

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.5 + phase: 0
(196 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
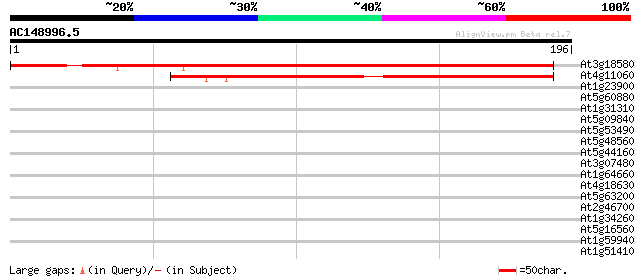
Score E
Sequences producing significant alignments: (bits) Value
At3g18580 hypothetical protein 215 1e-56
At4g11060 unknown protein 99 1e-21
At1g23900 gamma-adaptin, putative 33 0.13
At5g60880 putative protein 31 0.37
At1g31310 unknown protein 30 0.63
At5g09840 putative protein 29 1.4
At5g53490 thylakoid lumenal 17.4 kD protein, chloroplast precurs... 29 1.8
At5g48560 putative bHLH transcription factor (bHLH078) 28 2.4
At5g44160 unknown protein 28 3.1
At3g07480 Unknown protein (F21O3.19) 28 3.1
At1g64660 similar to O-succinylhomoserine sulfhydrylase 28 4.1
At4g18630 unknown protein 27 5.3
At5g63200 unknown protein 27 6.9
At2g46700 calcium/calmodulin-dependent protein kinase CaMK4 27 6.9
At1g34260 hypothetical protein 27 6.9
At5g16560 KANADI protein (KAN) 27 9.1
At1g59940 putative protein 27 9.1
At1g51410 cinnamyl alcohol dehydrogenase, putative 27 9.1
>At3g18580 hypothetical protein
Length = 217
Score = 215 bits (547), Expect = 1e-56
Identities = 112/199 (56%), Positives = 142/199 (71%), Gaps = 14/199 (7%)
Query: 1 MAKFSISRTSTLYRSLLSTPPPPLHHHRHIPLRFCT---TTTSLSDYDDADSAAPSPSPE 57
MA + + LYRSLLS P + FCT T+ SD+D+ +S + +
Sbjct: 1 MANSMATLSRRLYRSLLSNP-----RISQASMSFCTNNITSPEDSDFDELESPIEPKASD 55
Query: 58 QT------ERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIR 111
ER +RPLENGLD G+++AILVG+ GQ PLQKKLKSG VTL S+GTGGIR
Sbjct: 56 PVSRFSGEERVMEERPLENGLDSGIFKAILVGQVGQLPLQKKLKSGRTVTLFSVGTGGIR 115
Query: 112 NNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDP 171
NNRRPL +E+PREYA+R AVQWHRV+VYPERL +L++K V PG+ +Y+EGNLETK+F+DP
Sbjct: 116 NNRRPLINEDPREYASRSAVQWHRVSVYPERLADLVLKNVEPGTVIYLEGNLETKIFTDP 175
Query: 172 VTGLVRRVREVAVRRNGNL 190
VTGLVRR+REVA+RRNG +
Sbjct: 176 VTGLVRRIREVAIRRNGRV 194
>At4g11060 unknown protein
Length = 201
Score = 99.0 bits (245), Expect = 1e-21
Identities = 53/143 (37%), Positives = 86/143 (60%), Gaps = 15/143 (10%)
Query: 57 EQTERTFFDRP--LENGLDP-------GVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGT 107
E E +RP +G+DP GV+RAI+ GK GQ PLQK L++G VT+ ++GT
Sbjct: 43 EDFEENVTERPELQPHGVDPRKGWGFRGVHRAIICGKVGQAPLQKILRNGRTVTIFTVGT 102
Query: 108 GGIRNNRRPLDHENPREYANRCAVQWHRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKV 167
GG+ + R P+ QWHR+ V+ E LG+ ++ + S++YVEG++ET+V
Sbjct: 103 GGMFDQRLVGATNQPKP------AQWHRIAVHNEVLGSYAVQKLAKNSSVYVEGDIETRV 156
Query: 168 FSDPVTGLVRRVREVAVRRNGNL 190
++D ++ V+ + E+ VRR+G +
Sbjct: 157 YNDSISSEVKSIPEICVRRDGKI 179
>At1g23900 gamma-adaptin, putative
Length = 876
Score = 32.7 bits (73), Expect = 0.13
Identities = 36/138 (26%), Positives = 50/138 (36%), Gaps = 38/138 (27%)
Query: 6 ISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQT------ 59
++ ++ LS+P PP H+ TT+S +D D +PSPS E T
Sbjct: 710 VNNNPSIALDTLSSPAPP-----HV-----ATTSSTGMFDLLDGLSPSPSKEATNGPAYA 759
Query: 60 ------------ERTFFDRP----------LENGLDPGVYRAILVGKAGQKPLQKKLKSG 97
E TF P L P + + A K LQ L
Sbjct: 760 PIVAYESSSLKIEFTFSKTPGNLQTTNVQATFTNLSPNTFTDFIFQAAVPKFLQLHLDPA 819
Query: 98 TVVTLLSIGTGGIRNNRR 115
+ TLL+ G+G I N R
Sbjct: 820 SSNTLLASGSGAITQNLR 837
>At5g60880 putative protein
Length = 262
Score = 31.2 bits (69), Expect = 0.37
Identities = 32/109 (29%), Positives = 41/109 (37%), Gaps = 12/109 (11%)
Query: 14 RSLLSTPPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTF-FDRPLENGL 72
R L +PP HHH H+ + TTS S + P S E F FDR
Sbjct: 67 RKLSLSPPGTRHHHLHLRSSSVSPTTSGSQHRRLSWPQPPVSEESGFIVFCFDR------ 120
Query: 73 DPGVYRAILVGKAGQKPLQKKL-KSGTVVTLLSI----GTGGIRNNRRP 116
+ G + + GK +K + KS V I G GG N P
Sbjct: 121 EDGGFDVVKEGKQEKKETESSSEKSPRTVNRKLIYGDQGVGGTEKNNSP 169
>At1g31310 unknown protein
Length = 383
Score = 30.4 bits (67), Expect = 0.63
Identities = 33/129 (25%), Positives = 49/129 (37%), Gaps = 16/129 (12%)
Query: 12 LYRSLLSTPPPPLHHHRH--IPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLE 69
L R LL PPPP H +P + +T + S+Y D +P+ T P
Sbjct: 232 LPRPLLLPPPPPPSFHAQPILPTKDSSTDSDTSEYSD---TSPAKRRRTMPTTTTAGPSG 288
Query: 70 NGLDPGVYRAILVGKAGQKPLQKKLKSGTVVTLLSIGTGGIRNNRRPLDHENPREYANRC 129
G+D V + G+ K+ + TV LS I N R + R +
Sbjct: 289 GGVD--------VEEVGR---SKRDEETTVAAALSRSVSVIANAIRESEERQDRRHKEVM 337
Query: 130 AVQWHRVTV 138
VQ R+ +
Sbjct: 338 NVQERRLKI 346
>At5g09840 putative protein
Length = 924
Score = 29.3 bits (64), Expect = 1.4
Identities = 23/66 (34%), Positives = 31/66 (46%), Gaps = 10/66 (15%)
Query: 5 SISRTSTLYRSLLSTPPPPLHHHRHIPL--------RFCTTTTSLSDYDDADSAAPSPSP 56
S+ S L+R LS+ H R I L F TTTTS S++ S +PS P
Sbjct: 5 SLPSKSLLFR--LSSSSTTQSHRRTITLFITNIHSSPFSTTTTSGSNFVSGSSHSPSRRP 62
Query: 57 EQTERT 62
+Q E +
Sbjct: 63 QQDEES 68
>At5g53490 thylakoid lumenal 17.4 kD protein, chloroplast precursor
(P17.4) (sp|P81760)
Length = 236
Score = 28.9 bits (63), Expect = 1.8
Identities = 19/50 (38%), Positives = 25/50 (50%), Gaps = 4/50 (8%)
Query: 144 GNLLMKTVLPGSTLYVEGNLETKVFSDPVTG---LVRRVREVAVRRNGNL 190
G + TVL GST + E NLE VF D + G L + R ++ G L
Sbjct: 183 GAVFRNTVLSGST-FEEANLEDVVFEDTIIGYIDLQKICRNESINEEGRL 231
>At5g48560 putative bHLH transcription factor (bHLH078)
Length = 498
Score = 28.5 bits (62), Expect = 2.4
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 4/70 (5%)
Query: 6 ISRTSTLYRSLLSTPPPPLHHHRHIPLRFCTTTTSLS-DYDDADSAAPSP---SPEQTER 61
+SR+++ Y + +S+PPPP + + + + T T S D A+ AA S R
Sbjct: 116 MSRSASCYATPMSSPPPPTNSNSQMMMNRTTPLTEFSADPGFAERAARFSCFGSRSFNGR 175
Query: 62 TFFDRPLENG 71
T + P+ NG
Sbjct: 176 TNTNLPINNG 185
>At5g44160 unknown protein
Length = 466
Score = 28.1 bits (61), Expect = 3.1
Identities = 19/57 (33%), Positives = 26/57 (45%), Gaps = 15/57 (26%)
Query: 13 YRSLLSTPPPPL------------HHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPE 57
Y+ L+ T PPL HHH+H T++SLS + D A P P P+
Sbjct: 222 YQYLMGTFIPPLQPFVPQPQTNPNHHHQHFQP---PTSSSLSLWMGQDIAPPQPQPD 275
>At3g07480 Unknown protein (F21O3.19)
Length = 159
Score = 28.1 bits (61), Expect = 3.1
Identities = 15/52 (28%), Positives = 28/52 (53%), Gaps = 1/52 (1%)
Query: 49 SAAPSPSPEQTERTFFDRPLE-NGLDPGVYRAILVGKAGQKPLQKKLKSGTV 99
SA +PSP + DR ++ + +DP Y+ ++G +GQ L+ +G +
Sbjct: 27 SATSAPSPSLGSKKVSDRIVKLSAIDPDGYKQDIIGLSGQTLLRALTHTGLI 78
>At1g64660 similar to O-succinylhomoserine sulfhydrylase
Length = 441
Score = 27.7 bits (60), Expect = 4.1
Identities = 22/69 (31%), Positives = 32/69 (45%), Gaps = 10/69 (14%)
Query: 134 HRVTVYPERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVT------GLVRRVREVAVRRN 187
HR VY ER+ +L MK + PG +E + + K+F V GL+ E + N
Sbjct: 310 HRAQVYAERMRDLGMKVIYPG----LETHPQHKLFKGMVNRDYGYGGLLSIDMETEEKAN 365
Query: 188 GNLANSSNA 196
+A NA
Sbjct: 366 KLMAYLQNA 374
>At4g18630 unknown protein
Length = 479
Score = 27.3 bits (59), Expect = 5.3
Identities = 18/57 (31%), Positives = 30/57 (52%), Gaps = 3/57 (5%)
Query: 42 SDYDDADSAAPSPSPEQTERTFFDRPLENGLDPGVYRAILVGKAGQKPLQKKLKSGT 98
S+Y A S P P PE R++ R L ++P Y +++G+ G K L +++ T
Sbjct: 394 SEYTPALS--PPPLPETPSRSWLGRTLLPPVNPRPY-GVVLGQVGIKKLNQEVLEST 447
>At5g63200 unknown protein
Length = 404
Score = 26.9 bits (58), Expect = 6.9
Identities = 16/61 (26%), Positives = 28/61 (45%), Gaps = 7/61 (11%)
Query: 45 DDADSAAPSPSPEQTERTFFDRPLENG-------LDPGVYRAILVGKAGQKPLQKKLKSG 97
DD DS+ P P+P R + + G ++PG A + K G+ + K++S
Sbjct: 39 DDLDSSIPIPTPPPITRLSNEESHQEGGILTCKEVEPGEVEAKKISKVGKCRSRSKIESS 98
Query: 98 T 98
+
Sbjct: 99 S 99
>At2g46700 calcium/calmodulin-dependent protein kinase CaMK4
Length = 595
Score = 26.9 bits (58), Expect = 6.9
Identities = 14/39 (35%), Positives = 19/39 (47%)
Query: 51 APSPSPEQTERTFFDRPLENGLDPGVYRAILVGKAGQKP 89
+PSP+ T R FF RP +A L+ + G KP
Sbjct: 74 SPSPARTSTPRRFFRRPFPPPSPAKHIKASLIKRLGVKP 112
>At1g34260 hypothetical protein
Length = 1456
Score = 26.9 bits (58), Expect = 6.9
Identities = 10/28 (35%), Positives = 17/28 (60%)
Query: 46 DADSAAPSPSPEQTERTFFDRPLENGLD 73
+ ++ +PSPSP ++ D P+ NG D
Sbjct: 446 EIENFSPSPSPRESPSEAVDIPVSNGFD 473
>At5g16560 KANADI protein (KAN)
Length = 403
Score = 26.6 bits (57), Expect = 9.1
Identities = 14/28 (50%), Positives = 16/28 (57%), Gaps = 3/28 (10%)
Query: 5 SISRTSTLYRSLLSTPP---PPLHHHRH 29
S S + YRSL S+P PLHHH H
Sbjct: 150 SSSGYNNAYRSLQSSPRLKGVPLHHHHH 177
>At1g59940 putative protein
Length = 231
Score = 26.6 bits (57), Expect = 9.1
Identities = 16/51 (31%), Positives = 23/51 (44%), Gaps = 3/51 (5%)
Query: 20 PPPPLHHHRHIPLRFCTTTTSLSDYDDADSAAPSPSPEQTERTFFDRPLEN 70
PPPPL + T + LS DD DS SP ++ + D P+ +
Sbjct: 180 PPPPLSATSSMESSDSTVESPLSMVDDEDSLTMSP---ESATSLVDSPMRS 227
>At1g51410 cinnamyl alcohol dehydrogenase, putative
Length = 809
Score = 26.6 bits (57), Expect = 9.1
Identities = 14/42 (33%), Positives = 22/42 (52%)
Query: 141 ERLGNLLMKTVLPGSTLYVEGNLETKVFSDPVTGLVRRVREV 182
ER + T+L G EG E+KV D ++ LV ++ E+
Sbjct: 156 ERTLRSAVLTLLSGKERISEGTKESKVLEDEISYLVEKLNEL 197
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,734,018
Number of Sequences: 26719
Number of extensions: 215979
Number of successful extensions: 789
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 773
Number of HSP's gapped (non-prelim): 19
length of query: 196
length of database: 11,318,596
effective HSP length: 94
effective length of query: 102
effective length of database: 8,807,010
effective search space: 898315020
effective search space used: 898315020
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148996.5


