

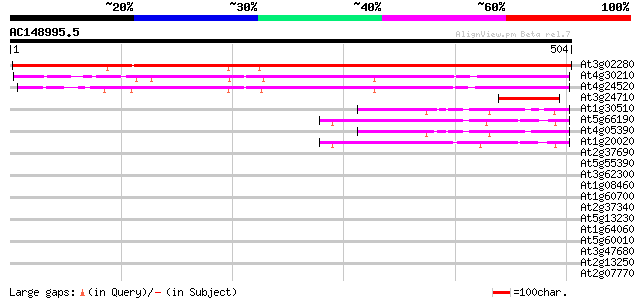
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.5 + phase: 0
(504 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g02280 NADPH-ferrihemoprotein reductase like protein 667 0.0
At4g30210 NADPH-ferrihemoprotein reductase (ATR2) 220 1e-57
At4g24520 NADPH-ferrihemoprotein reductase ATR1 213 2e-55
At3g24710 hypothetical protein 64 2e-10
At1g30510 ferrodoxin NADP oxidoreductase - like protein 61 1e-09
At5g66190 ferredoxin-NADP+ reductase 59 5e-09
At4g05390 ferredoxin-NADP+ reductase like protein 59 8e-09
At1g20020 ferredoxin--NADP reductase precursor, putative 52 6e-07
At2g37690 phosphoribosylaminoimidazole carboxylase like protein 33 0.37
At5g55390 putative protein 32 0.63
At3g62300 putative protein 32 0.82
At1g08460 unknown protein 31 1.8
At1g60700 unknown protein 30 2.4
At2g37340 unknown protein 30 4.1
At5g13230 putative protein 29 5.3
At1g64060 cytochrome b245 beta chain homolog RbohAp108 29 5.3
At5g60010 respiratory burst oxidase protein - like 29 6.9
At3g47680 unknown protein 29 6.9
At2g13250 putative Athila retroelement ORF1 protein 29 6.9
At2g07770 putative protein 29 6.9
>At3g02280 NADPH-ferrihemoprotein reductase like protein
Length = 623
Score = 667 bits (1720), Expect = 0.0
Identities = 327/515 (63%), Positives = 396/515 (76%), Gaps = 14/515 (2%)
Query: 3 FVTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPD 62
FV KKLDKRL DLG I+E+GLGDDQ PSGYEGTLDPW+ SLWR L I P+ P GPD
Sbjct: 110 FVAKKLDKRLSDLGATTIIEKGLGDDQHPSGYEGTLDPWMLSLWRTLYQINPKYFPKGPD 169
Query: 63 VLIQDTTLIDQPKVQITYHNIENV----VSHSSDTGSVRSMHPGKSSSNRNGYPDCFLKL 118
V I +ID+PK +I +H E + +S S R M PGK +++ PDCFLK+
Sbjct: 170 VKIPQDEVIDKPKYRILFHKQEKLEPKLLSDSDIIQRARGMSPGKLFKDKSK-PDCFLKM 228
Query: 119 VKNLPLTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSL 178
+N LTK S KDVRHFEF+FVS IEY+ GD++E+LP Q+S+ VDAFI RC LDP+S
Sbjct: 229 TRNEVLTKAESTKDVRHFEFQFVSSTIEYEVGDVVELLPSQNSSVVDAFIERCGLDPESF 288
Query: 179 ITVSPKGMDCNGHGSRM----PVKLRTFVELTMDVASASPRRYFFEARC----SEHERER 230
ITV P+ + + M P+KL+TFVELTMDV SASPRRYFFE +EHE+ER
Sbjct: 289 ITVGPRETENSSFSEEMITQIPIKLKTFVELTMDVTSASPRRYFFEIMSFYATAEHEKER 348
Query: 231 LEYFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLEWLIQLVPMLKKREFSISSSQS 290
L+YFASPEGRDDLY YNQKERR++LEVL+DFPSVQ+P +WL+QLVP LK R FSISSS
Sbjct: 349 LQYFASPEGRDDLYNYNQKERRSILEVLEDFPSVQIPFDWLVQLVPPLKPRAFSISSSPL 408
Query: 291 SHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVWFQKGSLPTPSPSLPL 350
+HP VHLTVS+VSW TPYKR +KGLCSSWLA+L P V++PVWF KGSLP PS SLPL
Sbjct: 409 AHPAAVHLTVSIVSWITPYKRTRKGLCSSWLASLAPEQEVNIPVWFHKGSLPAPSQSLPL 468
Query: 351 ILVGPGTGCAPFRGFIEERALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVL 410
ILVGPGTGCAPFRGFI ERA+Q+++ +AP++FFFGC N+D DFLY+DFW +H++ G+L
Sbjct: 469 ILVGPGTGCAPFRGFIAERAVQAQSSPVAPVMFFFGCRNKDTDFLYRDFWESHAREGGML 528
Query: 411 SESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAF 470
SE GGGFY AFSRDQP+KVYVQHK+RE S RVW+LL +GA+VY+AGS TKMP DV SAF
Sbjct: 529 SEGKGGGFYTAFSRDQPKKVYVQHKIREMSKRVWDLLCDGAAVYVAGSSTKMPCDVMSAF 588
Query: 471 EEIVSKE-NDVSKEDAVRWIRALEKCGKYHIEAWS 504
E+IVS+E SKE A RW++ALEK G+Y++EAWS
Sbjct: 589 EDIVSEETGGGSKEVASRWLKALEKTGRYNVEAWS 623
>At4g30210 NADPH-ferrihemoprotein reductase (ATR2)
Length = 711
Score = 220 bits (561), Expect = 1e-57
Identities = 158/523 (30%), Positives = 262/523 (49%), Gaps = 49/523 (9%)
Query: 4 VTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPDV 63
V K +D L++ G + +++ GLGDD Q E W +LW L+ I E
Sbjct: 215 VAKVVDDILVEQGAQRLVQVGLGDDDQC--IEDDFTAWREALWPELDTILRE-------- 264
Query: 64 LIQDTTLIDQPKVQITYHNIENVVS-HSSDTGSVRSMHPGKSSSNRNGYP--DCFLKLVK 120
+ T + P T +E VS H S+ ++ +N NGY D
Sbjct: 265 --EGDTAVATP---YTAAVLEYRVSIHDSEDAKFNDIN----MANGNGYTVFDAQHPYKA 315
Query: 121 NLPLTK----PNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPD 176
N+ + + P S + H EF+ + Y+TGD + VL S VD +R ++ PD
Sbjct: 316 NVAVKRELHTPESDRSCIHLEFDIAGSGLTYETGDHVGVLCDNLSETVDEALRLLDMSPD 375
Query: 177 SLITVSPKGMDCNGHGSRMP-----VKLRTFVELTMDVASASPRRYFFEARCSEH----E 227
+ ++ + D S +P LRT + + S SP++ A + E
Sbjct: 376 TYFSLHAEKEDGTPISSSLPPPFPPCNLRTALTRYACLLS-SPKKSALVALAAHASDPTE 434
Query: 228 RERLEYFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLE-WLIQLVPMLKKREFSIS 286
ERL++ ASP G+D+ ++ + +R++LEV+ +FPS + PL + + P L+ R +SIS
Sbjct: 435 AERLKHLASPAGKDEYSKWVVESQRSLLEVMAEFPSAKPPLGVFFAGVAPRLQPRFYSIS 494
Query: 287 SSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLP 342
SS ++H+T ++V P R KG+CS+W+ P + S P++ ++ +
Sbjct: 495 SSPKIAETRIHVTCALVYEKMPTGRIHKGVCSTWMKNAVPYEKSENCSSAPIFVRQSNFK 554
Query: 343 TPSPS-LPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFW 400
PS S +P+I++GPGTG APFRGF++ER AL + + P + FFGC N DF+Y++
Sbjct: 555 LPSDSKVPIIMIGPGTGLAPFRGFLQERLALVESGVELGPSVLFFGCRNRRMDFIYEEE- 613
Query: 401 LNHSQNNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLT 460
L +G L+E + VAFSR+ P K YVQHK+ + + +WN++++GA +Y+ G
Sbjct: 614 LQRFVESGALAELS-----VAFSREGPTKEYVQHKMMDKASDIWNMISQGAYLYVCGDAK 668
Query: 461 KMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
M DV + I ++ + A +++ L+ G+Y + W
Sbjct: 669 GMARDVHRSLHTIAQEQGSMDSTKAEGFVKNLQTSGRYLRDVW 711
>At4g24520 NADPH-ferrihemoprotein reductase ATR1
Length = 692
Score = 213 bits (542), Expect = 2e-55
Identities = 158/518 (30%), Positives = 250/518 (47%), Gaps = 46/518 (8%)
Query: 8 LDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLSSLWRMLNMIKPELLPSGPDVLIQD 67
LD+ L G K ++E GLGDD Q E + W SLW L D L++D
Sbjct: 199 LDEELCKKGAKRLIEVGLGDDDQ--SIEDDFNAWKESLWSEL------------DKLLKD 244
Query: 68 TTLIDQPKVQITYHNI---ENVVSHSSDTGSVRSMHPGKSSSNR--NGYPDCFLKLVKNL 122
D V Y + VV+H + +SM ++ N + + C + +
Sbjct: 245 E---DDKSVATPYTAVIPEYRVVTHDPRFTTQKSMESNVANGNTTIDIHHPCRVDVAVQK 301
Query: 123 PLTKPNSGKDVRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITVS 182
L S + H EF+ I Y+TGD + V V+ + D + ++
Sbjct: 302 ELHTHESDRSCIHLEFDISRTGITYETGDHVGVYAENHVEIVEEAGKLLGHSLDLVFSIH 361
Query: 183 PKGMDCNGHGSRM------PVKLRTFVELTMDVASASPRRYFFEARCS----EHERERLE 232
D + S + P L T + D+ + PR+ A + E E+L+
Sbjct: 362 ADKEDGSPLESAVPPPFPGPCTLGTGLARYADLLN-PPRKSALVALAAYATEPSEAEKLK 420
Query: 233 YFASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPL-EWLIQLVPMLKKREFSISSSQSS 291
+ SP+G+D+ Q+ +R++LEV+ FPS + PL + + P L+ R +SISSS
Sbjct: 421 HLTSPDGKDEYSQWIVASQRSLLEVMAAFPSAKPPLGVFFAAIAPRLQPRYYSISSSPRL 480
Query: 292 HPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDP----RDAVSLPVWFQKGSLPTPS-P 346
P++VH+T ++V TP R KG+CS+W+ P + P++ + + PS P
Sbjct: 481 APSRVHVTSALVYGPTPTGRIHKGVCSTWMKNAVPAEKSHECSGAPIFIRASNFKLPSNP 540
Query: 347 SLPLILVGPGTGCAPFRGFIEER-ALQSKTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQ 405
S P+++VGPGTG APFRGF++ER AL+ + + FFGC N DF+Y+D LN+
Sbjct: 541 STPIVMVGPGTGLAPFRGFLQERMALKEDGEELGSSLLFFGCRNRQMDFIYED-ELNNFV 599
Query: 406 NNGVLSESTGGGFYVAFSRDQPEKVYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTD 465
+ GV+SE +AFSR+ +K YVQHK+ E + +VW+L+ E +Y+ G M D
Sbjct: 600 DQGVISE-----LIMAFSREGAQKEYVQHKMMEKAAQVWDLIKEEGYLYVCGDAKGMARD 654
Query: 466 VTSAFEEIVSKENDVSKEDAVRWIRALEKCGKYHIEAW 503
V IV ++ VS +A ++ L+ G+Y + W
Sbjct: 655 VHRTLHTIVQEQEGVSSSEAEAIVKKLQTEGRYLRDVW 692
>At3g24710 hypothetical protein
Length = 126
Score = 63.9 bits (154), Expect = 2e-10
Identities = 33/55 (60%), Positives = 42/55 (76%)
Query: 440 SGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWIRALEK 494
S +VW+LL +GA VY+AGS T+MP+DV SA EIVS+E SKE A R ++ALEK
Sbjct: 2 SKKVWDLLCDGAVVYVAGSSTEMPSDVMSALGEIVSEETGGSKEVASRRLKALEK 56
>At1g30510 ferrodoxin NADP oxidoreductase - like protein
Length = 381
Score = 61.2 bits (147), Expect = 1e-09
Identities = 54/202 (26%), Positives = 94/202 (45%), Gaps = 26/202 (12%)
Query: 313 KKGLCSSWLAALDPRDAVSLPVWFQKGSL-PTPSPSLPLILVGPGTGCAPFRGFIEERAL 371
K G+CS++L P D + + K L P P+ I++ GTG AP+RG++ +
Sbjct: 195 KNGVCSNFLCDSKPGDKIQITGPSGKVMLLPESDPNATHIMIATGTGVAPYRGYLRRMFM 254
Query: 372 QS---KTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQPE 428
++ KT S +F G N D LY + + + +++ F A SR++
Sbjct: 255 ENVPNKTFSGLAWLFL-GVANTDS-LLYDEEFTKYLKDH-----PDNFRFDKALSREEKN 307
Query: 429 K----VYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKED 484
K +YVQ K+ E+S ++ LL GA +Y G MP ++ + + V++E
Sbjct: 308 KKGGKMYVQDKIEEYSDEIFKLLDNGAHIYFCGLKGMMP-----GIQDTLKR---VAEER 359
Query: 485 AVRW---IRALEKCGKYHIEAW 503
W + L K ++H+E +
Sbjct: 360 GESWDLKLSQLRKNKQWHVEVY 381
>At5g66190 ferredoxin-NADP+ reductase
Length = 360
Score = 59.3 bits (142), Expect = 5e-09
Identities = 58/237 (24%), Positives = 102/237 (42%), Gaps = 25/237 (10%)
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS V L V + +T KG+CS++L L P D +
Sbjct: 137 KLRLYSIASSAIGDFGDSKTVSLCVKRLVYTNDGGEIVKGVCSNFLCDLKPGDEAKITGP 196
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++G GTG APFR F+ + + + + + F
Sbjct: 197 VGKEMLMPKDPNATIIMLGTGTGIAPFRSFLWKMFFEEHEDYKFNGLAWLFLGVPTSSSL 256
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQP----EKVYVQHKLREHSGRVWNLL-AE 449
LYK+ + + N A SR+Q EK+Y+Q ++ E++ +W LL +
Sbjct: 257 LYKEEFEKMKEKN-----PDNFRLDFAVSREQTNEKGEKMYIQTRMAEYAEELWELLKKD 311
Query: 450 GASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWI---RALEKCGKYHIEAW 503
VY+ G L M + + +K D + W+ + L++ ++++E +
Sbjct: 312 NTFVYMCG-LKGMEKGIDDIMVSLAAK-------DGIDWLEYKKQLKRSEQWNVEVY 360
>At4g05390 ferredoxin-NADP+ reductase like protein
Length = 378
Score = 58.5 bits (140), Expect = 8e-09
Identities = 51/200 (25%), Positives = 90/200 (44%), Gaps = 22/200 (11%)
Query: 313 KKGLCSSWLAALDPRDAVSLPVWFQKGSL-PTPSPSLPLILVGPGTGCAPFRGFIEERAL 371
K G+CS++L P D V + K L P P I++ GTG AP+RG++ +
Sbjct: 192 KAGVCSNFLCNAKPGDKVKITGPSGKVMLLPEDDPKATHIMIATGTGVAPYRGYLRRMFM 251
Query: 372 QS----KTISIAPIIFFFGCWNEDGDFLYKDFWLNHSQNNGVLSESTGGGFYVAFSRDQP 427
++ K +A + F G N D LY + + + ++ + A SR++
Sbjct: 252 ENVPNFKFDGLAWL--FLGVANSDS-LLYDEEFAGYRKDY-----PENFRYDKALSREEK 303
Query: 428 EK----VYVQHKLREHSGRVWNLLAEGASVYIAGSLTKMPTDVTSAFEEIVSKENDVSKE 483
K +YVQ K+ E+S ++ LL GA +Y G MP ++ + + + E
Sbjct: 304 NKKGGKMYVQDKIEEYSDEIFKLLDNGAHIYFCGLKGMMP-----GIQDTLKRVAEERGE 358
Query: 484 DAVRWIRALEKCGKYHIEAW 503
+ + L K ++H+E +
Sbjct: 359 SWEQKLTQLRKNKQWHVEVY 378
>At1g20020 ferredoxin--NADP reductase precursor, putative
Length = 369
Score = 52.4 bits (124), Expect = 6e-07
Identities = 53/235 (22%), Positives = 100/235 (42%), Gaps = 21/235 (8%)
Query: 279 KKREFSISSS---QSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPRDAVSLPVW 335
K R +SI+SS + V L V + +T KG+CS++L L P V L
Sbjct: 146 KVRLYSIASSALGDLGNSETVSLCVKRLVYTNDQGETVKGVCSNFLCDLAPGSDVKLTGP 205
Query: 336 FQKGSLPTPSPSLPLILVGPGTGCAPFRGFIEERALQS-KTISIAPIIFFFGCWNEDGDF 394
K L P+ +I++ GTG APFR F+ + + + + F
Sbjct: 206 VGKEMLMPKDPNATVIMLATGTGIAPFRSFLWKMFFEKHDDYKFNGLAWLFLGVPTTSSL 265
Query: 395 LYKDFWLNHSQNNGVLSESTGGGFYVA--FSRDQPEKVYVQHKLREHSGRVWNLL-AEGA 451
LY++ + E+ + ++ + D+ EK+Y+Q ++ +++ +W LL +
Sbjct: 266 LYQE---EFDKMKAKAPENFRVDYAISREQANDKGEKMYIQTRMAQYAAELWELLKKDNT 322
Query: 452 SVYIAGSLTKMPTDVTSAFEEIVSKENDVSKEDAVRWI---RALEKCGKYHIEAW 503
VY+ G L M + + + D + W + L+K ++++E +
Sbjct: 323 FVYMCG-LKGMEKGIDDIMVSLAA-------NDGIDWFDYKKQLKKAEQWNVEVY 369
>At2g37690 phosphoribosylaminoimidazole carboxylase like protein
Length = 642
Score = 33.1 bits (74), Expect = 0.37
Identities = 29/120 (24%), Positives = 47/120 (39%), Gaps = 18/120 (15%)
Query: 160 DSAAVDAFIRRC--------NLDPDSLITVSPKGMDCNGHGSRMPVKLRTFVELTMDVAS 211
DSA V+ F +RC ++D D+L + +G+DC S + + +++
Sbjct: 141 DSATVEEFAKRCGVLTVEIEHVDVDTLEKLEKQGVDCQPKASTIRIIQDKYMQKVHFSQH 200
Query: 212 ASPRRYFFEARCSEHERERLEYFASP----------EGRDDLYQYNQKERRTVLEVLKDF 261
P F E E R+ E F P +GR + NQ E + + L F
Sbjct: 201 GIPLPEFMEISDIEGARKAGELFGYPLMIKSKRLAYDGRGNAVANNQDELSSAVTALGGF 260
>At5g55390 putative protein
Length = 1332
Score = 32.3 bits (72), Expect = 0.63
Identities = 15/42 (35%), Positives = 22/42 (51%)
Query: 2 QFVTKKLDKRLMDLGGKAILERGLGDDQQPSGYEGTLDPWLS 43
QF + + D G + L+RG+G + P Y G LDP L+
Sbjct: 1202 QFPLRNGPNEMFDFRGYSDLDRGIGQREYPQQYGGHLDPMLA 1243
>At3g62300 putative protein
Length = 708
Score = 32.0 bits (71), Expect = 0.82
Identities = 32/124 (25%), Positives = 59/124 (46%), Gaps = 9/124 (7%)
Query: 11 RLMDLGGKAILERGLGDDQQPSGYEGTLD-PWL--SSLWRMLNMIKP-ELLPSGPDVLIQ 66
R D+G ++E+ + + P G E T+ P++ S LW++L ++ +++P P
Sbjct: 498 RTPDIGLNTVVEKHVDIVETPPGRESTMVLPFVKKSQLWKVLESMEVFKVVPQSPHF--- 554
Query: 67 DTTLIDQPKVQITYHNIENVVSHSSDTGSVRSMHPGKSSSNRNGYPDCFLKLVKN-LPLT 125
+ L++ + I +V SS V ++ S+ N +CFLKL K+ +T
Sbjct: 555 -SPLLESEEECREGDAIGRMVMFSSLLEKVNNLQVDDPISSINRIDECFLKLEKHGFNVT 613
Query: 126 KPNS 129
P S
Sbjct: 614 TPRS 617
>At1g08460 unknown protein
Length = 377
Score = 30.8 bits (68), Expect = 1.8
Identities = 25/79 (31%), Positives = 36/79 (44%), Gaps = 1/79 (1%)
Query: 87 VSHSSDTGSVRSMHPGKSSSNRNGYPDCFLKLVKNLPLTKPNSGKDVRHFEFEFVSHAIE 146
VS + GS S HP K S + G D L N+PL + + E V A+
Sbjct: 202 VSLHMNHGSWGSSHPQKGSIDELG-EDVGLGYNLNVPLPNGTGDRGYEYAMNELVVPAVR 260
Query: 147 YDTGDILEVLPGQDSAAVD 165
D++ ++ GQDS+A D
Sbjct: 261 RFGPDMVVLVVGQDSSAFD 279
>At1g60700 unknown protein
Length = 525
Score = 30.4 bits (67), Expect = 2.4
Identities = 33/131 (25%), Positives = 53/131 (40%), Gaps = 17/131 (12%)
Query: 58 PSGPDVLIQDTTLIDQPKVQITYHNIENVVSHSSDTGSVRSMHPGKSSSNRNGYP---DC 114
P+G + L+ +T Q I +N+ H + G S P + R P C
Sbjct: 233 PNGCESLLLETCSRHQIDRCINEYNV-----HEDNIGLEISPIPLTKTLERPRRPWSVCC 287
Query: 115 FLKLVKNLPLTKPNSGKDVRHFEFEFVSHAIEYDTGDIL---------EVLPGQDSAAVD 165
+ ++NLPL + +SG E + + D L E + G++ +D
Sbjct: 288 SIAALRNLPLMEDSSGACTSLDEQLYANATTSEDRDAELSQPPSTLYQEEVDGEEEIDID 347
Query: 166 AFIRRCNLDPD 176
A IR+ NL PD
Sbjct: 348 AMIRKLNLVPD 358
>At2g37340 unknown protein
Length = 249
Score = 29.6 bits (65), Expect = 4.1
Identities = 24/77 (31%), Positives = 34/77 (43%), Gaps = 2/77 (2%)
Query: 182 SPKGMDCNGHGSRMPVKLRTFVELTMDVASASPRRYFFEARCSEHERERL--EYFASPEG 239
SPK + +G SR PV+ R+ S S R + +R RER E SP+
Sbjct: 98 SPKKLRRSGSYSRSPVRSRSPRRRRSPSRSLSRSRSYSRSRSPVRRRERSVEERSRSPKR 157
Query: 240 RDDLYQYNQKERRTVLE 256
DD ++R VL+
Sbjct: 158 MDDSLSPRARDRSPVLD 174
>At5g13230 putative protein
Length = 822
Score = 29.3 bits (64), Expect = 5.3
Identities = 17/57 (29%), Positives = 29/57 (50%), Gaps = 5/57 (8%)
Query: 133 VRHFEFEFVSHAIEYDTGDILEVLPGQDSAAVDAFIRRCNLDPDSLITVSPKGMDCN 189
+R F + A++ ++ D ++PG DS A A +RRC D +S K + C+
Sbjct: 23 IRQCGFSVKTAALDLESSD--SIIPGLDSHAYGAMLRRCIQKND---PISAKAIHCD 74
>At1g64060 cytochrome b245 beta chain homolog RbohAp108
Length = 944
Score = 29.3 bits (64), Expect = 5.3
Identities = 29/112 (25%), Positives = 40/112 (34%), Gaps = 21/112 (18%)
Query: 267 PLEWLIQLVPMLKKREFSISSSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCS------SW 320
P EW FSI+S+ +H+ + WT KR +C S
Sbjct: 667 PFEW----------HPFSITSAPEDDYISIHIR-QLGDWTQELKRVFSEVCEPPVGGKSG 715
Query: 321 LAALDPRDAVSLPVWFQKGSLPTPSPSLP----LILVGPGTGCAPFRGFIEE 368
L D SLP G P+ L+LVG G G PF +++
Sbjct: 716 LLRADETTKKSLPKLLIDGPYGAPAQDYRKYDVLLLVGLGIGATPFISILKD 767
>At5g60010 respiratory burst oxidase protein - like
Length = 839
Score = 28.9 bits (63), Expect = 6.9
Identities = 31/120 (25%), Positives = 45/120 (36%), Gaps = 28/120 (23%)
Query: 276 PMLKKRE---FSISSSQSSHPNQVHLTVSVVSWTTPYKRKKKGLCSSWLAALDPR----- 327
P L K E FSI+S+ VH+ ++ WTT + + C AA P+
Sbjct: 593 PDLSKFEWHPFSITSAPGDDYLSVHIR-ALGDWTTELRSRFAKTCEPTQAAAKPKPNSLM 651
Query: 328 ---------------DAVSLPVWFQKGSLPTPSPSLP----LILVGPGTGCAPFRGFIEE 368
V P F KG P+ + L+LVG G G PF +++
Sbjct: 652 RMETRAAGVNPHIEESQVLFPKIFIKGPYGAPAQNYQKFDILLLVGLGIGATPFISILKD 711
>At3g47680 unknown protein
Length = 302
Score = 28.9 bits (63), Expect = 6.9
Identities = 28/119 (23%), Positives = 51/119 (42%), Gaps = 12/119 (10%)
Query: 388 WNEDGDFLYKDFWLNHSQNNGVLSESTGGGFY---VAFSRDQPEKVYVQHKLREHSGRVW 444
W+ D + WLN S++ + +E G F+ A+ P+ V+ + H + W
Sbjct: 50 WSAGEDLVLVSAWLNTSKDAVIGNEQKGYAFWSRIAAYYGASPKLNGVEKRETGHIKQRW 109
Query: 445 NLLAEGASVYIAG--SLTKMPT------DVTSAFEEIVSKEN-DVSKEDAVRWIRALEK 494
+ EG ++ + TK + DV + EI + E+ + E A R +R +K
Sbjct: 110 TKINEGVGKFVGSYEAATKQKSSGQNDDDVVALAHEIYNSEHGKFTLEHAWRVLRFEQK 168
>At2g13250 putative Athila retroelement ORF1 protein
Length = 507
Score = 28.9 bits (63), Expect = 6.9
Identities = 22/82 (26%), Positives = 36/82 (43%), Gaps = 6/82 (7%)
Query: 85 NVVSHSSDTGSVRSMHPGKSSSNRNGYPDCFLKLVKNLPLTKPNSGKDVRHFEFEFVSHA 144
N HSS + S + + + D L LV+NL + N G+D ++
Sbjct: 207 NAKEHSSQSSSPQQERQASNGNFLGQDIDVGLSLVENLAQSDGNYGED-----YDRTPRE 261
Query: 145 IEYDTGDIL-EVLPGQDSAAVD 165
+ DT +L ++L GQ A+D
Sbjct: 262 TQSDTNALLRQILEGQGRGAID 283
>At2g07770 putative protein
Length = 518
Score = 28.9 bits (63), Expect = 6.9
Identities = 19/48 (39%), Positives = 28/48 (57%), Gaps = 2/48 (4%)
Query: 234 FASPEGRDDLYQYNQKERRTVLEVLKDFPSVQMPLEWL-IQLVPMLKK 280
FA E D++ Y K+ V+ K FPS MPLE+L + +P+L+K
Sbjct: 53 FAFEENVKDIF-YLIKKCNEVVGPQKVFPSFVMPLEYLAFEAIPVLRK 99
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,606,228
Number of Sequences: 26719
Number of extensions: 588834
Number of successful extensions: 1451
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1417
Number of HSP's gapped (non-prelim): 26
length of query: 504
length of database: 11,318,596
effective HSP length: 104
effective length of query: 400
effective length of database: 8,539,820
effective search space: 3415928000
effective search space used: 3415928000
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148995.5


