

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148995.10 - phase: 0
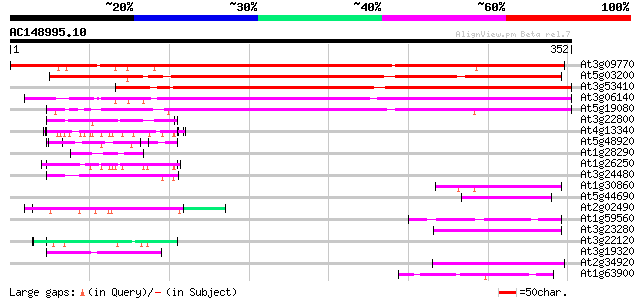
(352 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g09770 putative RING zinc finger protein 417 e-117
At5g03200 unknown protein 325 2e-89
At3g53410 unknown protein 322 2e-88
At3g06140 putative RING zinc finger protein 280 1e-75
At5g19080 unknown protein 270 1e-72
At3g22800 hypothetical protein 55 4e-08
At4g13340 extensin-like protein 55 8e-08
At5g48920 unknown protein 54 2e-07
At1g28290 proline-rich protein, putative 54 2e-07
At1g26250 hypothetical protein 52 4e-07
At3g24480 disease resistance protein, putative 52 5e-07
At1g30860 hypothetical protein 52 6e-07
At5g44690 putative protein 51 8e-07
At2g02490 hypothetical protein 51 1e-06
At1g59560 unknown protein 51 1e-06
At3g23280 unknown protein 50 2e-06
At3g22120 unknown protein 49 3e-06
At3g19320 unknown protein 49 3e-06
At2g34920 hypothetical protein 49 4e-06
At1g63900 putative RING zinc finger protein 49 4e-06
>At3g09770 putative RING zinc finger protein
Length = 388
Score = 417 bits (1072), Expect = e-117
Identities = 218/371 (58%), Positives = 263/371 (70%), Gaps = 25/371 (6%)
Query: 1 MGNISSTGAHNRRRHATASRRTHPPPPPP----VTPQP---EIAPHQFVYPGAAPYPNPP 53
MGNISS+G RRR PPPPPP + P P EI + V+ PYPNP
Sbjct: 1 MGNISSSGGEGRRRRRRNHTAAPPPPPPPPSSSLPPPPLPTEIQANPIVFAAVTPYPNPN 60
Query: 54 MHYPQYHYPGYY---PPPIPMP----HHHHPHPHPHPHMDPAW----VSRYYPCGPVVNQ 102
+ P Y YP Y PPP MP HH H PHP+ + +W ++RY G ++ Q
Sbjct: 61 PN-PVYQYPASYYHHPPPGAMPLPPYDHHLQHHPPHPYHNHSWAPVAMARYPYAGHMMAQ 119
Query: 103 PAPFVEHQKAVTIRNDVNIKKETIVISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDE 162
P P+VEHQKAVTIRNDVN+KKE++ + PD +NPG FLVSFTFDA VSG I++ FFAKE E
Sbjct: 120 PTPYVEHQKAVTIRNDVNLKKESLRLEPDPDNPGRFLVSFTFDATVSGRISVIFFAKESE 179
Query: 163 GCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVK-VGDVDVYPLAVK 221
C LT TKE L P+T+ F++GLGQKF+Q +G+GI+FS+FE+ +L K D ++YPLAVK
Sbjct: 180 DCKLTATKEDILPPITLDFEKGLGQKFKQSSGSGIDFSVFEDVELFKAAADTEIYPLAVK 239
Query: 222 ADASSDNHDGSNETETSSKPNSQITQAVFEKEKGEFRVKVVKQILSVNGMRYELQEIYGI 281
A+A+ + E + SK N+QITQAV+EK+KGE +++VVKQIL VNG RYELQEIYGI
Sbjct: 240 AEAAPSGGENEEEERSGSK-NAQITQAVYEKDKGEIKIRVVKQILWVNGTRYELQEIYGI 298
Query: 282 GNSVESDVDD----NEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICR 337
GN+VE D D N+ GKECVICLSEPRDT V PCRHMCMCSGCAKVLRFQTNRCPICR
Sbjct: 299 GNTVEGDDDSADDANDPGKECVICLSEPRDTTVLPCRHMCMCSGCAKVLRFQTNRCPICR 358
Query: 338 QPVERLLEIKV 348
QPVERLLEIKV
Sbjct: 359 QPVERLLEIKV 369
>At5g03200 unknown protein
Length = 337
Score = 325 bits (834), Expect = 2e-89
Identities = 174/331 (52%), Positives = 213/331 (63%), Gaps = 27/331 (8%)
Query: 26 PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMP----------HHH 75
PP T E+ P++FV+ PY NP +Y + PPP+ P HH+
Sbjct: 20 PPAMETAPLELPPNRFVFAAVPPYLNPNPNYVDQYPGNCLPPPVTEPPMLPYNFNHLHHY 79
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENP 135
P+ + PH P + YP P P HQKAVTIRNDVN+KK+T+ + PD ENP
Sbjct: 80 PPNSYQLPH--PLFHGGRYPI-----LPPPTYVHQKAVTIRNDVNLKKKTLTLIPDPENP 132
Query: 136 GFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGT 195
LVSFTFDA++ G IT+ FFA ED C L TKE L P+T F +GLGQKF Q +GT
Sbjct: 133 NRLLVSFTFDASMPGRITVVFFATEDAECNLRATKEDTLPPITFDFGEGLGQKFIQSSGT 192
Query: 196 GINFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEKEKG 255
GI+ + F++S+L K D DV+PLAVKA+A+ S T N QITQ V+ KEKG
Sbjct: 193 GIDLTAFKDSELFKEVDTDVFPLAVKAEATPAEEGKSGST------NVQITQVVYTKEKG 246
Query: 256 EFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRH 315
E +++VVKQIL VN RYEL EIYGI E+ VD +++GKECV+CLSEPRDT V PCRH
Sbjct: 247 EIKIEVVKQILWVNKRRYELLEIYGI----ENTVDGSDEGKECVVCLSEPRDTTVLPCRH 302
Query: 316 MCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
MCMCSGCAK LRFQTN CP+CRQPVE LLEI
Sbjct: 303 MCMCSGCAKALRFQTNLCPVCRQPVEMLLEI 333
>At3g53410 unknown protein
Length = 299
Score = 322 bits (825), Expect = 2e-88
Identities = 168/289 (58%), Positives = 207/289 (71%), Gaps = 14/289 (4%)
Query: 67 PPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEHQKAVTIRNDVNIKKETI 126
PP P P+ + +P P+ D A YP G + + P +VEHQ+AVTIRND+N+KKET+
Sbjct: 20 PPYPNPNAQYQGNYPSPYQDCA----RYPYGEMAS-PVQYVEHQEAVTIRNDINLKKETL 74
Query: 127 VISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLG 186
+ PDE+NPG FL+SFTFDA+V GSIT+ FFAKE + C L TKE V F +GL
Sbjct: 75 RLEPDEQNPGKFLLSFTFDASVPGSITVMFFAKEGKDCNLIATKEDLFPSTQVSFAKGLE 134
Query: 187 QKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKAD-ASSDNHDGSNETETSSKPNSQI 245
Q+F+Q GTGI+FS E+DLV+ + DVY +AVKA+ S D+H S PN QI
Sbjct: 135 QRFKQACGTGIDFSDMSEADLVEANETDVYHVAVKAEVVSEDDH------PESGTPNRQI 188
Query: 246 TQAVFEKE-KGEFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDD-NEQGKECVICLS 303
T V EK+ KGE++ +VVKQIL VNG RY LQEIYGIGN+V+ + +D NE+GKECVICLS
Sbjct: 189 THVVLEKDHKGEYKARVVKQILWVNGNRYVLQEIYGIGNTVDDNGEDANERGKECVICLS 248
Query: 304 EPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTEE 352
EPRDT V PCRHMCMCSGCAK+LRFQTN CPICRQPV+RLLEI V +
Sbjct: 249 EPRDTTVLPCRHMCMCSGCAKLLRFQTNLCPICRQPVDRLLEITVNNND 297
>At3g06140 putative RING zinc finger protein
Length = 546
Score = 280 bits (715), Expect = 1e-75
Identities = 159/366 (43%), Positives = 206/366 (55%), Gaps = 40/366 (10%)
Query: 10 HNRRRHATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY--PP 67
+NRRR R H PPPP P+ ++ P P P PP HY Y+Y Y+ PP
Sbjct: 195 NNRRRDNNNRRHLHHYPPPP--------PYYYLDPPPPPPPFPP-HY-DYNYSNYHLSPP 244
Query: 68 PIPMPH-------HHHPHPHPH------------PHMDPAWVSRYYPCGPVVNQPAPFVE 108
P P H+H HP P P M PA+ Y P P P P++E
Sbjct: 245 LPPQPQINSCSYGHYHYHPQPPQYFTTAQPNWWGPMMRPAY---YCPPQPQTQPPKPYLE 301
Query: 109 HQKAVTIRNDVNIKKETIVISPDEENPGFFLVSFTFDAAVSGSITIFFFAKEDEGCILTP 168
Q A +RNDVN+ ++T+ + D+ PG LVSF FDA GS TI FFAKE+ C + P
Sbjct: 302 QQNAKKVRNDVNVHRDTVRLEVDDLVPGHHLVSFVFDALFDGSFTITFFAKEEPNCTIIP 361
Query: 169 TKETHLAPVTVQFQQGLGQKFRQQAGTGINFSMFEESDLVKVGDVDVYPLAVKADASSDN 228
+P FQ+G GQKF Q +GTG + S F DL K + DVYPL + A+
Sbjct: 362 QFPEVYSPTRFHFQKGPGQKFLQPSGTGTDLSFFVLDDLSKPLEEDVYPLVISAETII-- 419
Query: 229 HDGSNETETSSKPNSQITQAVFEKEK-GEFRVKVVKQILSVNGMRYELQEIYG-IGNSVE 286
N S + Q+TQAV EK+ G F+VKVVKQIL + G+RYEL+E+YG
Sbjct: 420 --SPNSISEQSSVHKQVTQAVLEKDNDGSFKVKVVKQILWIEGVRYELRELYGSTTQGAA 477
Query: 287 SDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
S +D++ G ECVIC++E +DT V PCRH+CMCS CAK LR Q+N+CPICRQP+E LLEI
Sbjct: 478 SGLDESGSGTECVICMTEAKDTAVLPCRHLCMCSDCAKELRLQSNKCPICRQPIEELLEI 537
Query: 347 KVGTEE 352
K+ + +
Sbjct: 538 KMNSSD 543
>At5g19080 unknown protein
Length = 378
Score = 270 bits (689), Expect = 1e-72
Identities = 153/340 (45%), Positives = 194/340 (57%), Gaps = 32/340 (9%)
Query: 24 PPPP--PPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
PPPP PP + QP P Q Y PY YH YYP P + H
Sbjct: 57 PPPPAQPPSSSQPP--PSQISY---RPYGQ------NYHQNQYYPQQAPPYFTGYHHNGF 105
Query: 82 HPHMDPAWVSRYYPCGP----VVNQPAPFVEHQKAVTIRNDVNIKKETIVISPDEENPGF 137
+P M P + GP V+ PAP+VEHQ A ++NDVN+ K T+ + D+ NPG
Sbjct: 106 NPMMRPVYF------GPTPVAVMEPPAPYVEHQTAKKVKNDVNVNKATVRLVADDLNPGH 159
Query: 138 FLVSFTFDAAVSGSITIFFFAKEDEGCILTPTKETHLAPVTVQFQQGLGQKFRQQAGTGI 197
+LVSF FDA GS TI FF +E+ C + P P+ V FQ+G GQKF Q GTGI
Sbjct: 160 YLVSFVFDALFDGSFTIIFFGEEESKCTIVPHLPEAFPPIKVPFQKGAGQKFLQAPGTGI 219
Query: 198 NFSMFEESDLVKVGDVDVYPLAVKADASSDNHDGSNETETSSKPNSQITQAVFEK-EKGE 256
+ F DL K +VYPL + A+ S E + QITQAV EK G
Sbjct: 220 DLGFFSLDDLSKPSPEEVYPLVISAETVISPSSVSEEPLV----HKQITQAVLEKTNDGS 275
Query: 257 FRVKVVKQILSVNGMRYELQEIYGIGNSVESDVD----DNEQGKECVICLSEPRDTIVHP 312
F+VKV+KQIL + G RYELQE+YGI NS+ ++ GKECVICL+EP+DT V P
Sbjct: 276 FKVKVMKQILWIEGERYELQELYGIDNSITQGTAASGLEDTGGKECVICLTEPKDTAVMP 335
Query: 313 CRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEIKVGTEE 352
CRH+C+CS CA+ LRFQTN+CPICRQP+ L++IKV + +
Sbjct: 336 CRHLCLCSDCAEELRFQTNKCPICRQPIHELVKIKVESSD 375
>At3g22800 hypothetical protein
Length = 470
Score = 55.5 bits (132), Expect = 4e-08
Identities = 34/84 (40%), Positives = 38/84 (44%), Gaps = 10/84 (11%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYP--NPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
PPPPPP P P P +VYP P P PP YP P YPPP P P + +P P P
Sbjct: 392 PPPPPPPPPPPP-PPPPYVYPSPPPPPPSPPPYVYPPPPPPYVYPPP-PSPPYVYPPPPP 449
Query: 82 HPHMDPAWVSRYYPCGPVVNQPAP 105
P YP P + P P
Sbjct: 450 SPQ------PYMYPSPPCNDLPTP 467
Score = 51.6 bits (122), Expect = 6e-07
Identities = 31/81 (38%), Positives = 35/81 (42%), Gaps = 10/81 (12%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPP-IPMPHHHHPHPHPH 82
PPPPPP P P P P P PP P Y YP PPP P P+ + P P P+
Sbjct: 382 PPPPPPPPPPP---------PPPPPPPPPPPPPPPYVYPSPPPPPPSPPPYVYPPPPPPY 432
Query: 83 PHMDPAWVSRYYPCGPVVNQP 103
+ P YP P QP
Sbjct: 433 VYPPPPSPPYVYPPPPPSPQP 453
Score = 38.1 bits (87), Expect = 0.007
Identities = 23/69 (33%), Positives = 27/69 (38%), Gaps = 17/69 (24%)
Query: 24 PPPPPPVT----PQPEIAPHQFVYPGAAP-------------YPNPPMHYPQYHYPGYYP 66
PPPPPP P P +P +VYP P YP PP Y YP
Sbjct: 402 PPPPPPYVYPSPPPPPPSPPPYVYPPPPPPYVYPPPPSPPYVYPPPPPSPQPYMYPSPPC 461
Query: 67 PPIPMPHHH 75
+P P H+
Sbjct: 462 NDLPTPVHY 470
Score = 30.0 bits (66), Expect = 2.0
Identities = 15/37 (40%), Positives = 20/37 (53%), Gaps = 3/37 (8%)
Query: 23 HPPPPPP--VTPQPEIAPHQFVYPGAAPYPNP-PMHY 56
+PPPP P V P P +P ++YP P P+HY
Sbjct: 434 YPPPPSPPYVYPPPPPSPQPYMYPSPPCNDLPTPVHY 470
>At4g13340 extensin-like protein
Length = 760
Score = 54.7 bits (130), Expect = 8e-08
Identities = 34/85 (40%), Positives = 37/85 (43%), Gaps = 6/85 (7%)
Query: 24 PPPPPPV---TPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPH 80
PPPPPPV P P P VY P P PP P Y P PPP P P + P P
Sbjct: 442 PPPPPPVYSPPPPPPPPPPPPVYSPPPPPPPPPPPPPVYSPPPPSPPPPPPPVYSPPPPP 501
Query: 81 PHPHMDPAWVSRYYPCGPVVNQPAP 105
P P P + P PV + P P
Sbjct: 502 PPPPPPPVYSP---PPPPVYSSPPP 523
Score = 51.6 bits (122), Expect = 6e-07
Identities = 31/86 (36%), Positives = 40/86 (46%), Gaps = 10/86 (11%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPI----PMPHHHHPHP 79
PPPPPP P P +P P + P P PP++ P P PPP+ P P + P P
Sbjct: 468 PPPPPPPPPPPVYSPP----PPSPPPPPPPVYSPPPPPPPPPPPPVYSPPPPPVYSSPPP 523
Query: 80 HPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P P + +R P P + P P
Sbjct: 524 PPSPAPTPVYCTR--PPPPPPHSPPP 547
Score = 49.3 bits (116), Expect = 3e-06
Identities = 34/98 (34%), Positives = 41/98 (41%), Gaps = 16/98 (16%)
Query: 24 PPPPPPVTPQPEI--APHQFVY------PGAAPYP----NPPMHYPQYHYPGYYPPPIPM 71
PPPPPP P P + P VY P AP P PP P P + PP P
Sbjct: 497 PPPPPPPPPPPPVYSPPPPPVYSSPPPPPSPAPTPVYCTRPPPPPPHSPPPPQFSPPPPE 556
Query: 72 PHHHH----PHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P+++ PH P PH P S P P ++ P P
Sbjct: 557 PYYYSSPPPPHSSPPPHSPPPPHSPPPPIYPYLSPPPP 594
Score = 45.4 bits (106), Expect = 5e-05
Identities = 36/107 (33%), Positives = 48/107 (44%), Gaps = 22/107 (20%)
Query: 23 HPPPPPPVTPQPEIAPHQFVYPG---------AAPYPNPPMHY-----PQYHYPGYYPPP 68
+ PPPPP P VY ++P P PP+HY P+ HY + PPP
Sbjct: 627 YSPPPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPPPVHYSSPPPPEVHY--HSPPP 684
Query: 69 IPMPHHHHPHPHPHPHMD----PAWVSRYYPCGPVV-NQPAPFVEHQ 110
P+ H+ P P P + PA V + P P+V + P P V HQ
Sbjct: 685 SPV-HYSSPPPPPSAPCEESPPPAPVVHHSPPPPMVHHSPPPPVIHQ 730
Score = 43.5 bits (101), Expect = 2e-04
Identities = 30/91 (32%), Positives = 37/91 (39%), Gaps = 9/91 (9%)
Query: 24 PPPPPPV--------TPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
PPPP P+ +P P P P P P PP++ P P PPP+ P
Sbjct: 410 PPPPAPIFSTPPTLTSPPPPSPPPPVYSPPPPPPPPPPVYSPPPPPPPPPPPPVYSPPPP 469
Query: 76 HPHPHPHPHM-DPAWVSRYYPCGPVVNQPAP 105
P P P P + P S P PV + P P
Sbjct: 470 PPPPPPPPPVYSPPPPSPPPPPPPVYSPPPP 500
Score = 42.4 bits (98), Expect = 4e-04
Identities = 34/102 (33%), Positives = 39/102 (37%), Gaps = 22/102 (21%)
Query: 24 PPPPPPV----TPQPEIAPHQFVYPGAAPY----PNPPMHYPQYHYPG-----YYPPPIP 70
PPPPPPV P PE+ H P +P P PP P P ++ PP P
Sbjct: 661 PPPPPPVHYSSPPPPEVHYHS---PPPSPVHYSSPPPPPSAPCEESPPPAPVVHHSPPPP 717
Query: 71 MPHHHHPHPHPHPHMDPAWVSRYYPCGPVVN------QPAPF 106
M HH P P H P P PV+ P PF
Sbjct: 718 MVHHSPPPPVIHQSPPPPSPEYEGPLPPVIGVSYASPPPPPF 759
Score = 42.0 bits (97), Expect = 5e-04
Identities = 36/107 (33%), Positives = 39/107 (35%), Gaps = 24/107 (22%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHY-----PQYHY------PGYYPPPIPMP 72
PPPPPP P P P P P P +HY P +Y P YY P P P
Sbjct: 609 PPPPPPCIEPPPPPPCIEYSP---PPPPPVVHYSSPPPPPVYYSSPPPPPVYYSSPPPPP 665
Query: 73 HHHH---PHPHPHPHMDPAWVSRYY-----PCGPVVNQ--PAPFVEH 109
H+ P P H H P Y P P PAP V H
Sbjct: 666 PVHYSSPPPPEVHYHSPPPSPVHYSSPPPPPSAPCEESPPPAPVVHH 712
Score = 41.6 bits (96), Expect = 7e-04
Identities = 31/90 (34%), Positives = 39/90 (42%), Gaps = 18/90 (20%)
Query: 22 THPPPPP---PVTPQ---PEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHH 75
T PPPPP P PQ P P+ + P P+ +PP H P P + PP P+ +
Sbjct: 535 TRPPPPPPHSPPPPQFSPPPPEPYYYSSP-PPPHSSPPPHSP----PPPHSPPPPIYPYL 589
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P P P P P P PV + P P
Sbjct: 590 SPPPPPTPVSSP-------PPTPVYSPPPP 612
Score = 39.7 bits (91), Expect = 0.003
Identities = 28/81 (34%), Positives = 30/81 (36%), Gaps = 5/81 (6%)
Query: 25 PPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHPH 84
P PPP P P F P P PP P + P PPP P P + P P P P
Sbjct: 401 PLPPPSLPSPPPPAPIFSTPPTLTSPPPPSPPPPVYSPP-PPPPPPPPVYSPPPPPPPPP 459
Query: 85 MDPAWVSRYYPCGPVVNQPAP 105
P Y P P P P
Sbjct: 460 PPPV----YSPPPPPPPPPPP 476
Score = 38.9 bits (89), Expect = 0.004
Identities = 30/84 (35%), Positives = 35/84 (40%), Gaps = 15/84 (17%)
Query: 24 PP--PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
PP PPPP +P P I P+ P P +PP P Y PPP P P P P P
Sbjct: 571 PPHSPPPPHSPPPPIYPYLSPPPPPTPVSSPP------PTPVYSPPP-PPPCIEPPPPPP 623
Query: 82 HPHMDPAWVSRYYPCGPVVNQPAP 105
P P PVV+ +P
Sbjct: 624 CIEYSPP------PPPPVVHYSSP 641
Score = 38.1 bits (87), Expect = 0.007
Identities = 31/101 (30%), Positives = 37/101 (35%), Gaps = 12/101 (11%)
Query: 15 HATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHH 74
H+ + PPPP P PH P + P P+ P P YP PPP P P
Sbjct: 543 HSPPPPQFSPPPPEPYYYSSPPPPHSSPPPHSPPPPHSP---PPPIYPYLSPPPPPTPVS 599
Query: 75 HHP------HPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P P P P ++P PC P P V H
Sbjct: 600 SPPPTPVYSPPPPPPCIEPPPPP---PCIEYSPPPPPPVVH 637
>At5g48920 unknown protein
Length = 205
Score = 53.5 bits (127), Expect = 2e-07
Identities = 27/67 (40%), Positives = 33/67 (48%), Gaps = 4/67 (5%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHY-PGYYPPPIPMPHHH--HPHPH 80
PP P P P + P P+ +PP H P H+ P + PPP P PH H P P+
Sbjct: 13 PPSHQHPLPSPVPPPPSHISPPPPPF-SPPHHPPPPHFSPPHQPPPSPYPHPHPPPPSPY 71
Query: 81 PHPHMDP 87
PHPH P
Sbjct: 72 PHPHQPP 78
Score = 50.8 bits (120), Expect = 1e-06
Identities = 25/58 (43%), Positives = 28/58 (48%), Gaps = 8/58 (13%)
Query: 25 PPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPH 82
PPPPP +P P F + P+ PP YP H PPP P PH H P P PH
Sbjct: 33 PPPPPFSPPHHPPPPHF----SPPHQPPPSPYPHPH----PPPPSPYPHPHQPPPPPH 82
Score = 44.7 bits (104), Expect = 8e-05
Identities = 26/75 (34%), Positives = 33/75 (43%), Gaps = 7/75 (9%)
Query: 34 PEIAPHQFVYPGAAPYPNPPMHY---PQYHYPGYYPPPIPMPHHHHPHPHPHPHMDPAWV 90
P +P +P +P P PP H P P ++PPP H P P P+PH P
Sbjct: 9 PYYSPPSHQHPLPSPVPPPPSHISPPPPPFSPPHHPPPPHFSPPHQPPPSPYPHPHPPPP 68
Query: 91 SRYYPCGPVVNQPAP 105
S Y P +QP P
Sbjct: 69 SPY----PHPHQPPP 79
Score = 38.1 bits (87), Expect = 0.007
Identities = 23/60 (38%), Positives = 25/60 (41%), Gaps = 13/60 (21%)
Query: 23 HPPPP---PPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY----PPPIPMPHHH 75
HPPPP PP P P PH P P P +P P + PPP P P HH
Sbjct: 43 HPPPPHFSPPHQPPPSPYPHPH------PPPPSPYPHPHQPPPPPHVLPPPPPTPAPGHH 96
>At1g28290 proline-rich protein, putative
Length = 359
Score = 53.5 bits (127), Expect = 2e-07
Identities = 23/46 (50%), Positives = 27/46 (58%), Gaps = 9/46 (19%)
Query: 39 HQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHPH 84
H+ P AP P P YH+ ++P P PHHHHPHPHPHPH
Sbjct: 29 HKTQTPSLAPAPAP------YHHGHHHPHP---PHHHHPHPHPHPH 65
Score = 39.7 bits (91), Expect = 0.003
Identities = 19/53 (35%), Positives = 22/53 (40%), Gaps = 17/53 (32%)
Query: 31 TPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
T P +AP Y +P+PP H HH HPHPHPHP
Sbjct: 31 TQTPSLAPAPAPYHHGHHHPHPPHH-----------------HHPHPHPHPHP 66
Score = 30.4 bits (67), Expect = 1.5
Identities = 29/89 (32%), Positives = 35/89 (38%), Gaps = 10/89 (11%)
Query: 22 THPPPPPPVTP--QPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPP---PIPMPHHHH 76
T PP PPV+P +P + P VYP PP P P YPP P+ P
Sbjct: 104 TKPPVKPPVSPPAKPPVKPP--VYPPTKAPVKPPTKPPV--KPPVYPPTKAPVKPPTKPP 159
Query: 77 PHPHPHPHMD-PAWVSRYYPCGPVVNQPA 104
P +P P P P V+ PA
Sbjct: 160 VKPPVYPPTKAPVKPPTKPPVKPPVSPPA 188
Score = 28.1 bits (61), Expect = 7.5
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 6/50 (12%)
Query: 22 THPPPPPPVTP--QPEIAPHQFVYPGAAPYPNPPMHYPQYH--YPGYYPP 67
T PP PPV+P +P + P VYP PP+ P P YPP
Sbjct: 176 TKPPVKPPVSPPAKPPVKPP--VYPPTKAPVKPPVSPPTKPPVTPPVYPP 223
>At1g26250 hypothetical protein
Length = 443
Score = 52.4 bits (124), Expect = 4e-07
Identities = 33/98 (33%), Positives = 44/98 (44%), Gaps = 14/98 (14%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY----PNPPMHYPQYHYPG----YYPPPI-----P 70
PPPPP V P P+ + P PY P PP + Y P Y PPP P
Sbjct: 310 PPPPPYVYKSPPPPPYVYTSPPPPPYVYKSPPPPPYVDSYSPPPAPYVYKPPPYVYKPPP 369
Query: 71 MPHHHHPHPHPHPHMDPAWVSRYY-PCGPVVNQPAPFV 107
+++ P P P+ + P +V Y P P V +P P+V
Sbjct: 370 YVYNYSPPPAPYVYKPPPYVYSYSPPPAPYVYKPPPYV 407
Score = 47.8 bits (112), Expect = 9e-06
Identities = 34/99 (34%), Positives = 42/99 (42%), Gaps = 19/99 (19%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHY-----PQYHYPGYYPPPI---- 69
PPPPP V P P+ + P PY P PP Y P Y Y PPP
Sbjct: 270 PPPPPYVYKSPPPPPYVYSSPPPPPYVYSSPPPPPYVYSSPPPPPYVYKSPPPPPYVYTS 329
Query: 70 -PMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFV 107
P P + + P P P++D S P P V +P P+V
Sbjct: 330 PPPPPYVYKSPPPPPYVD----SYSPPPAPYVYKPPPYV 364
Score = 47.8 bits (112), Expect = 9e-06
Identities = 34/95 (35%), Positives = 41/95 (42%), Gaps = 13/95 (13%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHYPQYHYPGYY---PPPIPMPHHH 75
PPPPP V P P+ + P PY P PP Y P Y PPP P +
Sbjct: 290 PPPPPYVYSSPPPPPYVYSSPPPPPYVYKSPPPPPYVYTSPPPPPYVYKSPPPPPYVDSY 349
Query: 76 HPHPHPHPHMDPAWVSRYYPCGPVVN---QPAPFV 107
P P P+ + P +V Y P V N PAP+V
Sbjct: 350 SPPPAPYVYKPPPYV--YKPPPYVYNYSPPPAPYV 382
Score = 44.7 bits (104), Expect = 8e-05
Identities = 35/108 (32%), Positives = 45/108 (41%), Gaps = 24/108 (22%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHY-----PQYHYPGYYPPPI---- 69
PPPPP V P P+ + P PY P PP Y P Y Y PPP
Sbjct: 250 PPPPPYVYKSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYSSPPPPPYVYSS 309
Query: 70 -PMPHHHHPHPHPHPHM-----DPAWVSRYYPCGPVVNQ----PAPFV 107
P P + + P P P++ P +V + P P V+ PAP+V
Sbjct: 310 PPPPPYVYKSPPPPPYVYTSPPPPPYVYKSPPPPPYVDSYSPPPAPYV 357
Score = 44.3 bits (103), Expect = 1e-04
Identities = 33/102 (32%), Positives = 41/102 (39%), Gaps = 20/102 (19%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHY-----PQYHYPGYYPPPI---- 69
PPPPP V P P+ + P PY P PP Y P Y Y PPP
Sbjct: 70 PPPPPYVYSSPPPPPYVYNSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYKS 129
Query: 70 -PMPHHHHPHPHPHPHM-----DPAWVSRYYPCGPVVNQPAP 105
P P + + P P P++ P +V + P P V P P
Sbjct: 130 PPPPPYVYSSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSPPP 171
Score = 43.9 bits (102), Expect = 1e-04
Identities = 31/97 (31%), Positives = 40/97 (40%), Gaps = 13/97 (13%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPI-----PMPHHHHPH 78
PPPPP V P P+ + P PY P P Y Y PPP P P + +
Sbjct: 140 PPPPPYVYSSPPPPPYVYKSPPPPPYVYSPPPPPPYVYQSPPPPPYVYSSPPPPPYVYKS 199
Query: 79 PHPHPHM-----DPAWVSRYYPCGPVVNQ---PAPFV 107
P P P++ P +V + P P V P P+V
Sbjct: 200 PPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYV 236
Score = 42.0 bits (97), Expect = 5e-04
Identities = 31/93 (33%), Positives = 44/93 (46%), Gaps = 11/93 (11%)
Query: 21 RTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHY-----PGYYPPPIPMPHHH 75
++ PPPP + P AP+ + P PY P Y Y+Y P Y PP P + +
Sbjct: 338 KSPPPPPYVDSYSPPPAPYVYKPP---PYVYKPPPY-VYNYSPPPAPYVYKPP-PYVYSY 392
Query: 76 HPHPHPHPHMDPAWVSRYY-PCGPVVNQPAPFV 107
P P P+ + P +V Y P P V +P P+V
Sbjct: 393 SPPPAPYVYKPPPYVYSYSPPPAPYVYKPPPYV 425
Score = 41.2 bits (95), Expect = 9e-04
Identities = 33/97 (34%), Positives = 41/97 (42%), Gaps = 13/97 (13%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHYPQYHYPGY-YPPPIPMPH-HHH 76
PPPPP V P P+ + P PY P PP Y P Y Y P P P+ +
Sbjct: 100 PPPPPYVYKSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYSSPPPPPYVYKS 159
Query: 77 PHPHPH---PHMDPAWVSRYYPCGPVVNQ---PAPFV 107
P P P+ P P +V + P P V P P+V
Sbjct: 160 PPPPPYVYSPPPPPPYVYQSPPPPPYVYSSPPPPPYV 196
Score = 41.2 bits (95), Expect = 9e-04
Identities = 30/91 (32%), Positives = 37/91 (39%), Gaps = 11/91 (12%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPY-----PNPPMHYPQYHYPGY-YPPPIPMPH-HHH 76
PPPPP V P P+ + P PY P PP Y P Y Y P P P+ +
Sbjct: 200 PPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYKS 259
Query: 77 PHPHPHPHMDPAWVSRYYPCGPVVNQPAPFV 107
P P P+ + P Y P P P+V
Sbjct: 260 PPPPPYVYSSPPPPPYVYKSPP----PPPYV 286
Score = 38.5 bits (88), Expect = 0.006
Identities = 30/106 (28%), Positives = 41/106 (38%), Gaps = 24/106 (22%)
Query: 22 THPPPPPPVTPQPEIAPHQFVY-PGAAPY---------------------PNPPMHYPQY 59
T PPPPP V P P+ Y P APY P P ++ P
Sbjct: 328 TSPPPPPYVYKSPPPPPYVDSYSPPPAPYVYKPPPYVYKPPPYVYNYSPPPAPYVYKPPP 387
Query: 60 HYPGYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
+ Y PPP P+ + P P+ + + P Y P V + P+P
Sbjct: 388 YVYSYSPPP--APYVYKPPPYVYSYSPPPAPYVYKPPPYVYSSPSP 431
Score = 38.5 bits (88), Expect = 0.006
Identities = 34/110 (30%), Positives = 44/110 (39%), Gaps = 25/110 (22%)
Query: 23 HPPPPPP--VTPQPEIAPHQFVYPGAAPY-----PNPPMHY-----PQYHYPGYYPPPI- 69
+ PPPPP V P P+ + P PY P PP Y P Y Y PPP
Sbjct: 167 YSPPPPPPYVYQSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYKSPPPPPYV 226
Query: 70 ----PMPHHHHPHPHPHPHM-----DPAWVSRYYPCGPVVNQ---PAPFV 107
P P + + P P P++ P +V + P P V P P+V
Sbjct: 227 YSSPPPPPYVYKSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYV 276
Score = 38.1 bits (87), Expect = 0.007
Identities = 33/101 (32%), Positives = 42/101 (40%), Gaps = 21/101 (20%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAP----YPNPPMHYPQYHYPGYYPPPI-----PMPHH 74
PPPPP V P P +VY P Y +PP P Y Y PPP P P +
Sbjct: 240 PPPPPYVYSSPP--PPPYVYKSPPPPPYVYSSPPP--PPYVYKSPPPPPYVYSSPPPPPY 295
Query: 75 HHPHPHPHPHM-----DPAWVSRYYPCGPVV---NQPAPFV 107
+ P P P++ P +V + P P V P P+V
Sbjct: 296 VYSSPPPPPYVYSSPPPPPYVYKSPPPPPYVYTSPPPPPYV 336
Score = 35.8 bits (81), Expect = 0.036
Identities = 24/70 (34%), Positives = 32/70 (45%), Gaps = 7/70 (10%)
Query: 23 HPPPPPPVTPQPEIAPHQFVY-PGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
+ PPP P +P P+ + Y P APY P P Y Y Y PPP P + P+ +
Sbjct: 374 YSPPPAPYVYKPP--PYVYSYSPPPAPYVYKP---PPYVY-SYSPPPAPYVYKPPPYVYS 427
Query: 82 HPHMDPAWVS 91
P P + S
Sbjct: 428 SPSPPPYYSS 437
Score = 35.0 bits (79), Expect = 0.062
Identities = 28/94 (29%), Positives = 39/94 (40%), Gaps = 13/94 (13%)
Query: 23 HPPPPPPVTPQPEI---APHQFVYPGAAP----YPNPPMHYPQYHYPGY-YPPPIPMPH- 73
+ P P P +P P +P +VY +P Y PP Y P Y Y P P P+
Sbjct: 27 YSPTPTPYSPLPPYVYNSPPPYVYNSPSPPPYVYKPPPYIYSSPPPPPYVYSSPPPPPYV 86
Query: 74 HHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFV 107
++ P P P+ + P Y P P P+V
Sbjct: 87 YNSPPPPPYVYSSPPPPPYVYKSPP----PPPYV 116
Score = 33.5 bits (75), Expect = 0.18
Identities = 27/95 (28%), Positives = 36/95 (37%), Gaps = 13/95 (13%)
Query: 26 PPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPI-----PMPHHHHPHPH 80
PPP + P P+ + P PY P Y Y PPP P P + + P
Sbjct: 62 PPPYIYSSPPPPPYVYSSPPPPPYVYNSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPP 121
Query: 81 PHPHM-----DPAWVSRYYPCGPVVNQ---PAPFV 107
P P++ P +V P P V P P+V
Sbjct: 122 PPPYVYKSPPPPPYVYSSPPPPPYVYSSPPPPPYV 156
Score = 32.3 bits (72), Expect = 0.40
Identities = 30/100 (30%), Positives = 41/100 (41%), Gaps = 22/100 (22%)
Query: 28 PPVTPQPEIAPHQFVYPGAAPY----PNPPMHY---PQYHYPGYYPPPI-----PMPHHH 75
P TP + P +VY PY P+PP + P Y Y PPP P P +
Sbjct: 29 PTPTPYSPLPP--YVYNSPPPYVYNSPSPPPYVYKPPPYIYSSPPPPPYVYSSPPPPPYV 86
Query: 76 HPHPHPHPHM-----DPAWVSRYYPCGPVVNQ---PAPFV 107
+ P P P++ P +V + P P V P P+V
Sbjct: 87 YNSPPPPPYVYSSPPPPPYVYKSPPPPPYVYSSPPPPPYV 126
Score = 29.6 bits (65), Expect = 2.6
Identities = 19/56 (33%), Positives = 26/56 (45%), Gaps = 9/56 (16%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHP 79
PPP P V P P+ + Y +P P P ++ P P Y P P P++ P P
Sbjct: 394 PPPAPYVYKPP---PYVYSY---SPPPAPYVYKPP---PYVYSSPSPPPYYSSPSP 440
>At3g24480 disease resistance protein, putative
Length = 494
Score = 52.0 bits (123), Expect = 5e-07
Identities = 33/91 (36%), Positives = 39/91 (42%), Gaps = 16/91 (17%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
PPPPPP P P P +P P+PP+ P P Y PPP P H+ P P P
Sbjct: 411 PPPPPPSPPLPP--------PVYSPPPSPPVFSPPPSPPVYSPPPPPSIHYSSPPPPPVH 462
Query: 84 HMDPAWVSRYY--PCGPVVN------QPAPF 106
H P S + P PV+ P PF
Sbjct: 463 HSSPPPPSPEFEGPLPPVIGVSYASPPPPPF 493
Score = 28.9 bits (63), Expect = 4.4
Identities = 18/62 (29%), Positives = 22/62 (35%)
Query: 44 PGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQP 103
P P PP P P Y PPP P P P + P + P P V+
Sbjct: 405 PPIVALPPPPPPSPPLPPPVYSPPPSPPVFSPPPSPPVYSPPPPPSIHYSSPPPPPVHHS 464
Query: 104 AP 105
+P
Sbjct: 465 SP 466
>At1g30860 hypothetical protein
Length = 739
Score = 51.6 bits (122), Expect = 6e-07
Identities = 24/94 (25%), Positives = 49/94 (51%), Gaps = 15/94 (15%)
Query: 268 VNGMRYELQEIYG----IGNSVESDVD-----------DNEQGKECVICLSEPRDTIVHP 312
++GMR ++Q++ + +SV++ +D +N ++C +C + +++
Sbjct: 640 ISGMRSQIQQLQQEMSVLRDSVKTCLDANASLQHKAHQENPMKRKCCVCDETQVEAVLYR 699
Query: 313 CRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
C HMCMC CA L + +CPICR + ++ +
Sbjct: 700 CGHMCMCLKCANELHWSGGKCPICRAQIVDVVRV 733
>At5g44690 putative protein
Length = 684
Score = 51.2 bits (121), Expect = 8e-07
Identities = 20/57 (35%), Positives = 32/57 (56%)
Query: 284 SVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPV 340
S+E + + + C IC P D++++ C HMC C CA L++ +CPIC P+
Sbjct: 618 SLEFESLSDSLERNCSICFEMPIDSLLYRCGHMCTCLKCAHELQWSNMKCPICMAPI 674
>At2g02490 hypothetical protein
Length = 415
Score = 50.8 bits (120), Expect = 1e-06
Identities = 36/116 (31%), Positives = 49/116 (42%), Gaps = 16/116 (13%)
Query: 10 HNRRRHATASRRTHP---PPPPPVTPQPEIAPHQFV------YPGAAPYPNP----PMHY 56
H ++A S +P P P PV P P PHQ YPG P P+P P H
Sbjct: 60 HPPHQNAKISVNQYPSVFPIPHPVPPSPGHPPHQNTKISVNQYPGVFPIPHPVPPSPGHP 119
Query: 57 PQYHYP---GYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAPFVEH 109
P + YP P+PH P P PH + V++Y P+ + P ++H
Sbjct: 120 PHQNAKISVNQYPGVFPIPHPVSPSPGHPPHQNEISVNQYPHILPISHPVPPSLKH 175
Score = 46.6 bits (109), Expect = 2e-05
Identities = 41/145 (28%), Positives = 58/145 (39%), Gaps = 28/145 (19%)
Query: 15 HATASRRTHP---PPPPPVTPQPEIAPHQFV------YPGAAPYPNP----PMHYPQYH- 60
H T S +P P P PV P P PHQ YP P P+P P H P +
Sbjct: 36 HHTISVNQYPGVFPIPHPVPPSPGHPPHQNAKISVNQYPSVFPIPHPVPPSPGHPPHQNT 95
Query: 61 ------YPGYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYP-CGPVVNQPAP---FVEHQ 110
YPG +P P P+P P P PH + YP P+ + +P HQ
Sbjct: 96 KISVNQYPGVFPIPHPVP----PSPGHPPHQNAKISVNQYPGVFPIPHPVSPSPGHPPHQ 151
Query: 111 KAVTIRNDVNIKKETIVISPDEENP 135
+++ +I + + P ++P
Sbjct: 152 NEISVNQYPHILPISHPVPPSLKHP 176
Score = 38.5 bits (88), Expect = 0.006
Identities = 34/112 (30%), Positives = 43/112 (38%), Gaps = 16/112 (14%)
Query: 10 HNRRRHATASRRTHP---PPPPPVTPQPEIAPHQFV------YPGAAP--YPNPPM--HY 56
H ++A S +P P P PV P PHQ YP P +P PP H
Sbjct: 175 HPPHQNAKISMNQYPCVFPIPHPVPPSLGHPPHQNKKILVNQYPRILPISHPLPPSQGHP 234
Query: 57 PQYHYP---GYYPPPIPMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
PQ + YP P+PH P P PH + YP ++ P P
Sbjct: 235 PQQNVKISVKQYPDVFPIPHPVPPSPGHPPHQNKKIPVNQYPRILPISHPVP 286
Score = 37.4 bits (85), Expect = 0.012
Identities = 26/67 (38%), Positives = 31/67 (45%), Gaps = 6/67 (8%)
Query: 22 THPPPPPPVTPQPEIAPHQF-VYPGAAPYPNPPMHYPQYHYP---GYYPPPIPMPHHHHP 77
T P PPPP I+ +QF V P P P H P + YP IP+PH+ P
Sbjct: 340 TEPFPPPP-NQNENISVNQFQVIPHLPWPPKRPGHPPHQNAKVAVNQYPGIIPIPHYPEP 398
Query: 78 HPHP-HP 83
HP HP
Sbjct: 399 PKHPGHP 405
Score = 32.7 bits (73), Expect = 0.31
Identities = 22/55 (40%), Positives = 24/55 (43%), Gaps = 14/55 (25%)
Query: 23 HPPPPPPVTPQPEIAPHQFV------YPGAAP---YPNPPMH--YPQYHYPGYYP 66
H P PP +P PHQ YPG P YP PP H +P Y YP P
Sbjct: 363 HLPWPPK---RPGHPPHQNAKVAVNQYPGIIPIPHYPEPPKHPGHPPYQYPKISP 414
Score = 30.8 bits (68), Expect = 1.2
Identities = 22/71 (30%), Positives = 28/71 (38%), Gaps = 8/71 (11%)
Query: 22 THP-PPPPPVTPQPEIAPHQFVYPGAAPYPNP----PMHYPQYHYP---GYYPPPIPMPH 73
+HP PP PQ + YP P P+P P H P + YP +P+ H
Sbjct: 224 SHPLPPSQGHPPQQNVKISVKQYPDVFPIPHPVPPSPGHPPHQNKKIPVNQYPRILPISH 283
Query: 74 HHHPHPHPHPH 84
P P PH
Sbjct: 284 PVPPSPGHPPH 294
>At1g59560 unknown protein
Length = 338
Score = 50.8 bits (120), Expect = 1e-06
Identities = 29/96 (30%), Positives = 49/96 (50%), Gaps = 14/96 (14%)
Query: 251 EKEKGEFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIV 310
E+ + +F +K V V+ + + G G S + D D CV+CL + +T
Sbjct: 254 ERRRRQFALKRV-----VDAAARRAKPVTGGGTSRDGDTPDL-----CVVCLDQKYNTAF 303
Query: 311 HPCRHMCMCSGCAKVLRFQTNRCPICRQPVERLLEI 346
C HMC C+ C+ LR CP+CR+ ++++L+I
Sbjct: 304 VECGHMCCCTPCSLQLR----TCPLCRERIQQVLKI 335
>At3g23280 unknown protein
Length = 462
Score = 50.1 bits (118), Expect = 2e-06
Identities = 19/80 (23%), Positives = 40/80 (49%)
Query: 267 SVNGMRYELQEIYGIGNSVESDVDDNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAKVL 326
S++ +L + S E + ++ C ICL P + + PC H+ C C K +
Sbjct: 380 SIDSTPVDLPSAASLPASTEGERKEDGNTGTCAICLDAPSEAVCVPCGHVAGCMSCLKEI 439
Query: 327 RFQTNRCPICRQPVERLLEI 346
+ + CP+CR +++++++
Sbjct: 440 KSKNWGCPVCRAKIDQVIKL 459
>At3g22120 unknown protein
Length = 351
Score = 49.3 bits (116), Expect = 3e-06
Identities = 31/91 (34%), Positives = 34/91 (37%), Gaps = 11/91 (12%)
Query: 15 HATASRRTHPPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHH 74
H + HP P PP+ P P P P P+ P P P PP P P H
Sbjct: 118 HPKPPTKPHPHPKPPIVKPPTKPPPSTPKPPTKPPPSTPK--PPTTKP---PPSTPKPPH 172
Query: 75 HHPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
H P P P P P P PVV P P
Sbjct: 173 HKPPPTPCPPPTPT------PTPPVVTPPTP 197
Score = 47.8 bits (112), Expect = 9e-06
Identities = 32/96 (33%), Positives = 35/96 (36%), Gaps = 7/96 (7%)
Query: 15 HATASRRTHPPPPPPVTPQ--PEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYP---PPI 69
H PP PP V P P + P P P+P PP P P P PP
Sbjct: 54 HKPPKHPVKPPKPPAVKPPKPPAVKPPTPKPPTVKPHPKPPTVKPHPKPPTVKPHPKPPT 113
Query: 70 PMPHHHHPHPHPHPHMDPAWVSRYYPCGPVVNQPAP 105
P H P PHPH P V P P + P P
Sbjct: 114 VKPPHPKPPTKPHPHPKPPIVKP--PTKPPPSTPKP 147
Score = 43.5 bits (101), Expect = 2e-04
Identities = 30/89 (33%), Positives = 34/89 (37%), Gaps = 8/89 (8%)
Query: 24 PPPPPPVTPQ----PEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHP 79
PP PP V P P + PH P P+P PP P P PP P HPHP
Sbjct: 71 PPKPPAVKPPTPKPPTVKPHPKP-PTVKPHPKPPTVKPHPKPPTVKPPHPKPPTKPHPHP 129
Query: 80 HP---HPHMDPAWVSRYYPCGPVVNQPAP 105
P P P + P P + P P
Sbjct: 130 KPPIVKPPTKPPPSTPKPPTKPPPSTPKP 158
Score = 41.6 bits (96), Expect = 7e-04
Identities = 34/106 (32%), Positives = 40/106 (37%), Gaps = 17/106 (16%)
Query: 16 ATASRRTHPPP---------PPPVTPQP-EIAPHQFVYPGAAPYPNPPMHYPQYHYPGYY 65
+T T PPP PPP TP+P P P P P PP+ P P
Sbjct: 143 STPKPPTKPPPSTPKPPTTKPPPSTPKPPHHKPPPTPCPPPTPTPTPPVVTPPTPTPPVI 202
Query: 66 PPPIPMPHHHHPHPHPHPHM------DPAWVSRYYPCGPVVNQPAP 105
PP P P P P P P + P ++ P PVV P P
Sbjct: 203 TPPTPTPPVVTP-PTPTPPVITPPTPTPPVITPPTPTPPVVTPPTP 247
Score = 39.3 bits (90), Expect = 0.003
Identities = 30/88 (34%), Positives = 33/88 (37%), Gaps = 10/88 (11%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPP---PIPMPHHHHPHPH 80
PP P P +P P + P A P PP P P PP P P P PHP
Sbjct: 46 PPKPSPAPHKPPKHPVKPPKPPAVKPPKPPAVKP----PTPKPPTVKPHPKPPTVKPHPK 101
Query: 81 P---HPHMDPAWVSRYYPCGPVVNQPAP 105
P PH P V +P P P P
Sbjct: 102 PPTVKPHPKPPTVKPPHPKPPTKPHPHP 129
Score = 38.9 bits (89), Expect = 0.004
Identities = 28/84 (33%), Positives = 31/84 (36%), Gaps = 7/84 (8%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAA--PYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHP 81
P PPP TP P + P P P PP+ P P PP P P P
Sbjct: 179 PCPPPTPTPTPPVVTPPTPTPPVITPPTPTPPVVTPPTPTPPVITPPTPTPPVITP---- 234
Query: 82 HPHMDPAWVSRYYPCGPVVNQPAP 105
P P V+ P PVV P P
Sbjct: 235 -PTPTPPVVTPPTPTPPVVTPPTP 257
Score = 32.7 bits (73), Expect = 0.31
Identities = 23/72 (31%), Positives = 24/72 (32%), Gaps = 12/72 (16%)
Query: 22 THPPPPPPVTPQPEIAPHQFVYPGAAP------YPNPPMHYPQYHYPGYYPPPIPMPHHH 75
T P P PPV P P P P P PP+ P P PP P P
Sbjct: 203 TPPTPTPPVVTPPTPTPPVITPPTPTPPVITPPTPTPPVVTPPTPTPPVVTPPTPTP--- 259
Query: 76 HPHPHPHPHMDP 87
P P P P
Sbjct: 260 ---PTPIPETCP 268
>At3g19320 unknown protein
Length = 493
Score = 49.3 bits (116), Expect = 3e-06
Identities = 26/72 (36%), Positives = 29/72 (40%), Gaps = 3/72 (4%)
Query: 24 PPPPPPVTPQPEIAPHQFVYPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP 83
P PPPP TP P P P +P P HYP Y Y P P P P P P
Sbjct: 61 PLPPPPQTPPPPPPPQSLPPPSPSPEPE---HYPPPPYHHYITPSPPPPRPLPPPPPPPL 117
Query: 84 HMDPAWVSRYYP 95
H + + YP
Sbjct: 118 HFSSPLIKKVYP 129
Score = 32.3 bits (72), Expect = 0.40
Identities = 21/64 (32%), Positives = 23/64 (35%), Gaps = 1/64 (1%)
Query: 43 YPGAAPYPNPPMHYPQYHYPGYYPPPIPMPHHHHPHPHPHP-HMDPAWVSRYYPCGPVVN 101
Y AP P P + P P PPP P P P P P H P Y P
Sbjct: 47 YNSPAPSPEPEDYLPLPPPPQTPPPPPPPQSLPPPSPSPEPEHYPPPPYHHYITPSPPPP 106
Query: 102 QPAP 105
+P P
Sbjct: 107 RPLP 110
>At2g34920 hypothetical protein
Length = 785
Score = 48.9 bits (115), Expect = 4e-06
Identities = 21/84 (25%), Positives = 44/84 (52%), Gaps = 1/84 (1%)
Query: 266 LSVNGMRYELQEIYGIGNSVESDVD-DNEQGKECVICLSEPRDTIVHPCRHMCMCSGCAK 324
L ++ +R ++ + S++ V +N ++C +C +T+++ C HMC C CA
Sbjct: 698 LEMSELRDSVKTCLDVNASLQKSVHLENPFKRKCCVCNETQVETLLYRCGHMCTCLRCAN 757
Query: 325 VLRFQTNRCPICRQPVERLLEIKV 348
L++ +CPIC + ++ + V
Sbjct: 758 ELQYNGGKCPICHAKILDVVRVFV 781
>At1g63900 putative RING zinc finger protein
Length = 115
Score = 48.9 bits (115), Expect = 4e-06
Identities = 27/102 (26%), Positives = 50/102 (48%), Gaps = 14/102 (13%)
Query: 245 ITQAVFEKEKGEFRVKVVKQILSVNGMRYELQEIYGIGNSVESDVDDNEQGKE-----CV 299
+ +V E+ + R ++ K++L R EL+ N + D+ + ++ CV
Sbjct: 15 VIDSVLERRR---RRQLQKRVLDAAAKRAELESEGS--NGTRESISDSTKKEDAVPDLCV 69
Query: 300 ICLSEPRDTIVHPCRHMCMCSGCAKVLRFQTNRCPICRQPVE 341
ICL + + + PC HMC C+ C+ L CP+CR+ ++
Sbjct: 70 ICLEQEYNAVFVPCGHMCCCTACSSHL----TSCPLCRRRID 107
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,282,088
Number of Sequences: 26719
Number of extensions: 495290
Number of successful extensions: 8825
Number of sequences better than 10.0: 459
Number of HSP's better than 10.0 without gapping: 204
Number of HSP's successfully gapped in prelim test: 268
Number of HSP's that attempted gapping in prelim test: 3063
Number of HSP's gapped (non-prelim): 2067
length of query: 352
length of database: 11,318,596
effective HSP length: 100
effective length of query: 252
effective length of database: 8,646,696
effective search space: 2178967392
effective search space used: 2178967392
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148995.10


