

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.11 - phase: 0
(664 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
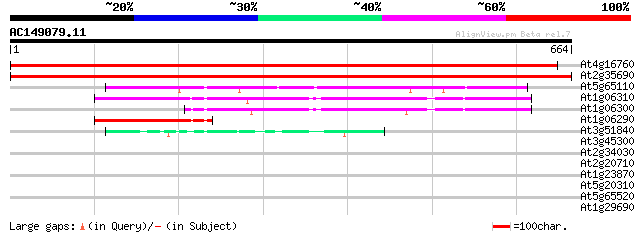
At4g16760 unknown protein 1089 0.0
At2g35690 putative acyl-CoA oxidase 1084 0.0
At5g65110 acyl-CoA oxidase (gb|AAC13497.1) 283 2e-76
At1g06310 hypothetical protein 211 8e-55
At1g06300 hypothetical protein, 5' partial 127 2e-29
At1g06290 acyl-CoA oxidase, putative, 3' partial 125 1e-28
At3g51840 Short-chain acyl CoA oxidase 63 5e-10
At3g45300 isovaleryl-CoA-dehydrogenase precursor (IVD) 41 0.002
At2g34030 unknown protein 32 1.5
At2g20710 unknown protein 32 1.5
At1g23870 trehalose 6-phosphate synthase, putative 32 1.5
At5g20310 putative protein 29 7.4
At5g65520 unknown protein 29 9.6
At1g29690 unknown protein 29 9.6
>At4g16760 unknown protein
Length = 675
Score = 1089 bits (2817), Expect = 0.0
Identities = 530/648 (81%), Positives = 586/648 (89%)
Query: 1 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 60
MEG DHLA ERNKAEFDV+ MKIVWAGS HA EVSDR++RLVASDP F K +R L RKE
Sbjct: 1 MEGIDHLADERNKAEFDVEDMKIVWAGSRHAFEVSDRIARLVASDPVFEKSNRARLSRKE 60
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 120
LFKSTL+K A+A+KRIIELRL EEEA LR F+D+PA+ DLHWGMF+PAIKGQGTEEQQ+
Sbjct: 61 LFKSTLRKCAHAFKRIIELRLNEEEAGRLRHFIDQPAYVDLHWGMFVPAIKGQGTEEQQK 120
Query: 121 KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG 180
KWL LA +MQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIH+PT T+SKWWPGGLG
Sbjct: 121 KWLSLANKMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHTPTQTASKWWPGGLG 180
Query: 181 KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNG 240
KVSTHAVVYARLIT+G+D+G+HGFIVQLRSL+DH PLP ITVGDIG K GNGAYN+MDNG
Sbjct: 181 KVSTHAVVYARLITNGKDYGIHGFIVQLRSLEDHSPLPNITVGDIGTKMGNGAYNSMDNG 240
Query: 241 VLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVSDASTAMSRAV 300
L F+HVRIPRDQMLMR+S+VTREG+YV S+VP+QLVYGTMVYVRQTIV+DAS A+SRAV
Sbjct: 241 FLMFDHVRIPRDQMLMRLSKVTREGEYVPSDVPKQLVYGTMVYVRQTIVADASNALSRAV 300
Query: 301 CIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFRFVGEWLKWLYTDVMK 360
CIA RYSAVRRQFG +NG +E+QVIDYKTQQ RLFPLLASAYAFRFVGEWLKWLYTDV +
Sbjct: 301 CIATRYSAVRRQFGAHNGGIETQVIDYKTQQNRLFPLLASAYAFRFVGEWLKWLYTDVTE 360
Query: 361 RLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRKLCGGHGYLCSSGLPELFAVYVPT 420
RL A+DF+TLPEAHACTAGLKSLTT+AT+DGIEECRKLCGGHGYL SGLPELFAVYVP
Sbjct: 361 RLAASDFATLPEAHACTAGLKSLTTTATADGIEECRKLCGGHGYLWCSGLPELFAVYVPA 420
Query: 421 CTYEGDNIVLLLQVARHLMKVISRLGSGKKPVGTTAYMGRVEQLLEARSDVQKAEDWLNP 480
CTYEGDN+VL LQVAR LMK +++LGSGK PVGTTAYMGR LL+ RS VQKAEDWLNP
Sbjct: 421 CTYEGDNVVLQLQVARFLMKTVAQLGSGKVPVGTTAYMGRAAHLLQCRSGVQKAEDWLNP 480
Query: 481 NTVLRAFEARAARMSVACAKNLTKFSNPEEGFQELLADLVDAARAHCQLIVVSKFIEKLQ 540
+ VL AFEARA RM+V CAKNL+KF N E+GFQELLADLV+AA AHCQLIVVSKFI KL+
Sbjct: 481 DVVLEAFEARALRMAVTCAKNLSKFENQEQGFQELLADLVEAAIAHCQLIVVSKFIAKLE 540
Query: 541 QDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNA 600
QDI GKGVK+QL LC IYAL+ LHKHLGDFLST CITPKQ SLANDQLRSLY+QVRPNA
Sbjct: 541 QDIGGKGVKKQLNNLCYIYALYLLHKHLGDFLSTNCITPKQASLANDQLRSLYTQVRPNA 600
Query: 601 IALVDAFNYTDHYLSSVLGRYDGNVYPKLYEEAWKDPLNDSVVPDGFQ 648
+ALVDAFNYTDHYL+SVLGRYDGNVYPKL+EEA KDPLNDSVVPDG++
Sbjct: 601 VALVDAFNYTDHYLNSVLGRYDGNVYPKLFEEALKDPLNDSVVPDGYK 648
>At2g35690 putative acyl-CoA oxidase
Length = 664
Score = 1084 bits (2804), Expect = 0.0
Identities = 525/664 (79%), Positives = 592/664 (89%)
Query: 1 MEGEDHLAFERNKAEFDVDAMKIVWAGSAHALEVSDRMSRLVASDPAFRKDHRPMLGRKE 60
ME DHLA ERNKAEF+VD MKIVWAGS HA +VS+RMSRLVA+DP F K R ++ RKE
Sbjct: 1 MERVDHLADERNKAEFNVDDMKIVWAGSRHAFDVSNRMSRLVANDPVFEKSKRAVMSRKE 60
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 120
LFK+TL+K+ +AWK I ELRL++EE LRSF+D+P F DLHWGMF+PAIKGQGTE+QQQ
Sbjct: 61 LFKNTLRKSVHAWKLINELRLSDEEGLKLRSFMDQPGFLDLHWGMFVPAIKGQGTEQQQQ 120
Query: 121 KWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSKWWPGGLG 180
KWL LA +MQIIGCYAQTELGHGSNVQGLETTATFDPKTD+F+IHSPT TSSKWWPGGLG
Sbjct: 121 KWLSLATKMQIIGCYAQTELGHGSNVQGLETTATFDPKTDQFIIHSPTQTSSKWWPGGLG 180
Query: 181 KVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNG 240
KVSTHAV+YARLIT+G+DHGVHGFIVQLRSLDDH PLPGITVGDIGMKFGNGAYN+MDNG
Sbjct: 181 KVSTHAVIYARLITNGKDHGVHGFIVQLRSLDDHSPLPGITVGDIGMKFGNGAYNSMDNG 240
Query: 241 VLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVSDASTAMSRAV 300
L F+H RIPRDQMLMR+S+VTREGKYV S+VPRQLVYGTMVYVRQ+IVS+ASTA++RAV
Sbjct: 241 FLMFDHFRIPRDQMLMRLSKVTREGKYVASDVPRQLVYGTMVYVRQSIVSNASTALARAV 300
Query: 301 CIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFRFVGEWLKWLYTDVMK 360
CIA RYSAVRRQFG ++G +E+QVI+YKTQQ RLFPLLASAYAFRFVGEWLKWLYTDV K
Sbjct: 301 CIATRYSAVRRQFGSHDGGIETQVINYKTQQNRLFPLLASAYAFRFVGEWLKWLYTDVTK 360
Query: 361 RLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRKLCGGHGYLCSSGLPELFAVYVPT 420
RL+A+DF+TLPEAHACTAGLKS+TTSATSDGIEECRKLCGGHGYL SGLPELFAVYVP
Sbjct: 361 RLEASDFATLPEAHACTAGLKSMTTSATSDGIEECRKLCGGHGYLWCSGLPELFAVYVPA 420
Query: 421 CTYEGDNIVLLLQVARHLMKVISRLGSGKKPVGTTAYMGRVEQLLEARSDVQKAEDWLNP 480
CTYEGDN+VL LQVAR LMK +S+LGSGK P GTTAYMGR + LL+ S V+ A DWLNP
Sbjct: 421 CTYEGDNVVLQLQVARFLMKTVSQLGSGKAPSGTTAYMGRAKHLLQCSSGVRNARDWLNP 480
Query: 481 NTVLRAFEARAARMSVACAKNLTKFSNPEEGFQELLADLVDAARAHCQLIVVSKFIEKLQ 540
VL AFEARA RM+V CA NL+KF N E+GF ELLADLV+AA AHCQLIVVSKFI K++
Sbjct: 481 GMVLEAFEARALRMAVTCANNLSKFENQEQGFSELLADLVEAATAHCQLIVVSKFIAKVE 540
Query: 541 QDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNA 600
DI GKGVK+QL+ LC IYAL+ LHKHLGDFLST +TP+Q SLAN QLRSLYSQVRPNA
Sbjct: 541 GDIEGKGVKKQLKNLCYIYALYLLHKHLGDFLSTNSVTPEQASLANQQLRSLYSQVRPNA 600
Query: 601 IALVDAFNYTDHYLSSVLGRYDGNVYPKLYEEAWKDPLNDSVVPDGFQEYIRPMLKQQLR 660
+ALVDAF+YTD YL SVLGRYDGNVYPKL+EEA KDPLNDSVVPDG++EYIRP++KQ+ R
Sbjct: 601 VALVDAFDYTDQYLGSVLGRYDGNVYPKLFEEALKDPLNDSVVPDGYREYIRPLIKQRFR 660
Query: 661 NARL 664
+A+L
Sbjct: 661 SAKL 664
>At5g65110 acyl-CoA oxidase (gb|AAC13497.1)
Length = 692
Score = 283 bits (724), Expect = 2e-76
Identities = 188/517 (36%), Positives = 264/517 (50%), Gaps = 26/517 (5%)
Query: 114 GTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSK 173
GT++ + K+ + GC+A TEL HGSNVQGL+TTATFDP DEFVI +P + K
Sbjct: 162 GTKKHRDKYFDGIDNLDYTGCFAMTELHHGSNVQGLQTTATFDPLKDEFVIDTPNDGAIK 221
Query: 174 WWPGGLGKVSTHAVVYARLITDGRDH------GVHGFIVQLRSLDDHLPLPGITVGDIGM 227
WW G A V+ARLI D GVH FIV +R + H LPG+ + D G
Sbjct: 222 WWIGNAAVHGKFATVFARLILPTHDSKGVSDMGVHAFIVPIRDMKTHQTLPGVEIQDCGH 281
Query: 228 KFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQS----NVPRQLVYGTMVY 283
K G N +DNG LRF VRIPRD +L R V+R+G Y S N G +V
Sbjct: 282 KVG---LNGVDNGALRFRSVRIPRDNLLNRFGDVSRDGTYTSSLPTINKRFGATLGELVG 338
Query: 284 VRQTIVSDASTAMSRAVCIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYA 343
R + + + + IA RYS +R+QFG E ++DY++QQ +L P+LAS YA
Sbjct: 339 GRVGLAYASVGVLKISATIAIRYSLLRQQFGPPK-QPEVSILDYQSQQHKLMPMLASTYA 397
Query: 344 FRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRKLCGGHG 403
+ F +L Y+++ K +D + + HA +AGLKS TS T+ + CR+ CGGHG
Sbjct: 398 YHFATVYLVEKYSEMKK---THDEQLVADVHALSAGLKSYVTSYTAKALSVCREACGGHG 454
Query: 404 YLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVISRLGSGKKPVGTTAYMGR-VE 462
Y + L + T+EGDN VLL QVA L+K G T +Y+ +
Sbjct: 455 YAAVNRFGSLRNDHDIFQTFEGDNTVLLQQVAADLLKRYKEKFQGGTLTVTWSYLRESMN 514
Query: 463 QLLEARSDVQ---KAEDWL-NPNTVLRAFEARAARMSVACAKNLTKFSNPEEGF---QEL 515
L + V + ED L +P L AF R +R+ A L K S GF
Sbjct: 515 TYLSQPNPVTARWEGEDHLRDPKFQLDAFRYRTSRLLQNVAARLQKHSKTLGGFGAWNRC 574
Query: 516 LADLVDAARAHCQLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLHKHLGDFLSTG 575
L L+ A +H + ++++KFIE + ++ P K L++ C++YAL + K +G + +
Sbjct: 575 LNHLLTLAESHIETVILAKFIEAV-KNCPDPSAKAALKLACDLYALDRIWKDIGTYRNVD 633
Query: 576 CITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDH 612
+ P + + L QVR A LVDAF DH
Sbjct: 634 YVAPNKAKAIHKLTEYLSFQVRNVAKELVDAFELPDH 670
>At1g06310 hypothetical protein
Length = 667
Score = 211 bits (538), Expect = 8e-55
Identities = 154/525 (29%), Positives = 250/525 (47%), Gaps = 30/525 (5%)
Query: 101 LHWGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTD 160
+H+ ++ A+K GT+ +KWL + + GC+A TELGHG+NV+G+ET T+DP T+
Sbjct: 152 VHFLLWGNAVKFFGTKRHHEKWLKDTEDYVVKGCFAMTELGHGTNVRGIETVTTYDPTTE 211
Query: 161 EFVIHSPTLTSSKWWPGGLGKVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGI 220
EFVI++P ++ K+W G + HA+V ++L +G + G+H FI Q+R D + P +
Sbjct: 212 EFVINTPCESAQKYWIGEAANHANHAIVISQLSMNGTNQGIHVFIAQIRDHDGN-TCPNV 270
Query: 221 TVGDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGT 280
+ D G K G N +DNG + F+++RIPR+ +L V+ V +GKYV S +G
Sbjct: 271 RIADCGHKIG---LNGVDNGRIWFDNLRIPRENLLNSVADVLADGKYVSSIKDPDQRFGA 327
Query: 281 ----MVYVRQTIVSDASTAMSRAVCIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFP 336
+ R TI S A + + +A RYS RR F E ++DY + Q RL P
Sbjct: 328 FLAPLTSGRVTIASSAIYSAKLGLAVAIRYSLSRRAFSVAANGPEVLLLDYPSHQRRLLP 387
Query: 337 LLASAYAFRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECR 396
LLA YA F LK +Y +KR T H ++G K++ T ++ECR
Sbjct: 388 LLAKTYAMSFAVNDLKMIY---VKRTP----ETNKAIHVVSSGFKAVLTWHNMRTLQECR 440
Query: 397 KLCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVISRLGSGKKPVGTTA 456
+ GG G + + L Y T+EGDN VL+ V++ L KP
Sbjct: 441 EAVGGQGLKTENRVGHLKGEYDVQTTFEGDNNVLMQLVSKALFAEYVSCKKRNKPFKGLG 500
Query: 457 --YMGRVEQLLEARSDVQKAEDWLNPNTVLRAFEARAARMSVACAKNLTKFSNPEEGFQE 514
+M +L + +L F + A + + F
Sbjct: 501 LEHMNSPRPVLPTQLTSSTLRCSQFQRDLLERFTSEVAEL---------QGRGESREFLF 551
Query: 515 LLADLV--DAARAHCQLIVVSKFIEKLQQDIPGKGVKRQLEILCNIYALFHLHKHLGDFL 572
LL + D ++A + ++ ++ + +P VK L ++ ++YAL L + L
Sbjct: 552 LLNHQLSEDLSKAFTEKAILQTVLD-AEAKLPPGSVKDVLGLVRSMYALISLEED-PSLL 609
Query: 573 STGCITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDHYLSSV 617
G ++ ++ L ++RP+A+ALV +F D +LS +
Sbjct: 610 RYGHLSRDNVGDVRKEVSKLCGELRPHALALVASFGIPDAFLSPI 654
>At1g06300 hypothetical protein, 5' partial
Length = 419
Score = 127 bits (320), Expect = 2e-29
Identities = 116/427 (27%), Positives = 187/427 (43%), Gaps = 38/427 (8%)
Query: 207 QLRSLDDHLPLPGITVGDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGK 266
Q+R D + P I + D G K G N +DNG + F+++RIPR+ +L V+ V+ +GK
Sbjct: 2 QIRDQDGSI-CPNIRIADCGHKIG---LNGVDNGRIWFDNLRIPRENLLNAVADVSSDGK 57
Query: 267 YVQSNVPRQLVYGTMVYV----RQTIVSDASTAMSRAVCIAARYSAVRRQFGGNNGSLES 322
YV S +G + R TI S A + + IA RYS RR F E
Sbjct: 58 YVSSIKDPDQRFGAFMAPLTSGRVTIASSAIYSAKVGLSIAIRYSLSRRAFSVTANGPEV 117
Query: 323 QVIDYKTQQARLFPLLASAYAFRFVGEWLKWLYTDVMKRLQANDFSTLPEAHACTAGLKS 382
++DY + Q RL PLLA YA F LK +Y +KR T H ++G K+
Sbjct: 118 LLLDYPSHQRRLLPLLAKTYAMSFAANELKMIY---VKRTP----ETNKAIHVVSSGFKA 170
Query: 383 LTTSATSDGIEECRKLCGGHGYLCSSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVI 442
+ T ++ECR+ GG G + + +L + T+EGDN VL+ QV++ L
Sbjct: 171 VLTWHNMHTLQECREAVGGQGVKTENLVGQLKGEFDVQTTFEGDNNVLMQQVSKALFAEY 230
Query: 443 SRLGSGKKPVGTTA--YMGRVEQLLEAR--------SDVQKAEDWLNPNTVLRAFEARAA 492
KP +M +L + S Q L +L F + A
Sbjct: 231 VSCKKRNKPFKGLGLEHMNSPRPVLPTQLTSSTLRCSQFQTNVFCLRERDLLEQFTSEVA 290
Query: 493 RMSVACAKNLTKFSNPEEGFQELLADLV--DAARAHCQLIVVSKFIEKLQQDIPGKGVKR 550
++ + F LL+ + D +A + ++ ++ + +P VK
Sbjct: 291 QL---------QGRGESREFSFLLSHQLAEDLGKAFTEKAILQTILD-AEAKLPTGSVKD 340
Query: 551 QLEILCNIYALFHLHKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYT 610
L ++ ++YAL L + L G ++ ++ L ++RP+A+ALV +F
Sbjct: 341 VLGLVRSMYALISLEED-PSLLRYGYLSQDNVGDVRREVSKLCGELRPHALALVTSFGIP 399
Query: 611 DHYLSSV 617
D +LS +
Sbjct: 400 DSFLSPI 406
>At1g06290 acyl-CoA oxidase, putative, 3' partial
Length = 289
Score = 125 bits (313), Expect = 1e-28
Identities = 60/140 (42%), Positives = 90/140 (63%), Gaps = 4/140 (2%)
Query: 101 LHWGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTD 160
+H+ ++ A+K GT+ +KWL + + GC+A TELGHGSNV+G+ET T+DPKT+
Sbjct: 152 VHFFLWGNAVKFFGTKRHHEKWLKNTEDYVVKGCFAMTELGHGSNVRGIETVTTYDPKTE 211
Query: 161 EFVIHSPTLTSSKWWPGGLGKVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGI 220
EFVI++P ++ K+W GG +TH +V+++L +G + GVH FI Q+R D + P I
Sbjct: 212 EFVINTPCESAQKYWIGGAANHATHTIVFSQLHINGTNQGVHAFIAQIRDQDGSI-CPNI 270
Query: 221 TVGDIGMKFGNGAYNTMDNG 240
+ D G K G N +DNG
Sbjct: 271 RIADCGHKIG---LNGVDNG 287
>At3g51840 Short-chain acyl CoA oxidase
Length = 436
Score = 63.2 bits (152), Expect = 5e-10
Identities = 84/337 (24%), Positives = 133/337 (38%), Gaps = 67/337 (19%)
Query: 114 GTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDEFVIHSPTLTSSK 173
G+E Q++K+LP ++ + C+A TE +GS+ GL TTAT V + K
Sbjct: 151 GSEAQKEKYLPSLAQLNTVACWALTEPDNGSDASGLGTTAT-------KVEGGWKINGQK 203
Query: 174 WWPGGLGKVSTHA---VVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGITVGDIGMKFG 230
W G ST A +++AR T + ++GFIV+ + PG+ I K G
Sbjct: 204 RWIGN----STFADLLIIFARNTTTNQ---INGFIVKKDA-------PGLKATKIPNKIG 249
Query: 231 NGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYVQSNVPRQLVYGTMVYVRQTIVS 290
+ NG + ++V +P + L V+ K + V R +V + + I
Sbjct: 250 ---LRMVQNGDILLQNVFVPDEDRLPGVNSFQDTSKVLA--VSRVMVAWQPIGISMGIYD 304
Query: 291 DASTAMSRAVCIAARYSAVRRQFGGNNGSLESQVIDYKTQQARLFPLLASAYAFRFVGEW 350
+ RY R+QFG + + ++ Q +L +L + A +G
Sbjct: 305 -----------MCHRYLKERKQFG-------APLAAFQLNQQKLVQMLGNVQAMFLMGWR 346
Query: 351 LKWLYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGIEEC----RKLCGGHGYLC 406
L LY E T G SL + S E R+L GG+G L
Sbjct: 347 LCKLY----------------ETGQMTPGQASLGKAWISSKARETASLGRELLGGNGILA 390
Query: 407 SSGLPELFAVYVPTCTYEGDNIVLLLQVARHLMKVIS 443
+ + F P TYEG + L R + + S
Sbjct: 391 DFLVAKAFCDLEPIYTYEGTYDINTLVTGREVTGIAS 427
>At3g45300 isovaleryl-CoA-dehydrogenase precursor (IVD)
Length = 409
Score = 40.8 bits (94), Expect = 0.002
Identities = 43/167 (25%), Positives = 66/167 (38%), Gaps = 24/167 (14%)
Query: 102 HWGMFIPAIKGQGTEEQQQKWLPLAQRMQIIGCYAQTELGHGSNVQGLETTATFDPKTDE 161
H + I + GT Q++K+LP + +G A +E GS+V G++ A E
Sbjct: 118 HSNLCINQLVRNGTAAQKEKYLPKLISGEHVGALAMSEPNAGSDVVGMKCKA-------E 170
Query: 162 FVIHSPTLTSSKWWPGGLGKVSTHAVVYARLITDGRDHGVHGFIVQLRSLDDHLPLPGIT 221
V L +K W G + VVYA+ T G+ FI++ G+T
Sbjct: 171 KVDGGYILNGNKMWCTN-GPSAETLVVYAKTDTKAGSKGITAFIIE----------KGMT 219
Query: 222 VGDIGMKFGNGAYNTMDNGVLRFEHVRIPRDQMLMRVSQVTREGKYV 268
K D L FE+ +P + +L +EGK V
Sbjct: 220 GFSTAQKLDKLGMRGSDTCELVFENCFVPEENIL------DKEGKGV 260
>At2g34030 unknown protein
Length = 423
Score = 31.6 bits (70), Expect = 1.5
Identities = 28/97 (28%), Positives = 43/97 (43%), Gaps = 5/97 (5%)
Query: 564 LHKHLGDFLSTGCITPKQGSLANDQLRSLYSQVRPNAIALVDAFNYTDHYLS-SVLGR-- 620
+HKHL F I K G L+ + L+SL+ + N + D + S GR
Sbjct: 209 VHKHLKRFSPKHLI--KDGELSKESLKSLFKKTDKNKDGKIQISELKDLTIELSNFGRMR 266
Query: 621 YDGNVYPKLYEEAWKDPLNDSVVPDGFQEYIRPMLKQ 657
YD N K + E + + + + F+E I +LKQ
Sbjct: 267 YDINELAKAFLEDFDGDNDGELEENEFEEGIARLLKQ 303
>At2g20710 unknown protein
Length = 490
Score = 31.6 bits (70), Expect = 1.5
Identities = 37/148 (25%), Positives = 58/148 (39%), Gaps = 22/148 (14%)
Query: 461 VEQLLEARSDVQKAEDWLNPNTVLRAFEARAARMSVACAKNLTKFSNPEEGFQELLAD-- 518
VE+LL D D NT L A+ V+ + + KF E Q L D
Sbjct: 193 VEKLLREMEDETVKPDIFTVNTRLHAYSV------VSDVEGMEKFLMRCEADQGLHLDWR 246
Query: 519 -LVDAARAHCQLIVVSKFIEKL---QQDIPGKGVKRQLEILCN----------IYALFHL 564
D A + + + K +E L +Q + + K E+L + +Y L+ L
Sbjct: 247 TYADTANGYIKAGLTEKALEMLRKSEQMVNAQKRKHAYEVLMSFYGAAGKKEEVYRLWSL 306
Query: 565 HKHLGDFLSTGCITPKQGSLANDQLRSL 592
+K L F +TG I+ L D + +
Sbjct: 307 YKELDGFYNTGYISVISALLKMDDIEEV 334
>At1g23870 trehalose 6-phosphate synthase, putative
Length = 867
Score = 31.6 bits (70), Expect = 1.5
Identities = 17/59 (28%), Positives = 24/59 (39%)
Query: 354 LYTDVMKRLQANDFSTLPEAHACTAGLKSLTTSATSDGIEECRKLCGGHGYLCSSGLPE 412
++ ++ + D PE ACT G K D + + KL GG SS PE
Sbjct: 791 MFESILSTVTNPDLPMPPEIFACTVGRKPSKAKYFLDDVSDVLKLLGGLAAATSSSKPE 849
>At5g20310 putative protein
Length = 394
Score = 29.3 bits (64), Expect = 7.4
Identities = 15/72 (20%), Positives = 39/72 (53%), Gaps = 3/72 (4%)
Query: 61 LFKSTLKKAAYAWKRIIELRLTEEEASMLRSFVDEPAFTDLHWGMFIPAIKGQGTEEQQQ 120
++ + K+A +R +EL + +++A + +E A + M + A+KG+G+++++
Sbjct: 299 MYHAACKEALMEKERAVELVMWKKKAELRIQMAEETARMAI---MEMKAVKGKGSDDRKM 355
Query: 121 KWLPLAQRMQII 132
W L + ++
Sbjct: 356 VWDTLGESHTVV 367
>At5g65520 unknown protein
Length = 206
Score = 28.9 bits (63), Expect = 9.6
Identities = 12/32 (37%), Positives = 18/32 (55%)
Query: 329 TQQARLFPLLASAYAFRFVGEWLKWLYTDVMK 360
T ARLF LL Y F+ + E +W + + +K
Sbjct: 166 TDTARLFRLLGECYEFKGLKEKAQWAFNEALK 197
>At1g29690 unknown protein
Length = 561
Score = 28.9 bits (63), Expect = 9.6
Identities = 19/57 (33%), Positives = 25/57 (43%), Gaps = 8/57 (14%)
Query: 74 KRIIELRLTEEEASM------LRSFVDEPAFTDLHWGMFIP--AIKGQGTEEQQQKW 122
K + LRL+ E + L+ V P HW +P A K QG EEQ +W
Sbjct: 360 KPVTGLRLSLEGSKQNRLSIHLQHLVSLPKILQPHWDSHVPIGAPKWQGPEEQDSRW 416
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,527,554
Number of Sequences: 26719
Number of extensions: 609422
Number of successful extensions: 1322
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1296
Number of HSP's gapped (non-prelim): 17
length of query: 664
length of database: 11,318,596
effective HSP length: 106
effective length of query: 558
effective length of database: 8,486,382
effective search space: 4735401156
effective search space used: 4735401156
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC149079.11


