

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149079.10 - phase: 0
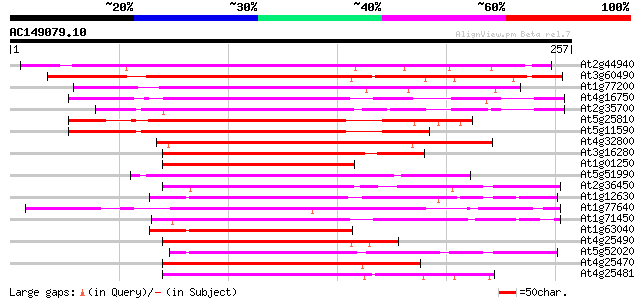
(257 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g44940 putative AP2 domain transcription factor 189 1e-48
At3g60490 transcription factor - like protein 183 9e-47
At1g77200 hypothetical protein 170 8e-43
At4g16750 apetala2 domain TINY like protein 169 2e-42
At2g35700 putative AP2 domain transcription factor 168 2e-42
At5g25810 transcription factor TINY 159 1e-39
At5g11590 transcription factor like protein 157 4e-39
At4g32800 transcription factor TINY homolog 152 2e-37
At3g16280 AP2 domain transcription factor like protein 151 4e-37
At1g01250 transcription factor TINY like protein 127 4e-30
At5g51990 AP2 domain transcription factor-like protein 119 2e-27
At2g36450 putative AP2 domain transcription factor 117 5e-27
At1g12630 hypothetical protein 111 3e-25
At1g77640 hypothetical protein 110 6e-25
At1g71450 putative TINY 110 7e-25
At1g63040 transcription factor DREB1A, putative 110 7e-25
At4g25490 transcriptional activator CBF1/ CRT/CRE binding factor 1 108 3e-24
At5g52020 unknown protein 106 1e-23
At4g25470 DRE/CRT-binding protein DREB1C 106 1e-23
At4g25481 DREB1A 105 3e-23
>At2g44940 putative AP2 domain transcription factor
Length = 295
Score = 189 bits (480), Expect = 1e-48
Identities = 118/270 (43%), Positives = 155/270 (56%), Gaps = 35/270 (12%)
Query: 6 SVLIHNSLCPQPTTSLSSYIIASTSSNSSSSLEEEEEEEEATNTKCK--------EKKKK 57
S+ SL PTTS SS +SS +S+ +EE+ + +A+ + +
Sbjct: 29 SITASASLSSSPTTSSSS-----SSSTNSNFIEEDNSKRKASRRSLSSLVSVEDDDDQNG 83
Query: 58 KVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAA 117
G K N +PTYRGVRMR WGKWVSEIREPRKKSRIWLGT+PT +MAARAHDVAA
Sbjct: 84 GGGKRRKTNGGDKHPTYRGVRMRSWGKWVSEIREPRKKSRIWLGTYPTAEMAARAHDVAA 143
Query: 118 LTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAAT---VYNHAITDQIELEAEAE 174
L IKG++AYLNFP+LA LPR + SPKDIQ AA+ AA N ++ EAE
Sbjct: 144 LAIKGTTAYLNFPKLAGELPRPVTNSPKDIQAAASLAAVNWQDSVNDVSNSEVAEIVEAE 203
Query: 175 PSQAV------SSSSSSSSSSSSSHNEGSSLK--------GEDDMFLNLPDLTLDLRHS- 219
PS+AV S +S+++++ S ++E S E++ +LPDL D
Sbjct: 204 PSRAVVAQLFSSDTSTTTTTQSQEYSEASCASTSACTDKDSEEEKLFDLPDLFTDENEMM 263
Query: 220 -GDDGFYYSSSAWLVNGAQHIDSSFRLEDP 248
+D F Y SS W + GA D+ FRLE+P
Sbjct: 264 IRNDAFCYYSSTWQLCGA---DAGFRLEEP 290
>At3g60490 transcription factor - like protein
Length = 256
Score = 183 bits (464), Expect = 9e-47
Identities = 111/244 (45%), Positives = 153/244 (62%), Gaps = 19/244 (7%)
Query: 18 TTSLSSYIIASTSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGV 77
T ++SS + ++SS+S S+ + ++ + K ++ N +D KN +YRGV
Sbjct: 23 TATISSSSVVTSSSDSWSTSKRSLVQDNDSGGKRRKS--------NVSDDNKNPTSYRGV 74
Query: 78 RMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLP 137
RMR WGKWVSEIREPRKKSRIWLGT+PT +MAARAHDVAAL IKG+S +LNFPEL+ +LP
Sbjct: 75 RMRSWGKWVSEIREPRKKSRIWLGTYPTAEMAARAHDVAALAIKGNSGFLNFPELSGLLP 134
Query: 138 RAASASPKDIQVAAAKAA-ATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSS---SSSH 193
R S SPKDIQ AA KAA AT ++ + D+ +L E S+ +S++ SS+SSS SS
Sbjct: 135 RPVSCSPKDIQAAATKAAEATTWHKPVIDK-KLADELSHSELLSTAQSSTSSSFVFSSDT 193
Query: 194 NEGSSLKGE--DDMFLNLPDLTLDLRHSGDDGFYYSSS--AWLVNGAQHIDSSFRLEDPI 249
+E SS E ++ +LPDL D + +D F + W + G + D FR E+P
Sbjct: 194 SETSSTDKESNEETVFDLPDLFTDGLMNPNDAFCLCNGTFTWQLYGEE--DVGFRFEEPF 251
Query: 250 LWDS 253
W +
Sbjct: 252 NWQN 255
>At1g77200 hypothetical protein
Length = 244
Score = 170 bits (430), Expect = 8e-43
Identities = 104/216 (48%), Positives = 128/216 (59%), Gaps = 20/216 (9%)
Query: 30 SSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEI 89
S SS + EEE+ + KK+V +++P YRGVRMR WGKWVSEI
Sbjct: 8 SVKQSSPVPEEEDHHHHQQDSHRTNTKKRV---------RSDPGYRGVRMRTWGKWVSEI 58
Query: 90 REPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQV 149
REPRKKSRIWLGTF TP+MAARAHD AALTIKG+SA LNFPELA LPR AS+SP+D+Q
Sbjct: 59 REPRKKSRIWLGTFSTPEMAARAHDAAALTIKGTSAVLNFPELATYLPRPASSSPRDVQA 118
Query: 150 AAAKAAATVYNHA-----ITDQIELEAEAEPSQAVSS-SSSSSSSSSSSHNEGSSLKGED 203
AAA AAA ++ + ++D + A AE + SS S+ +SSS S S E +S E
Sbjct: 119 AAAVAAAMDFSPSSSSLVVSDPTTVIAPAETQLSSSSYSTCTSSSLSPSSEEAASTAEEL 178
Query: 204 DMFLNLPDLTLDLRHSGD-----DGFYYSSSAWLVN 234
+ LP L S D Y SS W +N
Sbjct: 179 SEIVELPSLETSYDESLSEFVYVDSAYPPSSPWYIN 214
>At4g16750 apetala2 domain TINY like protein
Length = 179
Score = 169 bits (427), Expect = 2e-42
Identities = 107/228 (46%), Positives = 130/228 (56%), Gaps = 53/228 (23%)
Query: 28 STSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVS 87
S+S S +L E+ ++K K ++K V+K P +RGVRMRQWGKWVS
Sbjct: 4 SSSHESQRNLRSPVPEKTGKSSKTKNEQKG----VSK------QPNFRGVRMRQWGKWVS 53
Query: 88 EIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDI 147
EIREPRKKSRIWLGTF TP+MAARAHDVAAL IKG SA+LNFPELA LPR ASA PKDI
Sbjct: 54 EIREPRKKSRIWLGTFSTPEMAARAHDVAALAIKGGSAHLNFPELAYHLPRPASADPKDI 113
Query: 148 QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFL 207
Q AAA AAA ++ +A PS V+SS + DD F
Sbjct: 114 QEAAAAAAA----------VDWKAPESPSSTVTSSPVA-----------------DDAFS 146
Query: 208 NLPDLTLDLR-HSGDDGFYYSSSAWLVNGAQHIDSSFRLEDPILWDSY 254
+LPDL LD+ H+ +DGF+ SF EDP ++Y
Sbjct: 147 DLPDLLLDVNDHNKNDGFW---------------DSFPYEDPFFLENY 179
>At2g35700 putative AP2 domain transcription factor
Length = 194
Score = 168 bits (426), Expect = 2e-42
Identities = 107/218 (49%), Positives = 130/218 (59%), Gaps = 36/218 (16%)
Query: 40 EEEEEEATNTKCKEKKKKKVGMVNKVNDRK---NNPTYRGVRMRQWGKWVSEIREPRKKS 96
++ + T + K K KKK +D K +P +RGVRMRQWGKWVSEIREP+KKS
Sbjct: 10 QDSPAQTTERRVKYKPKKK--RAKDDDDEKVVSKHPNFRGVRMRQWGKWVSEIREPKKKS 67
Query: 97 RIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAA 156
RIWLGTF T +MAARAHDVAAL IKG SA+LNFPELA LPR ASA PKDIQ AAA AAA
Sbjct: 68 RIWLGTFSTAEMAARAHDVAALAIKGGSAHLNFPELAYHLPRPASADPKDIQAAAAAAAA 127
Query: 157 TVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFLNLPDLTLDL 216
V AI +E + + PS V+ +SS + + S DD F +LPDL L++
Sbjct: 128 AV---AIDMDVETSSPS-PSPTVTETSSPAMIALS-----------DDAFSDLPDLLLNV 172
Query: 217 RHSGDDGFYYSSSAWLVNGAQHIDSSFRLEDPILWDSY 254
H+ DGF+ SF E+P L SY
Sbjct: 173 NHN-IDGFW---------------DSFPYEEPFLSQSY 194
>At5g25810 transcription factor TINY
Length = 218
Score = 159 bits (402), Expect = 1e-39
Identities = 101/192 (52%), Positives = 118/192 (60%), Gaps = 39/192 (20%)
Query: 28 STSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVS 87
ST S +S++ +E EEE KKK V D +P YRGVR R WGKWVS
Sbjct: 6 STKSWEASAVRQENEEE-----------KKK-----PVKDSGKHPVYRGVRKRNWGKWVS 49
Query: 88 EIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDI 147
EIREPRKKSRIWLGTFP+P+MAARAHDVAAL+IKG+SA LNFP+LA PR +S SP+DI
Sbjct: 50 EIREPRKKSRIWLGTFPSPEMAARAHDVAALSIKGASAILNFPDLAGSFPRPSSLSPRDI 109
Query: 148 QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSS---SSSSSSSSHNE--GSSLKGE 202
QVAA KA A E SQ+ SSSSS SSS SSSS SS G
Sbjct: 110 QVAALKA----------------AHMETSQSFSSSSSLTFSSSQSSSSLESLVSSSATGS 153
Query: 203 DDM--FLNLPDL 212
+++ + LP L
Sbjct: 154 EELGEIVELPSL 165
>At5g11590 transcription factor like protein
Length = 236
Score = 157 bits (398), Expect = 4e-39
Identities = 88/165 (53%), Positives = 106/165 (63%), Gaps = 18/165 (10%)
Query: 28 STSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVS 87
S S + L E ++ + K K ++ +K + D +P YRGVRMR WGKWVS
Sbjct: 7 SLRSERVTQLLVPNSESDSVSDKSKAEQSEK--KTKRGRDSGKHPVYRGVRMRNWGKWVS 64
Query: 88 EIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELAAVLPRAASASPKDI 147
EIREPRKKSRIWLGTFPTP+MAARAHDVAAL+IKG++A LNFPELA PR S SP+DI
Sbjct: 65 EIREPRKKSRIWLGTFPTPEMAARAHDVAALSIKGTAAILNFPELADSFPRPVSLSPRDI 124
Query: 148 QVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSS 192
Q AA KA A EP+ + SSS+SSSSS SS+
Sbjct: 125 QTAALKA----------------AHMEPTTSFSSSTSSSSSLSST 153
>At4g32800 transcription factor TINY homolog
Length = 221
Score = 152 bits (384), Expect = 2e-37
Identities = 85/164 (51%), Positives = 106/164 (63%), Gaps = 10/164 (6%)
Query: 68 RKNN--PTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSA 125
++NN P YRGVRMR WGKWVSEIREPRKKSRIWLGTFPT +MA RAHDVAA++IKG+SA
Sbjct: 11 KENNKQPVYRGVRMRSWGKWVSEIREPRKKSRIWLGTFPTAEMAMRAHDVAAMSIKGTSA 70
Query: 126 YLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSS-- 183
LNFPEL+ +LPR S SP+D++ AA KAA ++ +E S +S SS
Sbjct: 71 ILNFPELSKLLPRPVSLSPRDVRAAATKAALMDFDTTAFRSDTETSETTTSNKMSESSES 130
Query: 184 ------SSSSSSSSSHNEGSSLKGEDDMFLNLPDLTLDLRHSGD 221
SSSS SS + E S++ + D + LP L L S +
Sbjct: 131 NETVSFSSSSWSSVTSIEESTVSDDLDEIVKLPSLGTSLNESNE 174
>At3g16280 AP2 domain transcription factor like protein
Length = 181
Score = 151 bits (381), Expect = 4e-37
Identities = 79/120 (65%), Positives = 90/120 (74%), Gaps = 5/120 (4%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVRMR WGKWVSEIR+PRKK+RIWLGTF T DMAARAHDVAALTIKGSSA LNFP
Sbjct: 3 HPVYRGVRMRSWGKWVSEIRQPRKKTRIWLGTFVTADMAARAHDVAALTIKGSSAVLNFP 62
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSS 190
ELA++ PR AS+SP DIQ AAA+AAA V + LE + P SS SS ++ S
Sbjct: 63 ELASLFPRPASSSPHDIQTAAAEAAAMVVEEKL-----LEKDEAPEAPPSSESSYVAAES 117
>At1g01250 transcription factor TINY like protein
Length = 192
Score = 127 bits (320), Expect = 4e-30
Identities = 60/88 (68%), Positives = 71/88 (80%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P Y GVR R+WGKWVSEIREPRKKSRIWLG+FP P+MAA+A+DVAA +KG A LNFP
Sbjct: 42 HPVYHGVRKRRWGKWVSEIREPRKKSRIWLGSFPVPEMAAKAYDVAAFCLKGRKAQLNFP 101
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATV 158
E LPR ++ +P+DIQVAAAKAA V
Sbjct: 102 EEIEDLPRPSTCTPRDIQVAAAKAANAV 129
>At5g51990 AP2 domain transcription factor-like protein
Length = 224
Score = 119 bits (298), Expect = 2e-27
Identities = 68/156 (43%), Positives = 88/156 (55%), Gaps = 5/156 (3%)
Query: 56 KKKVGMVNKVNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDV 115
KK+ G K +P YRGVR R GKWV E+REP KKSRIWLGTFPT +MAARAHDV
Sbjct: 38 KKRAG--RKKFRETRHPIYRGVRQRNSGKWVCEVREPNKKSRIWLGTFPTVEMAARAHDV 95
Query: 116 AALTIKGSSAYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEP 175
AAL ++G SA LNF + A L + PK+IQ AA++AA N T+ + AEAE
Sbjct: 96 AALALRGRSACLNFADSAWRLRIPETTCPKEIQKAASEAAMAFQNETTTEGSKTAAEAEE 155
Query: 176 SQAVSSSSSSSSSSSSSHNEGSSLKGEDDMFLNLPD 211
+ + + G +D+ L +P+
Sbjct: 156 A---AGEGVREGERRAEEQNGGVFYMDDEALLGMPN 188
>At2g36450 putative AP2 domain transcription factor
Length = 184
Score = 117 bits (294), Expect = 5e-27
Identities = 76/185 (41%), Positives = 106/185 (57%), Gaps = 17/185 (9%)
Query: 71 NPTYRGVRMRQ-WGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNF 129
+P YRGVR R+ KWVSEIREPRK +RIWLGTF TP+MAA A+DVAAL +KGS A LNF
Sbjct: 12 HPLYRGVRQRKNSNKWVSEIREPRKPNRIWLGTFSTPEMAAIAYDVAALALKGSQAELNF 71
Query: 130 PELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSS 189
P + LP S SP DIQ AAA AAA A D I + A ++S +S +
Sbjct: 72 PNSVSSLPAPTSMSPADIQAAAASAAAAF--GAARDAIVM--------ANNNSQTSGVAC 121
Query: 190 SSSHNEGSSLKG--EDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQHIDSSFRLED 247
+S + +++ G ++D+ ++P++ +++ +G S V A + F D
Sbjct: 122 MNSSYDNTNMNGFMDEDLVFDMPNVLMNMA----EGMLLSPPRPTVFDAAYDADGFPGGD 177
Query: 248 PILWD 252
LW+
Sbjct: 178 DYLWN 182
>At1g12630 hypothetical protein
Length = 188
Score = 111 bits (278), Expect = 3e-25
Identities = 74/194 (38%), Positives = 102/194 (52%), Gaps = 20/194 (10%)
Query: 65 VNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSS 124
+ R+ +P YRG+R R GKWVSEIREPRK +RIWLGT+P +MAA A+DVAA+ +KG
Sbjct: 6 LGSRRKDPVYRGIRCRS-GKWVSEIREPRKTTRIWLGTYPMAEMAAAAYDVAAMALKGRE 64
Query: 125 AYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSS 184
A LNFP P S S DI+ AAA AA A+ E E E + + SSSS
Sbjct: 65 AVLNFPGSVGSYPVPESTSAADIRAAAAAAA------AMKGCEEGEEEKKAKEKKSSSSK 118
Query: 185 SSSSSSSSHNE-------GSSLKGEDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQ 237
S + N+ G+ E+++ LN+P+L ++ +G + +W+
Sbjct: 119 SRARECHVDNDVGSSSWCGTEFMDEEEV-LNMPNLLANMA----EGMMVAPPSWM-GSRP 172
Query: 238 HIDSSFRLEDPILW 251
DS D LW
Sbjct: 173 SDDSPENSNDEDLW 186
>At1g77640 hypothetical protein
Length = 244
Score = 110 bits (276), Expect = 6e-25
Identities = 85/251 (33%), Positives = 117/251 (45%), Gaps = 55/251 (21%)
Query: 8 LIHNSLCPQPTTSLSSYIIASTSSNSSSSLEEEEEEEEATNTKCKEKKKKKVGMVNKVND 67
++ L Q TTS SS +S+SS+SS+S + CK K KK
Sbjct: 1 MVKQELKIQVTTSSSSLSHSSSSSSSSTSALRHQS--------CKNKIKK---------- 42
Query: 68 RKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYL 127
Y+GVRMR WG WV+EIR P +K+RIWLG++ T + AARA+D A L +KG A L
Sbjct: 43 ------YKGVRMRSWGSWVTEIRAPNQKTRIWLGSYSTAEAAARAYDAALLCLKGPKANL 96
Query: 128 NFPELAAVLP------RAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSS 181
NFP + P SPK IQ AA+AA + +H E + + +
Sbjct: 97 NFPNITTTSPFLMNIDEKTLLSPKSIQKVAAQAANSSSDHFTPPSDENDHDHDDGLDHHP 156
Query: 182 SSSSSSSSSSSHNEGSSLKGEDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQHIDS 241
S+SSS++SS PD D H+ DDG S V+ +H+
Sbjct: 157 SASSSAASSP------------------PD---DDHHNDDDGDLVSLMESFVDYNEHVS- 194
Query: 242 SFRLEDPILWD 252
L DP L++
Sbjct: 195 ---LMDPSLYE 202
>At1g71450 putative TINY
Length = 183
Score = 110 bits (275), Expect = 7e-25
Identities = 73/188 (38%), Positives = 99/188 (51%), Gaps = 21/188 (11%)
Query: 66 NDRKNNPT-YRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSS 124
ND K P+ YRGVR R+WGKWVSEIREP K+RIWLG+F TP+MAA A+DVAA +G
Sbjct: 15 NDGKGVPSAYRGVRKRKWGKWVSEIREPGTKNRIWLGSFETPEMAATAYDVAAFHFRGRE 74
Query: 125 AYLNFPELAAVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSS 184
A LNFPELA+ LPR A +S I++A +A E + V SSSS
Sbjct: 75 ARLNFPELASSLPRPADSSSDSIRMAVHEATL----------CRTTEGTESAMQVDSSSS 124
Query: 185 SSSSSSSSHNEGSSLKGEDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQHIDSSFR 244
S+ + + ++ ++ L P + HS D +++ + N + S F
Sbjct: 125 SNVAPTMVRLSPREIQAINESTLGSP---TTMMHSTYDPMEFANDVEM-NAWETYQSDF- 179
Query: 245 LEDPILWD 252
LWD
Sbjct: 180 -----LWD 182
>At1g63040 transcription factor DREB1A, putative
Length = 248
Score = 110 bits (275), Expect = 7e-25
Identities = 54/93 (58%), Positives = 67/93 (71%), Gaps = 1/93 (1%)
Query: 65 VNDRKNNPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSS 124
++ RK NP YRG+R R GKWVSEIREP+K +R+WLGT+PTP+MAA A+DVAAL +KG
Sbjct: 81 ISRRKKNPVYRGIRCRS-GKWVSEIREPKKTTRVWLGTYPTPEMAAAAYDVAALALKGGD 139
Query: 125 AYLNFPELAAVLPRAASASPKDIQVAAAKAAAT 157
LNFP+ P S+S I+ AAA AAAT
Sbjct: 140 TLLNFPDSLGSYPIPLSSSAAHIRCAAAAAAAT 172
>At4g25490 transcriptional activator CBF1/ CRT/CRE binding factor 1
Length = 213
Score = 108 bits (270), Expect = 3e-24
Identities = 62/118 (52%), Positives = 75/118 (63%), Gaps = 10/118 (8%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWVSE+REP KK+RIWLGTF T +MAARAHDVAAL ++G SA LNF
Sbjct: 45 HPIYRGVRQRNSGKWVSEVREPNKKTRIWLGTFQTAEMAARAHDVAALALRGRSACLNFA 104
Query: 131 ELAAVLPRAASASPKDIQVAAAKAA---------ATVYNHAI-TDQIELEAEAEPSQA 178
+ A L S KDIQ AAA+AA T NH + ++ +EA P Q+
Sbjct: 105 DSAWRLRIPESTCAKDIQKAAAEAALAFQDETCDTTTTNHGLDMEETMVEAIYTPEQS 162
>At5g52020 unknown protein
Length = 232
Score = 106 bits (264), Expect = 1e-23
Identities = 70/178 (39%), Positives = 92/178 (51%), Gaps = 13/178 (7%)
Query: 74 YRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFPELA 133
+RG+R+R GKWVSEIREPRK +RIWLGT+P P+MAA A+DVAAL +KG A LNFP LA
Sbjct: 65 FRGIRLRN-GKWVSEIREPRKTTRIWLGTYPVPEMAAAAYDVAALALKGPDAVLNFPGLA 123
Query: 134 AVLPRAASASPKDIQVAAAKAAATVYNHAITDQIELEAEAEPSQAVSSSSSSSSSSSSSH 193
S S DI+ AA++AA DQ E EP Q S +S
Sbjct: 124 LTYVAPVSNSAADIRAAASRAAEMKQ----PDQGGDEKVLEPVQPGKEEELEEVSCNSCS 179
Query: 194 NEGSSLKGEDDMFLNLPDLTLDLRHSGDDGFYYSSSAWLVNGAQHIDSSFRLEDPILW 251
E +++ LN+P L ++ +G S +++ DS E LW
Sbjct: 180 LEFM----DEEAMLNMPTLLTEMA----EGMLMSPPRMMIHPTMEDDSPENHEGDNLW 229
>At4g25470 DRE/CRT-binding protein DREB1C
Length = 216
Score = 106 bits (264), Expect = 1e-23
Identities = 59/121 (48%), Positives = 75/121 (61%), Gaps = 3/121 (2%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWV E+REP KK+RIWLGTF T +MAARAHDVAA+ ++G SA LNF
Sbjct: 48 HPIYRGVRQRNSGKWVCELREPNKKTRIWLGTFQTAEMAARAHDVAAIALRGRSACLNFA 107
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYN---HAITDQIELEAEAEPSQAVSSSSSSSS 187
+ A L S K+IQ AAA+AA + H TD L+ E +A+ + S
Sbjct: 108 DSAWRLRIPESTCAKEIQKAAAEAALNFQDEMCHMTTDAHGLDMEETLVEAIYTPEQSQD 167
Query: 188 S 188
+
Sbjct: 168 A 168
>At4g25481 DREB1A
Length = 216
Score = 105 bits (261), Expect = 3e-23
Identities = 71/162 (43%), Positives = 91/162 (55%), Gaps = 11/162 (6%)
Query: 71 NPTYRGVRMRQWGKWVSEIREPRKKSRIWLGTFPTPDMAARAHDVAALTIKGSSAYLNFP 130
+P YRGVR R GKWV E+REP KK+RIWLGTF T +MAARAHDVAAL ++G SA LNF
Sbjct: 48 HPIYRGVRRRNSGKWVCEVREPNKKTRIWLGTFQTAEMAARAHDVAALALRGRSACLNFA 107
Query: 131 ELAAVLPRAASASPKDIQVAAAKAAATVYNH---AITDQIELEAEAEPSQAVSSSSSSSS 187
+ A L S KDIQ AAA+AA + A TD + E +A+ ++ S +
Sbjct: 108 DSAWRLRIPESTCAKDIQKAAAEAALAFQDEMCDATTDH-GFDMEETLVEAIYTAEQSEN 166
Query: 188 S---SSSSHNEGSSLKGE--DDMFLNLPDLTLDLRH--SGDD 222
+ + E SL + M L LP + + H GDD
Sbjct: 167 AFYMHDEAMFEMPSLLANMAEGMLLPLPSVQWNHNHEVDGDD 208
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.308 0.122 0.343
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,191,849
Number of Sequences: 26719
Number of extensions: 280759
Number of successful extensions: 5280
Number of sequences better than 10.0: 302
Number of HSP's better than 10.0 without gapping: 250
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 3929
Number of HSP's gapped (non-prelim): 800
length of query: 257
length of database: 11,318,596
effective HSP length: 97
effective length of query: 160
effective length of database: 8,726,853
effective search space: 1396296480
effective search space used: 1396296480
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 59 (27.3 bits)
Medicago: description of AC149079.10


