 YGEKNGRNMSMGKSIEEGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDS 408
+GE+ + EE E+E+E+EEE+E +E+ ++E E+ E EE +
Sbjct: 4 WGEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 63
Query: 409 DEDE 412
+E++
Sbjct: 64 EEED 67
Score = 35.4 bits (80), Expect = 0.071
Identities = 18/64 (28%), Positives = 35/64 (54%)
Query: 365 EGTNEKEDEDEEEKEYSTSSMQEQTDKERFIEKRVILEAEETDSDEDEDVGELKQSVKSV 424
E E+E+E+EEE+E +E+ ++E E+ E EE + +E+E+ E ++ +
Sbjct: 6 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 65
Query: 425 KTEE 428
+ E
Sbjct: 66 EDRE 69
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.126 0.342
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,455,785
Number of Sequences: 26719
Number of extensions: 568461
Number of successful extensions: 9336
Number of sequences better than 10.0: 521
Number of HSP's better than 10.0 without gapping: 192
Number of HSP's successfully gapped in prelim test: 347
Number of HSP's that attempted gapping in prelim test: 5797
Number of HSP's gapped (non-prelim): 2010
length of query: 483
length of database: 11,318,596
effective HSP length: 103
effective length of query: 380
effective length of database: 8,566,539
effective search space: 3255284820
effective search space used: 3255284820
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC149079.6


