

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148996.1 - phase: 0
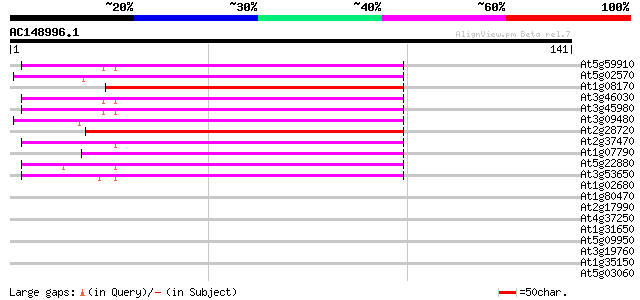
(141 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g59910 histone H2B - like protein 55 1e-08
At5g02570 putative protein 54 2e-08
At1g08170 hypothetical protein 54 2e-08
At3g46030 histone H2B -like protein 54 4e-08
At3g45980 histone H2B 54 4e-08
At3g09480 putative histone H2B 53 5e-08
At2g28720 putative histone H2B 53 6e-08
At2g37470 putative histone H2B 52 1e-07
At1g07790 histone H2B 51 2e-07
At5g22880 histone H2B like protein (emb|CAA69025.1) 50 5e-07
At3g53650 histone H2B - like protein 50 5e-07
At1g02680 transcription factor TFIID, putative 32 0.11
At1g80470 hypothetical protein 28 1.2
At2g17990 unknown protein 28 1.6
At4g37250 receptor kinase like protein 28 2.1
At1g31650 unknown protein 28 2.1
At5g09950 selenium-binding protein-like 27 2.7
At3g19760 RNA helicase, putative 27 2.7
At1g35150 hypothetical protein 27 2.7
At5g03060 putative protein 27 3.6
>At5g59910 histone H2B - like protein
Length = 150
Score = 55.1 bits (131), Expect = 1e-08
Identities = 30/101 (29%), Positives = 58/101 (56%), Gaps = 5/101 (4%)
Query: 4 KKGKNIILEVGSSSGTKRR----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDM 58
K GK + E G+ K++ +VE +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 36 KAGKKLPKEAGAGGDKKKKMKKKSVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDI 95
Query: 59 LHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ +E +K + + R++ V+ + P LA+ A
Sbjct: 96 FEKLAQEASKLARYNKKPTITSREIQTAVRLVLPGELAKHA 136
>At5g02570 putative protein
Length = 132
Score = 54.3 bits (129), Expect = 2e-08
Identities = 31/104 (29%), Positives = 57/104 (54%), Gaps = 6/104 (5%)
Query: 2 ASKKGKNIILEVGSSS------GTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIM 55
A K K I E G+S TK+ T +K +I++V K+VHPD+ I+ +++ +MN +
Sbjct: 15 APKAEKKIAKEGGTSEIVKKKKKTKKSTETYKIYIFKVLKQVHPDIGISGKAMGIMNSFI 74
Query: 56 NDMLHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
ND+ K+ +E ++ + + R++ V+ + P LA+ A
Sbjct: 75 NDIFEKLAQESSRLARYNKKPTITSREIQTAVRLVLPGELAKHA 118
>At1g08170 hypothetical protein
Length = 243
Score = 54.3 bits (129), Expect = 2e-08
Identities = 25/75 (33%), Positives = 47/75 (62%)
Query: 25 EFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLVERDLW 84
E+++++Y+V K+VHPDL IT +++ ++N M DM +I +E A+ ++ L R++
Sbjct: 150 EYRRYVYKVMKQVHPDLGITSKAMTVVNMFMGDMFERIAQEAARLSDYTKRRTLSSREIE 209
Query: 85 VGVKELYPKYLARCA 99
V+ + P L+R A
Sbjct: 210 AAVRLVLPGELSRHA 224
>At3g46030 histone H2B -like protein
Length = 145
Score = 53.5 bits (127), Expect = 4e-08
Identities = 30/101 (29%), Positives = 57/101 (55%), Gaps = 5/101 (4%)
Query: 4 KKGKNIILEVGSSSGTKRR----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDM 58
K GK + E G+ K++ +VE +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 31 KAGKKLPKEAGAGGDKKKKMKKKSVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDI 90
Query: 59 LHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ E +K + + R++ V+ + P LA+ A
Sbjct: 91 FEKLASESSKLARYNKKPTITSREIQTAVRLVLPGELAKHA 131
>At3g45980 histone H2B
Length = 150
Score = 53.5 bits (127), Expect = 4e-08
Identities = 30/101 (29%), Positives = 57/101 (55%), Gaps = 5/101 (4%)
Query: 4 KKGKNIILEVGSSSGTKRR----TVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDM 58
K GK + E G+ K++ +VE +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 36 KAGKKLPKEAGAGGDKKKKMKKKSVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDI 95
Query: 59 LHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ E +K + + R++ V+ + P LA+ A
Sbjct: 96 FEKLASESSKLARYNKKPTITSREIQTAVRLVLPGELAKHA 136
>At3g09480 putative histone H2B
Length = 126
Score = 53.1 bits (126), Expect = 5e-08
Identities = 31/100 (31%), Positives = 54/100 (54%), Gaps = 2/100 (2%)
Query: 2 ASKKGKNIILEVGSS--SGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDML 59
A K K I E GS TK+ T +K ++++V K+VHPD+ I+ +++ +MN +ND
Sbjct: 13 APKADKKITKEGGSERKKKTKKSTETYKIYLFKVLKQVHPDIGISGKAMGIMNSFINDTF 72
Query: 60 HKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
KI E ++ + + R++ V+ + P LA+ A
Sbjct: 73 EKIALESSRLARYNKKPTITSREIQTAVRLVLPGELAKHA 112
>At2g28720 putative histone H2B
Length = 151
Score = 52.8 bits (125), Expect = 6e-08
Identities = 24/80 (30%), Positives = 48/80 (60%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLV 79
K+ T +K +I++V K+VHPD+ I+ +++ +MN +ND+ K+ +E +K + +
Sbjct: 58 KKSTETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQEASKLARYNKKPTIT 117
Query: 80 ERDLWVGVKELYPKYLARCA 99
R++ V+ + P LA+ A
Sbjct: 118 SREIQTAVRLVLPGELAKHA 137
>At2g37470 putative histone H2B
Length = 138
Score = 52.0 bits (123), Expect = 1e-07
Identities = 28/100 (28%), Positives = 56/100 (56%), Gaps = 4/100 (4%)
Query: 4 KKGKNIILEVGSSSGTKRRTVE----FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDML 59
K K I + G S K+++ + +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 26 KAEKKISKDAGGSEKKKKKSKKSVETYKIYIFKVLKQVHPDVGISGKAMGIMNSFINDIF 85
Query: 60 HKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ +E +K + + R++ V+ + P LA+ A
Sbjct: 86 EKLAQESSKLARYNKKPTITSREIQTAVRLVLPGELAKHA 125
>At1g07790 histone H2B
Length = 148
Score = 50.8 bits (120), Expect = 2e-07
Identities = 23/81 (28%), Positives = 48/81 (58%)
Query: 19 TKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLL 78
+K+ +K +I++V K+VHPD+ I+ +++ +MN +ND+ K+ +E +K + +
Sbjct: 54 SKKNVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDIFEKLAQESSKLARYNKKPTI 113
Query: 79 VERDLWVGVKELYPKYLARCA 99
R++ V+ + P LA+ A
Sbjct: 114 TSREIQTAVRLVLPGELAKHA 134
>At5g22880 histone H2B like protein (emb|CAA69025.1)
Length = 145
Score = 49.7 bits (117), Expect = 5e-07
Identities = 29/101 (28%), Positives = 56/101 (54%), Gaps = 5/101 (4%)
Query: 4 KKGKNIILE-VGSSSGTKRRTVE----FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDM 58
K GK + E G+ K+R+ + +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 31 KAGKKLPKEPAGAGDKKKKRSKKNVETYKIYIFKVLKQVHPDIGISSKAMGIMNSFINDI 90
Query: 59 LHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ E +K + + R++ V+ + P LA+ A
Sbjct: 91 FEKLAGESSKLARYNKKPTITSREIQTAVRLVLPGELAKHA 131
>At3g53650 histone H2B - like protein
Length = 138
Score = 49.7 bits (117), Expect = 5e-07
Identities = 28/99 (28%), Positives = 56/99 (56%), Gaps = 3/99 (3%)
Query: 4 KKGKNIILEVGSSSGTKR--RTVE-FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLH 60
K K I E GS K+ + +E +K +I++V K+VHPD+ I+ +++ +MN +ND+
Sbjct: 26 KAEKKITKEGGSEKKKKKSKKNIETYKIYIFKVLKQVHPDIGISGKAMGIMNSFINDIFE 85
Query: 61 KIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCA 99
K+ +E ++ + + R++ V+ + P L++ A
Sbjct: 86 KLAQESSRLARYNKKPTITSREIQTAVRLVLPGELSKHA 124
>At1g02680 transcription factor TFIID, putative
Length = 127
Score = 32.0 bits (71), Expect = 0.11
Identities = 24/90 (26%), Positives = 47/90 (51%), Gaps = 2/90 (2%)
Query: 14 GSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAAL 73
G+S ++R F+K + + + + ES+ ++ DI+ + + +V A+ + +
Sbjct: 19 GTSQPQEKRKTLFQKELQHMMYGFGDEQNPLPESVALVEDIVVEYVTDLVTHKAQEIGSK 78
Query: 74 NQKLLVERDLWVGVKELYPKYLARCANLHA 103
+LLV+ L++ K+L PK L RC L A
Sbjct: 79 RGRLLVDDFLYLIRKDL-PK-LNRCRELLA 106
>At1g80470 hypothetical protein
Length = 464
Score = 28.5 bits (62), Expect = 1.2
Identities = 13/49 (26%), Positives = 22/49 (44%)
Query: 57 DMLHKIVEEVAKNVAALNQKLLVERDLWVGVKELYPKYLARCANLHAYF 105
+M+H VE + V + + E++P +L C NLH+ F
Sbjct: 302 EMIHDYVESETELVPQFSNLFCLHASFSESSWEMFPTFLGCCPNLHSLF 350
>At2g17990 unknown protein
Length = 338
Score = 28.1 bits (61), Expect = 1.6
Identities = 16/82 (19%), Positives = 42/82 (50%), Gaps = 3/82 (3%)
Query: 5 KGKNIILEVGSSS---GTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHK 61
KG N++ + G SS G + E +++ + + + ++SIEM+ D+++ ++ +
Sbjct: 104 KGLNLVEKDGGSSSSDGARNTNPETRRYSGSLGVEDGAYTNEMLQSIEMVTDVLDSLVRR 163
Query: 62 IVEEVAKNVAALNQKLLVERDL 83
+ +++ + LL E ++
Sbjct: 164 VTVAESESAVQKERALLGEEEI 185
>At4g37250 receptor kinase like protein
Length = 768
Score = 27.7 bits (60), Expect = 2.1
Identities = 11/32 (34%), Positives = 20/32 (62%)
Query: 10 ILEVGSSSGTKRRTVEFKKHIYRVAKKVHPDL 41
+ +G + ++RR +F+ HI + K VHP+L
Sbjct: 476 VRRLGENGLSQRRFKDFEPHIRAIGKLVHPNL 507
>At1g31650 unknown protein
Length = 548
Score = 27.7 bits (60), Expect = 2.1
Identities = 12/33 (36%), Positives = 21/33 (63%)
Query: 61 KIVEEVAKNVAALNQKLLVERDLWVGVKELYPK 93
K+V +V K A+N+ +L+E + + +KE PK
Sbjct: 281 KVVYQVFKATKAINENILLEMPVPIVIKEAIPK 313
>At5g09950 selenium-binding protein-like
Length = 995
Score = 27.3 bits (59), Expect = 2.7
Identities = 16/56 (28%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 2 ASKKGKNIILEVGSSSGTKRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMND 57
A KK K+ +V +G T++ H++ K HPD D+ + ++ +N M D
Sbjct: 847 ARKKMKDA--DVKKEAGYSWVTMKDGVHMFVAGDKSHPDADVIYKKLKELNRKMRD 900
>At3g19760 RNA helicase, putative
Length = 408
Score = 27.3 bits (59), Expect = 2.7
Identities = 20/68 (29%), Positives = 35/68 (51%), Gaps = 11/68 (16%)
Query: 26 FKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLLVERD--L 83
FK IY V + + PDL + + S + ++I+ +M K + E K+LV+RD
Sbjct: 194 FKDQIYDVYRYLPPDLQVCLVSATLPHEIL-EMTSKFMTEPV--------KILVKRDELT 244
Query: 84 WVGVKELY 91
G+K+ +
Sbjct: 245 LEGIKQFF 252
>At1g35150 hypothetical protein
Length = 459
Score = 27.3 bits (59), Expect = 2.7
Identities = 14/59 (23%), Positives = 31/59 (51%), Gaps = 5/59 (8%)
Query: 20 KRRTVEFKKHIYRVAKKVHPDLDITVESIEMMNDIMNDMLHKIVEEVAKNVAALNQKLL 78
K+RT+ KKH V++K ++D++ E D + ++++ K +A ++ L+
Sbjct: 386 KKRTIRRKKHFDEVSEKGDENMDLSPE-----EDFRINYFFTLIDQALKLTSASDESLM 439
>At5g03060 putative protein
Length = 292
Score = 26.9 bits (58), Expect = 3.6
Identities = 21/81 (25%), Positives = 41/81 (49%), Gaps = 7/81 (8%)
Query: 2 ASKKGKNIILEVGSSSGTKRRTVEFKKHI----YRVAKKVHPDLDITVESIEMMNDIMND 57
++K GKN I V + + RR + ++ I + + K+ + +L+ +SIE MN+
Sbjct: 9 SNKSGKNKISSVATPT---RRNLVVQEEILATKHEILKENYENLEKDYKSIEESFKQMNE 65
Query: 58 MLHKIVEEVAKNVAALNQKLL 78
M + + K + L +K+L
Sbjct: 66 MNEIMKFQYQKQIKELEEKIL 86
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,859,551
Number of Sequences: 26719
Number of extensions: 103981
Number of successful extensions: 396
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 376
Number of HSP's gapped (non-prelim): 27
length of query: 141
length of database: 11,318,596
effective HSP length: 89
effective length of query: 52
effective length of database: 8,940,605
effective search space: 464911460
effective search space used: 464911460
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148996.1


