

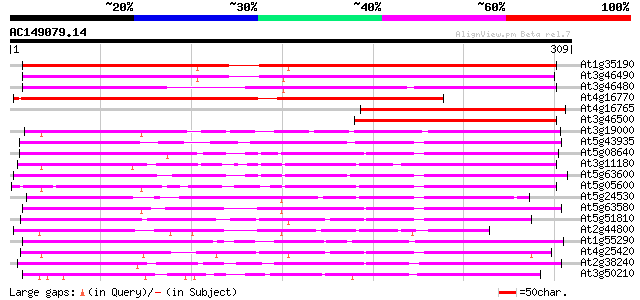
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.14 - phase: 0
(309 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g35190 hyoscyamine 6-dioxygenase hydroxylase like protein 263 1e-70
At3g46490 putative protein 235 2e-62
At3g46480 putative protein 223 1e-58
At4g16770 gibberellin oxidase-like protein 217 7e-57
At4g16765 unknown protein 182 2e-46
At3g46500 unknown protein 139 1e-33
At3g19000 unknown protein 122 2e-28
At5g43935 putative protein 117 6e-27
At5g08640 flavonol synthase (FLS) (sp|Q96330) 114 5e-26
At3g11180 putative leucoanthocyanidin dioxygenase 110 1e-24
At5g63600 1-aminocyclopropane-1-carboxylic acid oxidase-like pro... 109 2e-24
At5g05600 leucoanthocyanidin dioxygenase-like protein 109 2e-24
At5g24530 flavanone 3-hydroxylase-like protein 108 4e-24
At5g63580 flavonol synthase 106 1e-23
At5g51810 gibberellin 20-oxidase (emb|CAA58294.1) 103 9e-23
At2g44800 putative flavonol synthase 103 9e-23
At1g55290 leucoanthocyanidin dioxygenase 2, putative 103 1e-22
At4g25420 gibberellin 20-oxidase - Arabidopsis thaliana 101 4e-22
At2g38240 putative anthocyanidin synthase 101 4e-22
At3g50210 flavonol synthase - like protein 100 8e-22
>At1g35190 hyoscyamine 6-dioxygenase hydroxylase like protein
Length = 329
Score = 263 bits (671), Expect = 1e-70
Identities = 139/308 (45%), Positives = 187/308 (60%), Gaps = 30/308 (9%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL++ D + S+ QAC++ GFFY++NHG+ E+F+ F+QS F+LP+E+K+
Sbjct: 11 SLNCIDLANDDLNHSVVSLKQACLDCGFFYVINHGISEEFMDDVFEQSKKLFALPLEEKM 70
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIG-------PLTDTTSVRLNQWPSNG 119
K+ R E +RGYTP+ E LDP + GD KE YYIG P D N WP
Sbjct: 71 KVLRNEKHRGYTPVLDELLDPKNQINGDHKEGYYIGIEVPKDDPHWDKPFYGPNPWPDAD 130
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKEL------LSLIALSLNLDENYFEKISAL 173
V P W KY + L L+AL+L+LD YF++ L
Sbjct: 131 VL----------------PGWRETMEKYHQEALRVSMAIARLLALALDLDVGYFDRTEML 174
Query: 174 NKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
KP A +RLLRY G P++ I AHSD+GM+TLL T+GV GLQICK+K PQ WE
Sbjct: 175 GKPIATMRLLRYQGISDPSKGIYACGAHSDFGMMTLLATDGVMGLQICKDKNAMPQKWEY 234
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
V ++GA IVN+GDM+ERW+N ++STLHRV+ G+ERYS+ FF++P DC+VEC +C
Sbjct: 235 VPPIKGAFIVNLGDMLERWSNGFFKSTLHRVLGNGQERYSIPFFVEPNHDCLVECLPTCK 294
Query: 294 SESSPPRY 301
SES P+Y
Sbjct: 295 SESELPKY 302
>At3g46490 putative protein
Length = 306
Score = 235 bits (600), Expect = 2e-62
Identities = 124/307 (40%), Positives = 180/307 (58%), Gaps = 30/307 (9%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL + D +A + QAC++ GFFY++NHG+ E+ +AF+ S FF+LP+E+K+
Sbjct: 16 SLTCIDLDNSDLHQSAVLLKQACLDSGFFYVINHGISEELKDEAFEHSKKFFALPLEEKM 75
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIG-------PLTDTTSVRLNQWPSNG 119
K+ R E YRGY P + LDP + +GD KE + IG P D N WP+
Sbjct: 76 KVLRNEKYRGYAPFHDSLLDPENQVRGDYKEGFTIGFEGSKDGPHWDKPFHSPNIWPNPD 135
Query: 120 VFSFFPAMTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISAL 173
V P W KY K + ++AL+L+LD +YF L
Sbjct: 136 VL----------------PGWRETMEKYYQEALRVCKSIAKIMALALDLDVDYFNTPEML 179
Query: 174 NKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWED 233
P A + L Y G+ P++ I AHSD+GM++LL T+GV GLQICK+K +PQ WE
Sbjct: 180 GNPIADMVLFHYEGKSDPSKGIYACGAHSDFGMMSLLATDGVMGLQICKDKDVKPQKWEY 239
Query: 234 VSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCC 293
++GA IVN+GD++ERW+N ++STLHRV+ G++RYS+ FF+ P DC++EC +C
Sbjct: 240 TPSIKGAYIVNLGDLLERWSNGYFKSTLHRVLGNGQDRYSIPFFLKPSHDCIIECLPTCQ 299
Query: 294 SESSPPR 300
SE++ P+
Sbjct: 300 SENNLPK 306
>At3g46480 putative protein
Length = 297
Score = 223 bits (567), Expect = 1e-58
Identities = 121/301 (40%), Positives = 176/301 (58%), Gaps = 52/301 (17%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
SL IDL++P+ +A S+ QAC++ GFFY+ NHG+ E+ +AF+QS FF+LP+E+K+
Sbjct: 16 SLTCIDLANPNFQQSAVSLKQACLDCGFFYVTNHGISEELKDEAFEQSKKFFALPLEEKM 75
Query: 68 KLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFPA 126
K+ R E +RGY+P+ + LDP
Sbjct: 76 KVLRNEKHRGYSPVLDQILDP--------------------------------------- 96
Query: 127 MTLQNYSQIGDPQWNPYFGKYSG------KELLSLIALSLNLDENYFEKISALNKPEAFL 180
+N + P W KY K + L+AL+L+LD NYF+K L P A +
Sbjct: 97 ---ENQVDVVLPGWRATMEKYHQEALRVCKAIARLLALALDLDTNYFDKPEMLGNPIAVM 153
Query: 181 RLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGA 240
RLLRY G P + I G AHSDYGM+TLL T+ V GLQ +K +P+ WE V ++GA
Sbjct: 154 RLLRYEGMSDPLKGIFGCGAHSDYGMLTLLATDSVTGLQ---DKDVKPRKWEYVPSIKGA 210
Query: 241 IIVNIGDMMERWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPR 300
IVN+GD++ERW+N +++STLHRV+ G++RYS+ FF++P DC+VEC +C SE++ P+
Sbjct: 211 YIVNLGDLLERWSNGIFKSTLHRVLGNGQDRYSIPFFIEPSHDCLVECLPTCQSENNLPK 270
Query: 301 Y 301
Y
Sbjct: 271 Y 271
>At4g16770 gibberellin oxidase-like protein
Length = 243
Score = 217 bits (552), Expect = 7e-57
Identities = 114/239 (47%), Positives = 158/239 (65%), Gaps = 13/239 (5%)
Query: 3 GTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
GTA L LPIIDLSSP+++ST++ I QAC+++GFFYL NHGV E+ ++ +S FSLP
Sbjct: 2 GTA-LKLPIIDLSSPEKLSTSRLIRQACLDHGFFYLTNHGVSEELMEGVLIESKKLFSLP 60
Query: 63 IEDKIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTT-SVRLNQWPSNGVF 121
+++K+ + R +RGY+PLY EKL+ SS S GD KE + G + N+WP +
Sbjct: 61 LDEKMVMARHGFRGYSPLYDEKLESSSTSIGDSKEMFTFGSSEGVLGQLYPNKWPLEELL 120
Query: 122 SFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLR 181
+ Y + D GK+L L+AL+LNL+ENYFE++ A N A +R
Sbjct: 121 PLWRPTMECYYKNVMD----------VGKKLFGLVALALNLEENYFEQVGAFNDQAAVVR 170
Query: 182 LLRYPGELGPN-EEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEG 239
LLRY GE + EE CGASAHSD+GMITLL T+GV GLQ+C++K K+P+VWEDV+ ++G
Sbjct: 171 LLRYSGESNSSGEETCGASAHSDFGMITLLATDGVAGLQVCRDKDKEPKVWEDVAGIKG 229
>At4g16765 unknown protein
Length = 201
Score = 182 bits (462), Expect = 2e-46
Identities = 81/113 (71%), Positives = 96/113 (84%)
Query: 194 EICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWT 253
E GASAHSDYGM+TLLLT+GVPGLQ+C++K KQP +WEDV ++GA IVNIGDMMERWT
Sbjct: 74 ETYGASAHSDYGMVTLLLTDGVPGLQVCRDKSKQPHIWEDVPGIKGAFIVNIGDMMERWT 133
Query: 254 NCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKVLS 306
N L+RSTLHRVMP GKERYSV FF+DP DC V+C ESCCSE+ PPR+ +L+
Sbjct: 134 NGLFRSTLHRVMPVGKERYSVVFFLDPNPDCNVKCLESCCSETCPPRFPPILA 186
>At3g46500 unknown protein
Length = 139
Score = 139 bits (351), Expect = 1e-33
Identities = 59/111 (53%), Positives = 87/111 (78%)
Query: 191 PNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMME 250
P++ I G HSD+GM+TLL T+ V GLQICK++ +P+ WE + ++GA IVNIGD++E
Sbjct: 4 PSKGIYGCGPHSDFGMMTLLGTDSVMGLQICKDRDVKPRKWEYILSIKGAYIVNIGDLLE 63
Query: 251 RWTNCLYRSTLHRVMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRY 301
RW+N +++STLHRV+ G++RYS+AFF+ P DC+VEC +C SE++PP+Y
Sbjct: 64 RWSNGIFKSTLHRVLGNGQDRYSIAFFLQPSHDCIVECLPTCQSENNPPKY 114
>At3g19000 unknown protein
Length = 352
Score = 122 bits (306), Expect = 2e-28
Identities = 97/317 (30%), Positives = 148/317 (46%), Gaps = 48/317 (15%)
Query: 9 LPIIDLSS-----PDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPI 63
+P IDLSS D+ + AK I +AC +GFF ++NHG+ + +A FF+L
Sbjct: 32 IPTIDLSSLEDTHHDKTAIAKEIAEACKRWGFFQVINHGLPSALRHRVEKTAAEFFNLTT 91
Query: 64 EDKIKLNR--------------KEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTS 109
E+K K+ R K R + ++ L S++ P+ DT
Sbjct: 92 EEKRKVKRDEVNPMGYHDEEHTKNVRDWKEIFDFFLQDSTIVPASPEPE-------DTEL 144
Query: 110 VRL-NQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFE 168
+L NQWP N S F + + ++ + LL L+++SL L +
Sbjct: 145 RKLTNQWPQNP--SHFREVCQEYAREV----------EKLAFRLLELVSISLGLPGD--R 190
Query: 169 KISALNKPEAFLRLLRYPGELGPNEEIC-GASAHSDYGMITLLLTNGVPGLQICKEKLKQ 227
N+ +FLR YP PN E+ G H D G +T+L + V GLQ+ + Q
Sbjct: 191 LTGFFNEQTSFLRFNHYPP--CPNPELALGVGRHKDGGALTVLAQDSVGGLQVSRRSDGQ 248
Query: 228 PQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVV 286
W V + A+I+N+G+ ++ WTN Y S HR V+ T KER+S+ FF P + +
Sbjct: 249 ---WIPVKPISDALIINMGNCIQVWTNDEYWSAEHRVVVNTSKERFSIPFFFFPSHEANI 305
Query: 287 ECFESCCSESSPPRYRK 303
E E SE +PP Y+K
Sbjct: 306 EPLEELISEENPPCYKK 322
>At5g43935 putative protein
Length = 293
Score = 117 bits (294), Expect = 6e-27
Identities = 84/300 (28%), Positives = 133/300 (44%), Gaps = 39/300 (13%)
Query: 6 TLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIED 65
T +PI+DLS P A ++ +A E+G F LVNHG+ + +++ + FF LP +
Sbjct: 16 TKKIPIVDLSDPSDELVAHAVVKASEEWGIFQLVNHGIPAELMRRLQEVGRQFFELPASE 75
Query: 66 KIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFP 125
K + R A+ D DPK +L W + + + +P
Sbjct: 76 KESVTRP---------ADSQDIEGFFSKDPK--------------KLKAWDDHLIHNIWP 112
Query: 126 AMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLLRY 185
S I W YSG + N EKI +K + +R+ Y
Sbjct: 113 P------SSINYRYWPNNPSDYSGDGFREVTKEYTRNVTNLTEKIVGGDKAQYVMRINYY 166
Query: 186 PGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNI 245
P P++ GA AH+D+ + LL++N VPGLQ+ K+ W DV ++ A+IV I
Sbjct: 167 P----PSDSAIGAPAHTDFCGLALLVSNEVPGLQVFKD-----DHWFDVEYINSAVIVLI 217
Query: 246 GDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKV 304
GD + R +N Y++ LHR +M K R S ++P VV + +PP++ +
Sbjct: 218 GDQIMRMSNGKYKNVLHRSIMDAKKTRMSWPILVEPKRGLVVGPLPELTGDENPPKFESL 277
>At5g08640 flavonol synthase (FLS) (sp|Q96330)
Length = 336
Score = 114 bits (286), Expect = 5e-26
Identities = 84/300 (28%), Positives = 143/300 (47%), Gaps = 24/300 (8%)
Query: 6 TLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIED 65
T ++P++DLS PD S +++ +A E+G F +VNHG+ + I++ D FF LP +
Sbjct: 40 TPAIPVVDLSDPDEESVRRAVVKASEEWGLFQVVNHGIPTELIRRLQDVGRKFFELPSSE 99
Query: 66 KIKLNRKEYRGYTPLYAEKL--DPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSF 123
K + + E Y KL DP + I P + V WP N
Sbjct: 100 KESVAKPEDSKDIEGYGTKLQKDPEGKKAWVDHLFHRIWP---PSCVNYRFWPKNP---- 152
Query: 124 FPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLL 183
Y ++ + ++ + K S + LL +++ L L + ++ E +++
Sbjct: 153 ------PEYREVNE-EYAVHVKKLS-ETLLGILSDGLGLKRDALKEGLGGEMAEYMMKIN 204
Query: 184 RYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIV 243
YP P+ + G AH+D ITLL+ N VPGLQ+ K+ W D ++ A+IV
Sbjct: 205 YYPPCPRPDLAL-GVPAHTDLSGITLLVPNEVPGLQVFKD-----DHWFDAEYIPSAVIV 258
Query: 244 NIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPRYR 302
+IGD + R +N Y++ LHR + K R S F++PP + +V + +PP+++
Sbjct: 259 HIGDQILRLSNGRYKNVLHRTTVDKEKTRMSWPVFLEPPREKIVGPLPELTGDDNPPKFK 318
>At3g11180 putative leucoanthocyanidin dioxygenase
Length = 400
Score = 110 bits (275), Expect = 1e-24
Identities = 92/302 (30%), Positives = 147/302 (48%), Gaps = 27/302 (8%)
Query: 5 ATLSLPIIDLSS--PDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
A +++PIIDL S K I +AC E+GFF ++NHGV + + A + +FF+LP
Sbjct: 91 ADINIPIIDLDSLFSGNEDDKKRISEACREWGFFQVINHGVKPELMDAARETWKSFFNLP 150
Query: 63 IEDK--IKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGV 120
+E K + + Y G Y +L + D + YY+ L N+WPS
Sbjct: 151 VEAKEVYSNSPRTYEG----YGSRLGVEKGAILDWNDYYYLHFL-PLALKDFNKWPS--- 202
Query: 121 FSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFL 180
P+ N ++ D ++ K G+ L+++++ +L L ++ A L
Sbjct: 203 ---LPS----NIREMND-EYGKELVKLGGR-LMTILSSNLGLRAEQLQEAFGGEDVGACL 253
Query: 181 RLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGA 240
R+ YP P E G S HSD G +T+LL P Q+ +++ W V+ + A
Sbjct: 254 RVNYYPKCPQP-ELALGLSPHSDPGGMTILL----PDDQVVGLQVRHGDTWITVNPLRHA 308
Query: 241 IIVNIGDMMERWTNCLYRSTLHRVMPTG-KERYSVAFFMDPPSDCVVECFESCCSESSPP 299
IVNIGD ++ +N Y+S HRV+ KER S+AFF +P SD ++ + + + PP
Sbjct: 309 FIVNIGDQIQILSNSKYKSVEHRVIVNSEKERVSLAFFYNPKSDIPIQPMQQLVTSTMPP 368
Query: 300 RY 301
Y
Sbjct: 369 LY 370
>At5g63600 1-aminocyclopropane-1-carboxylic acid oxidase-like
protein
Length = 325
Score = 109 bits (272), Expect = 2e-24
Identities = 81/307 (26%), Positives = 142/307 (45%), Gaps = 28/307 (9%)
Query: 3 GTATLSLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLP 62
G + + +P++DLS D + + +A E+G F +VNHG+ + ++Q FF LP
Sbjct: 27 GGSAVDVPVVDLSVSDEDFLVREVVKASEEWGVFQVVNHGIPTELMRQLQMVGTQFFELP 86
Query: 63 IEDKIKLNRKE-YRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVF 121
+K + ++E + GY Y L + + + L+ + + WP N
Sbjct: 87 DAEKETVAKEEDFEGYKKNY--------LGGINNWDEHLFHRLSPPSIINYKYWPKNP-- 136
Query: 122 SFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLR 181
Y ++ + ++ + + + K +L ++ L L F + + E LR
Sbjct: 137 --------PQYREVTE-EYTKHMKRLTEK-ILGWLSEGLGLQRETFTQSIGGDTAEYVLR 186
Query: 182 LLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAI 241
+ YP E + GA+AHSD G I LL+ N VPGLQ K+ + W D+ +++ A+
Sbjct: 187 VNFYP-PTQDTELVIGAAAHSDMGAIALLIPNEVPGLQAFKD-----EQWLDLDYIDSAV 240
Query: 242 IVNIGDMMERWTNCLYRSTLHRV-MPTGKERYSVAFFMDPPSDCVVECFESCCSESSPPR 300
+V IGD + R TN ++ LHR K R S F+ P +D V + +PP+
Sbjct: 241 VVIIGDQLMRMTNGRLKNVLHRAKSDKDKLRISWPVFVAPRADMSVGPLPEFTGDENPPK 300
Query: 301 YRKVLSN 307
+ ++ N
Sbjct: 301 FETLIYN 307
>At5g05600 leucoanthocyanidin dioxygenase-like protein
Length = 371
Score = 109 bits (272), Expect = 2e-24
Identities = 103/312 (33%), Positives = 151/312 (48%), Gaps = 38/312 (12%)
Query: 2 AGTATLSLPIIDLSS-------PDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQ 54
A TAT ++PIIDL D + A+ I +AC +GFF +VNHGV + + A +
Sbjct: 56 APTAT-NIPIIDLEGLFSEEGLSDDVIMAR-ISEACRGWGFFQVVNHGVKPELMDAAREN 113
Query: 55 SANFFSLPIEDKIKLNR--KEYRGY-TPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVR 111
FF +P+ K + + Y GY + L EK +SL D YY L
Sbjct: 114 WREFFHMPVNAKETYSNSPRTYEGYGSRLGVEK--GASLDWSD----YYFLHLLPHHLKD 167
Query: 112 LNQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKIS 171
N+WPS FP + + G+ K SG+ ++ +++ +L L E+ F++
Sbjct: 168 FNKWPS------FPPTIREVIDEYGEE-----LVKLSGR-IMRVLSTNLGLKEDKFQEAF 215
Query: 172 ALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNG-VPGLQICKEKLKQPQV 230
A LR+ YP P E G S HSD G +T+LL + V GLQ+ K+
Sbjct: 216 GGENIGACLRVNYYPKCPRP-ELALGLSPHSDPGGMTILLPDDQVFGLQVRKD-----DT 269
Query: 231 WEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRV-MPTGKERYSVAFFMDPPSDCVVECF 289
W V A IVNIGD ++ +N Y+S HRV + + KER S+AFF +P SD ++
Sbjct: 270 WITVKPHPHAFIVNIGDQIQILSNSTYKSVEHRVIVNSDKERVSLAFFYNPKSDIPIQPL 329
Query: 290 ESCCSESSPPRY 301
+ S +PP Y
Sbjct: 330 QELVSTHNPPLY 341
>At5g24530 flavanone 3-hydroxylase-like protein
Length = 341
Score = 108 bits (270), Expect = 4e-24
Identities = 87/285 (30%), Positives = 135/285 (46%), Gaps = 38/285 (13%)
Query: 10 PIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKIKL 69
P+IDLSS DR + IHQAC +GFF ++NHGV++ I + + FFS+ +E+K+K
Sbjct: 39 PLIDLSSTDRSFLIQQIHQACARFGFFQVINHGVNKQIIDEMVSVAREFFSMSMEEKMK- 97
Query: 70 NRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFPAMTL 129
LY++ DP ++ + + +N W +P
Sbjct: 98 ----------LYSD----------DPTKTTRLSTSFNVKKEEVNNWRDYLRLHCYPIHKY 137
Query: 130 QNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISALNKPEAFLRLL 183
N P + KYS G ++ LI+ SL L+++Y +K+ L + + +
Sbjct: 138 VNEWPSNPPSFKEIVSKYSREVREVGFKIEELISESLGLEKDYMKKV--LGEQGQHMAVN 195
Query: 184 RYPGELGPNEEICGASAHSDYGMITLLLTN-GVPGLQICKEKLKQPQVWEDVSHVEGAII 242
YP P E G AH+D +T+LL + V GLQI + W V+ A +
Sbjct: 196 YYPPCPEP-ELTYGLPAHTDPNALTILLQDTTVCGLQILID-----GQWFAVNPHPDAFV 249
Query: 243 VNIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVV 286
+NIGD ++ +N +Y+S HR V T R SVA F+ P+DC V
Sbjct: 250 INIGDQLQALSNGVYKSVWHRAVTNTENPRLSVASFL-CPADCAV 293
>At5g63580 flavonol synthase
Length = 310
Score = 106 bits (265), Expect = 1e-23
Identities = 80/308 (25%), Positives = 137/308 (43%), Gaps = 42/308 (13%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
++PIIDLS+ D A ++ + E+G F++VNHG+ D I++ D FF LP +K
Sbjct: 18 TIPIIDLSNLDEELVAHAVVKGSEEWGIFHVVNHGIPMDLIQRLKDVGTQFFELPETEKK 77
Query: 68 KLNR----KEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSF 123
+ + K++ GYT KG+ + T + + WP N
Sbjct: 78 AVAKQDGSKDFEGYT-------TNLKYVKGEVWTENLFHRIWPPTCINFDYWPKN----- 125
Query: 124 FPAMTLQNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDENYFEKISALNKPE 177
PQ+ +Y+ + +L ++ L L + E
Sbjct: 126 -------------PPQYREVIEEYTKETKKLSERILGYLSEGLGLPSEALIQGLGGESTE 172
Query: 178 AFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHV 237
+R+ YP + P+ + G H+D IT+++TN VPGLQI K+ W DV ++
Sbjct: 173 YVMRINNYPPDPKPDLTL-GVPEHTDIIGITIIITNEVPGLQIFKD-----DHWLDVHYI 226
Query: 238 EGAIIVNIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVVECFESCCSES 296
+I VNIGD + R +N Y++ LHR ++ +R S F+D D V+ +
Sbjct: 227 PSSITVNIGDQIMRLSNGKYKNVLHRAIVDKENQRMSWPVFVDANPDVVIGPLPELITGD 286
Query: 297 SPPRYRKV 304
+P +++ +
Sbjct: 287 NPSKFKPI 294
>At5g51810 gibberellin 20-oxidase (emb|CAA58294.1)
Length = 378
Score = 103 bits (258), Expect = 9e-23
Identities = 88/285 (30%), Positives = 132/285 (45%), Gaps = 22/285 (7%)
Query: 7 LSLPIIDLSSPDR-ISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIED 65
L++P IDLSS D + + I +AC ++GFF +VNHGV E I A +FF +P+
Sbjct: 61 LNVPFIDLSSQDSTLEAPRVIAEACTKHGFFLVVNHGVSESLIADAHRLMESFFDMPLAG 120
Query: 66 KIKLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFP 125
K K RK G + YA +K KE+ D + R Q +F
Sbjct: 121 KQKAQRKP--GESCGYASSFTGRFSTKLPWKETLSFQFSNDNSGSRTVQ-------DYFS 171
Query: 126 AMTLQNYSQIGDPQWNPYFGKYSGKEL--LSLIALSLNLDENYFEKISALNKPEAFLRLL 183
Q + Q G + Y S L + L+ LSL ++ +YF N ++ +RL
Sbjct: 172 DTLGQEFEQFG-KVYQDYCEAMSSLSLKIMELLGLSLGVNRDYFRGFFEEN--DSIMRLN 228
Query: 184 RYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIV 243
YP P+ + G H D +T+L + V GLQ+ + W+ + A +V
Sbjct: 229 HYPPCQTPDLTL-GTGPHCDPSSLTILHQDHVNGLQVFVD-----NQWQSIRPNPKAFVV 282
Query: 244 NIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVVE 287
NIGD +N +++S LHR V+ R S+AFF+ P D VV+
Sbjct: 283 NIGDTFMALSNGIFKSCLHRAVVNRESARKSMAFFLCPKKDKVVK 327
>At2g44800 putative flavonol synthase
Length = 352
Score = 103 bits (258), Expect = 9e-23
Identities = 85/281 (30%), Positives = 133/281 (47%), Gaps = 57/281 (20%)
Query: 3 GTATLSLPIIDLS---SPDRISTA-KSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANF 58
GT+ +LP+IDLS P S A I AC E+GFF ++NHG+ + A D + F
Sbjct: 41 GTSETTLPVIDLSLLHQPFLRSLAIHEISMACKEFGFFQVINHGIPSSVVNDALDAATQF 100
Query: 59 FSLPIEDKIKLNRKEYRGYTPLYAEKLDP----SSLSKGDPKESY---YIGPLTDTTSVR 111
F LP+E+K+ L + A +P +SL+ + Y +I + S
Sbjct: 101 FDLPVEEKMLL----------VSANVHEPVRYGTSLNHSTDRVHYWRDFIKHYSHPLSKW 150
Query: 112 LNQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYS------GKELLSLIALSLNLDEN 165
++ WPSN P + GKY+ K+L+ I+ SL L++N
Sbjct: 151 IDMWPSN------------------PPCYKDKVGKYAEATHLLHKQLIEAISESLGLEKN 192
Query: 166 YFEKISALNKPEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQI--CKE 223
Y ++ + + + + YP P E G HSD+ +T+LL + GLQI C +
Sbjct: 193 YLQE--EIEEGSQVMAVNCYPACPEP-EMALGMPPHSDFSSLTILLQSS-KGLQIMDCNK 248
Query: 224 KLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRV 264
W V ++EGA+IV +GD +E +N +Y+S +HRV
Sbjct: 249 N------WVCVPYIEGALIVQLGDQVEVMSNGIYKSVIHRV 283
>At1g55290 leucoanthocyanidin dioxygenase 2, putative
Length = 361
Score = 103 bits (257), Expect = 1e-22
Identities = 80/300 (26%), Positives = 141/300 (46%), Gaps = 25/300 (8%)
Query: 8 SLPIIDLSSPDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPIEDKI 67
S+P+ID+S+ D S +K++ A E+GFF ++NHGV + ++ + FF LP+E+K
Sbjct: 61 SIPVIDISNLDEKSVSKAVCDAAEEWGFFQVINHGVSMEVLENMKTATHRFFGLPVEEKR 120
Query: 68 KLNRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRLNQWPSNGVFSFFPAM 127
K +R++ + P + + K+ + +++ + +L WP + +
Sbjct: 121 KFSREKSLSTNVRFGTSFSPHAEKALEWKDYLSLFFVSEAEASQL--WPDS-----CRSE 173
Query: 128 TLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDENYFEKISALNKPEAFLRLLRYPG 187
TL+ ++ K K+LL + +LN+ E K S + L YP
Sbjct: 174 TLEYMNET----------KPLVKKLLRFLGENLNVKELDKTKESFF-MGSTRINLNYYP- 221
Query: 188 ELGPNEEI-CGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIG 246
+ PN E+ G HSD +T+LL + + GL + + W V + G++++NIG
Sbjct: 222 -ICPNPELTVGVGRHSDVSSLTILLQDEIGGLHV---RSLTTGRWVHVPPISGSLVINIG 277
Query: 247 DMMERWTNCLYRSTLHRVMPTGK-ERYSVAFFMDPPSDCVVECFESCCSESSPPRYRKVL 305
D M+ +N Y+S HRV+ G R SV F+ P + V+ P Y+ +L
Sbjct: 278 DAMQIMSNGRYKSVEHRVLANGSYNRISVPIFVSPKPESVIGPLLEVIENGEKPVYKDIL 337
>At4g25420 gibberellin 20-oxidase - Arabidopsis thaliana
Length = 377
Score = 101 bits (252), Expect = 4e-22
Identities = 88/308 (28%), Positives = 139/308 (44%), Gaps = 38/308 (12%)
Query: 7 LSLPIIDLSS-----PDRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSL 61
L +P+IDL + + ++ I +AC ++GFF +VNHG+ E+ I A + ++ FF +
Sbjct: 59 LDVPLIDLQNLLSDPSSTLDASRLISEACKKHGFFLVVNHGISEELISDAHEYTSRFFDM 118
Query: 62 PIEDKIKLNRK--EYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVRL--NQWPS 117
P+ +K ++ RK E GY + + P +T S R + S
Sbjct: 119 PLSEKQRVLRKSGESVGYASSFTGRFSTKL-------------PWKETLSFRFCDDMSRS 165
Query: 118 NGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKEL--LSLIALSLNLDENYFEKISALNK 175
V +F + G + Y S L + L+ LSL + +YF + N
Sbjct: 166 KSVQDYFCDALGHGFQPFG-KVYQEYCEAMSSLSLKIMELLGLSLGVKRDYFREFFEEN- 223
Query: 176 PEAFLRLLRYPGELGPNEEICGASAHSDYGMITLLLTNGVPGLQICKEKLKQPQVWEDVS 235
++ +RL YP + P+ + G H D +T+L + V GLQ+ E W +
Sbjct: 224 -DSIMRLNYYPPCIKPDLTL-GTGPHCDPTSLTILHQDHVNGLQVFVE-----NQWRSIR 276
Query: 236 HVEGAIIVNIGDMMERWTNCLYRSTLHR-VMPTGKERYSVAFFMDPPSDCVV----ECFE 290
A +VNIGD +N Y+S LHR V+ + ER S+AFF+ P D VV E +
Sbjct: 277 PNPKAFVVNIGDTFMALSNDRYKSCLHRAVVNSESERKSLAFFLCPKKDRVVTPPRELLD 336
Query: 291 SCCSESSP 298
S S P
Sbjct: 337 SITSRRYP 344
>At2g38240 putative anthocyanidin synthase
Length = 353
Score = 101 bits (252), Expect = 4e-22
Identities = 89/307 (28%), Positives = 149/307 (47%), Gaps = 31/307 (10%)
Query: 5 ATLSLPIIDLSSP-DRISTAKSIHQACVEYGFFYLVNHGVDEDFIKQAFDQSANFFSLPI 63
A + +P++D++ + + + AC E+GFF +VNHGV +++ FF LP+
Sbjct: 44 AGIEIPVLDMNDVWGKPEGLRLVRSACEEWGFFQMVNHGVTHSLMERVRGAWREFFELPL 103
Query: 64 EDKIKL--NRKEYRGYTPLYAEKLDPSSLSKGDPKESYYIGPLTDTTSVR-LNQWPSNGV 120
E+K K + Y G Y +L +K D + +++ L +S+R ++WPS
Sbjct: 104 EEKRKYANSPDTYEG----YGSRLGVVKDAKLDWSDYFFLNYL--PSSIRNPSKWPSQ-- 155
Query: 121 FSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIALSLNLDEN-YFEKISALNKPEAF 179
P + + G+ + + L ++ SL L N + + +K A
Sbjct: 156 ----PPKIRELIEKYGEEV------RKLCERLTETLSESLGLKPNKLMQALGGGDKVGAS 205
Query: 180 LRLLRYPGELGPNEEICGASAHSDYGMITLLLTN-GVPGLQICKEKLKQPQVWEDVSHVE 238
LR YP P + G S+HSD G IT+LL + V GLQ+ ++ W + V
Sbjct: 206 LRTNFYPKCPQPQLTL-GLSSHSDPGGITILLPDEKVAGLQV-----RRGDGWVTIKSVP 259
Query: 239 GAIIVNIGDMMERWTNCLYRSTLHRV-MPTGKERYSVAFFMDPPSDCVVECFESCCSESS 297
A+IVNIGD ++ +N +Y+S H+V + +G ER S+AFF +P SD V E + +
Sbjct: 260 NALIVNIGDQLQILSNGIYKSVEHQVIVNSGMERVSLAFFYNPRSDIPVGPIEELVTANR 319
Query: 298 PPRYRKV 304
P Y+ +
Sbjct: 320 PALYKPI 326
>At3g50210 flavonol synthase - like protein
Length = 332
Score = 100 bits (250), Expect = 8e-22
Identities = 95/320 (29%), Positives = 145/320 (44%), Gaps = 62/320 (19%)
Query: 8 SLPIIDLS-------SPDR---ISTAKSIHQ---ACVEYGFFYLVNHGVDEDFIKQAFDQ 54
SLP+ID+S PD + A+ + Q AC + GFFY++ HG+ ED I + +
Sbjct: 7 SLPVIDISRLLLKCDDPDMAEDVGVAEVVQQLDKACRDAGFFYVIGHGISEDVINKVREI 66
Query: 55 SANFFSLPIEDKIKLNRKE---YRGYTPLYAEKLDPSSLSKGDP------------KESY 99
+ FF LP E+K+K+ YRGY + +++KG P K+
Sbjct: 67 TREFFKLPYEEKLKIKMTPAAGYRGYQRI------GENVTKGIPDIHEAIDCYREIKQGK 120
Query: 100 Y--IGPLTDTTSVRLNQWPSNGVFSFFPAMTLQNYSQIGDPQWNPYFGKYSGKELLSLIA 157
Y IG + + NQWP N Q + ++ + +++L I+
Sbjct: 121 YGDIGKVMEGP----NQWPENP----------QEFKELMEEYIK--LCTDLSRKILRGIS 164
Query: 158 LSLNLDENYFEKISALNKPEAFLRLLRYPG---ELGPNEEICGASAHSDYGMITLL-LTN 213
L+L FE A P +RL+ YPG G E G AH+DYG++TL+ +
Sbjct: 165 LALAGSPYEFEGKMA-GDPFWVMRLIGYPGAEFTNGQPENDIGCGAHTDYGLLTLVNQDD 223
Query: 214 GVPGLQICKEKLKQPQVWEDVSHVEGAIIVNIGDMMERWTNCLYRSTLHRVMPTGKE-RY 272
LQ+ W + G+ + NIGDM++ +N +Y STLHRV+ + R
Sbjct: 224 DKTALQV----RNLGGEWISAIPIPGSFVCNIGDMLKILSNGVYESTLHRVINNSPQYRV 279
Query: 273 SVAFFMDPPSDCVVECFESC 292
VAFF + D VVE + C
Sbjct: 280 CVAFFYETNFDAVVEPLDIC 299
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.419
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,585,495
Number of Sequences: 26719
Number of extensions: 333398
Number of successful extensions: 948
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 655
Number of HSP's gapped (non-prelim): 153
length of query: 309
length of database: 11,318,596
effective HSP length: 99
effective length of query: 210
effective length of database: 8,673,415
effective search space: 1821417150
effective search space used: 1821417150
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC149079.14


