

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.4 - phase: 0
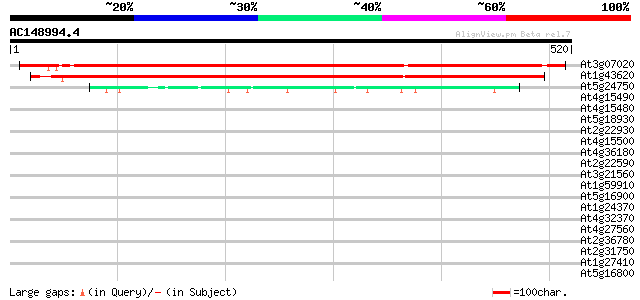
(520 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g07020 UDP-glucose:sterol glucosyltransferase 721 0.0
At1g43620 unknown protein 573 e-163
At5g24750 sterol glucosyltransferase - like protein 86 4e-17
At4g15490 indole-3-acetate beta-glucosyltransferase like protein 37 0.026
At4g15480 indole-3-acetate beta-glucosyltransferase like protein 35 0.10
At5g18930 S-adenosyl-L-methionine decarboxylase - like protein 33 0.38
At2g22930 putative flavonol 3-O-glucosyltransferase 32 0.65
At4g15500 indole-3-acetate beta-glucosyltransferase like protein 32 0.85
At4g36180 receptor protein kinase like protein 32 1.1
At2g22590 putative anthocyanidin-3-glucoside rhamnosyltransferase 31 1.5
At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferas... 31 1.9
At1g59910 hypothetical protein 31 1.9
At5g16900 receptor protein kinase -like protein 30 2.5
At1g24370 hypothetical protein 30 2.5
At4g32370 putative protein 30 4.2
At4g27560 UDP rhamnose-anthocyanidin-3-glucoside rhamnosyltransf... 30 4.2
At2g36780 putative glucosyl transferase 30 4.2
At2g31750 putative glucosyltransferase 30 4.2
At1g27410 unknown protein (At1g27420) 30 4.2
At5g16800 unknown protein 29 5.5
>At3g07020 UDP-glucose:sterol glucosyltransferase
Length = 637
Score = 721 bits (1861), Expect = 0.0
Identities = 354/511 (69%), Positives = 410/511 (79%), Gaps = 15/511 (2%)
Query: 10 QLRWLKNAFMVKDDGTVEIDVPGNI--KPLALENG---TGVIDPSDESCNETINEDIEPI 64
+L+ L VK DGTVE +VP + +P+ ++ G GV +DES + D++ I
Sbjct: 136 KLKLLNRIATVKHDGTVEFEVPADAIPQPIVVDRGESKNGVC--ADESIDGV---DLQYI 190
Query: 65 RPQQIAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPK 124
P QI MLIVGTRGDVQPFVAI KRLQ GHRVRLATH NF++FVL+AGLEFYPLGGDPK
Sbjct: 191 PPMQIVMLIVGTRGDVQPFVAIAKRLQDYGHRVRLATHANFKEFVLTAGLEFYPLGGDPK 250
Query: 125 VLAEYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPA 184
VLA YMVKNKGFLPSGPSEI +QR+Q++ II+SLLPAC P+S FKADAIIANPPA
Sbjct: 251 VLAGYMVKNKGFLPSGPSEIPIQRNQMKDIIYSLLPACKEPDPDSGISFKADAIIANPPA 310
Query: 185 YGHTHVAEYLNVPLHIFFTMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLI 244
YGHTHVAE L +P+H+FFTMPWTPTS+FPHPLSRV+QP GYRLSYQIVD+LIWLGIRD++
Sbjct: 311 YGHTHVAEALKIPIHVFFTMPWTPTSEFPHPLSRVKQPAGYRLSYQIVDSLIWLGIRDMV 370
Query: 245 NEFRKKKLKLRAVTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASN 304
N+ RKKKLKLR VTYL G+ ++P+GY+WSPHLVPKPKDWGP ID+VGFC+LDLASN
Sbjct: 371 NDLRKKKLKLRPVTYLSGTQGSGSNIPHGYMWSPHLVPKPKDWGPQIDVVGFCYLDLASN 430
Query: 305 YEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNL 364
YEPP LV+WLE G+ PIY+GFGSLP+QEPEKMT IIV+AL++T QRGIINKGWGGLGNL
Sbjct: 431 YEPPAELVEWLEAGDKPIYIGFGSLPVQEPEKMTEIIVEALQRTKQRGIINKGWGGLGNL 490
Query: 365 AELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGE 424
E VYLLDN PHDWLFPRC AVVHHGGAGTTAAGL+A CPTT+VPFFGDQPFWGE
Sbjct: 491 KE--PKDFVYLLDNVPHDWLFPRCKAVVHHGGAGTTAAGLKASCPTTIVPFFGDQPFWGE 548
Query: 425 RVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAGAVNAFYK 484
RVHARGVGP+PI V+EF+L +L DAI FML+ +VK A LA AMK+EDGVAGAV AF+K
Sbjct: 549 RVHARGVGPSPIPVDEFSLHKLEDAINFMLDDKVKSSAETLAKAMKDEDGVAGAVKAFFK 608
Query: 485 HYPREKPDTEAEPRPVPSVHKHLSIRGCFGC 515
H P K + P+P LS R CFGC
Sbjct: 609 HLPSAKQNIS---DPIPEPSGFLSFRKCFGC 636
>At1g43620 unknown protein
Length = 615
Score = 573 bits (1476), Expect = e-163
Identities = 268/485 (55%), Positives = 355/485 (72%), Gaps = 20/485 (4%)
Query: 20 VKDDGTVEIDVPGNIKPLALENGTGVID---------PSDESCNETINEDIEPIRPQQIA 70
+++DGTVE+ ++NGT V + S + +++ E I +IA
Sbjct: 110 IQNDGTVEV----------IDNGTPVSELWEFEPTKGQSTITYEKSLTESFRSIPRLKIA 159
Query: 71 MLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLSAGLEFYPLGGDPKVLAEYM 130
+L+VGTRGDVQPF+A+ KRLQ GHRVRLATH NF FV +AG+EFYPLGGDP+ LA YM
Sbjct: 160 ILVVGTRGDVQPFLAMAKRLQEFGHRVRLATHANFRSFVRAAGVEFYPLGGDPRELAAYM 219
Query: 131 VKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYGHTHV 190
+NKG +PSGPSEI QR Q++AII SLLPAC E+ F+A AIIANPPAYGH HV
Sbjct: 220 ARNKGLIPSGPSEISKQRKQLKAIIESLLPACIEPDLETATSFRAQAIIANPPAYGHVHV 279
Query: 191 AEYLNVPLHIFFTMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIRDLINEFRKK 250
AE L VP+HIFFTMPWTPT++FPHPL+RV Q Y LSY +VD ++W IR IN+FRK+
Sbjct: 280 AEALGVPIHIFFTMPWTPTNEFPHPLARVPQSAAYWLSYIVVDLMVWWSIRTYINDFRKR 339
Query: 251 KLKLRAVTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKS 310
KL L + Y + +P GY+WSPH+VPKP DWGP +D+VG+CFL+L S Y+P +
Sbjct: 340 KLNLAPIAYFSTYHGSISHLPTGYMWSPHVVPKPSDWGPLVDVVGYCFLNLGSKYQPREE 399
Query: 311 LVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNTS 370
+ W+E G P+Y+GFGS+PL +P++ II++ L+ T QRGI+++GWGGLGNLA
Sbjct: 400 FLHWIERGSPPVYIGFGSMPLDDPKQTMDIILETLKDTEQRGIVDRGWGGLGNLA-TEVP 458
Query: 371 KSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARG 430
++V+L+++CPHDWLFP+C+AVVHHGGAGTTA GL+A CPTT+VPFFGDQ FWG+R++ +G
Sbjct: 459 ENVFLVEDCPHDWLFPQCSAVVHHGGAGTTATGLKAGCPTTIVPFFGDQFFWGDRIYEKG 518
Query: 431 VGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVELANAMKNEDGVAGAVNAFYKHYPREK 490
+GPAPI + + ++E L +IRFML PEVK + +ELA ++NEDGVA AV+AF++H P E
Sbjct: 519 LGPAPIPIAQLSVENLSSSIRFMLQPEVKSQVMELAKVLENEDGVAAAVDAFHRHLPPEL 578
Query: 491 PDTEA 495
P E+
Sbjct: 579 PLPES 583
>At5g24750 sterol glucosyltransferase - like protein
Length = 517
Score = 86.3 bits (212), Expect = 4e-17
Identities = 109/482 (22%), Positives = 182/482 (37%), Gaps = 99/482 (20%)
Query: 75 GTRGDVQPFVAIGK---RLQADGHRVRLA--THKNFEDFVLSAGLEFYPLGGDPKVLAEY 129
GT+GDV P AI R+Q V ++ H+N + A + ++P+ P + E
Sbjct: 4 GTKGDVYPLAAIAAVFARVQQHYTVVLMSHLAHENLSSHLSKAKVSYFPINSPPALSNE- 62
Query: 130 MVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYPESNEPFKADAIIANPPAYGHTH 189
P G + S + + E + F+ I P
Sbjct: 63 --------PQGTQNV--TDSLRKMFLEEKERIKREHRQECHSAFRT--IFGKDPCMEGWS 110
Query: 190 VAEYLNVPLHIF--FTMPWTPTSDFPHPLSRV---------RQPIGYRLSYQIVDALIWL 238
+AE + + + +P++P S F + PIG ++S+ V +W
Sbjct: 111 LAEVFQIRCVVAAPYVVPYSPPSGFERQFRKELPDLYKYLKEAPIG-KVSWSDVTHWMWP 169
Query: 239 GIRDLINEFRKKKLKLRA------VTYLRGSYTFPPDMPYGYIWSPHLVPKPKDWGPNID 292
+ +R ++L L VT L + PP Y +S +V P W ++
Sbjct: 170 LFTEEWGSWRYEELNLSCYPFADPVTDLPIWHIRPPSPLVLYGFSKEIVECPDYWPLSVR 229
Query: 293 IVGFCFLD-------------------------LASNYEPPKSLVDWLEEGENPIYVGFG 327
+ GF FL SN+ + + E PI+VG
Sbjct: 230 VCGFWFLPNEWQFSCNECGDNPFAGRLGTDDSHTCSNHTELYTFISSCEPAL-PIFVGLS 288
Query: 328 SLP----LQEPEKMTRIIVQALEQTGQRGII-NKGWGGL-------------GNLAELNT 369
S+ +++P R++ ++ TG R II +G L L+
Sbjct: 289 SVGSMGFVRDPIAFLRVLQSVIQITGYRFIIFTASYGPLDAAIRTIANGSDSSEKQPLHA 348
Query: 370 SKSVY------LLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWG 423
S++ P++W+F CAA +HHGG+G+ AA L+A P + PF DQ +W
Sbjct: 349 GISIFNGKLFCFSGMVPYNWMFRTCAAAIHHGGSGSVAAALQAGIPQIICPFMLDQFYWA 408
Query: 424 ERVHARGVGPAPIRVEEFTLERLVD-------------AIRFMLNPEVKKRAVELANAMK 470
E++ GV P P++ LE D AI L+ + + RA+E+A +
Sbjct: 409 EKMSWLGVAPQPLKRNHLLLEDSNDEKNITEAAQVVAKAIYDALSAKTRARAMEIAEILS 468
Query: 471 NE 472
E
Sbjct: 469 LE 470
>At4g15490 indole-3-acetate beta-glucosyltransferase like protein
Length = 479
Score = 37.0 bits (84), Expect = 0.026
Identities = 44/180 (24%), Positives = 73/180 (40%), Gaps = 30/180 (16%)
Query: 306 EPPKSLVDWLEEGE--NPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGL-- 361
EP ++WL+ E + +Y+ FG++ + E+M I G+++ G L
Sbjct: 265 EPASDCMEWLDSREPSSVVYISFGTIANLKQEQMEEIA---------HGVLSSGLSVLWV 315
Query: 362 ------GNLAELNT-----SKSVYLLDNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAEC 408
G E + + +++ CP + + P A + H G +T L A
Sbjct: 316 VRPPMEGTFVEPHVLPRELEEKGKIVEWCPQERVLAHPAIACFLSHCGWNSTMEALTAGV 375
Query: 409 PTTVVPFFGDQ---PFWGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVEL 465
P P +GDQ + V GV EE + R V A + +L V ++AVEL
Sbjct: 376 PVVCFPQWGDQVTDAVYLADVFKTGVRLGRGAAEEMIVSREVVAEK-LLEATVGEKAVEL 434
>At4g15480 indole-3-acetate beta-glucosyltransferase like protein
Length = 490
Score = 35.0 bits (79), Expect = 0.10
Identities = 94/470 (20%), Positives = 173/470 (36%), Gaps = 71/470 (15%)
Query: 52 SCNETINEDIEPIRPQQIAMLIVGTRGDVQPFVAIGKRLQADGHRVRLATHKNFEDFVLS 111
S +E + E P + ++ +G V P + +GK + + G V T + + +
Sbjct: 3 SISEMVFETCPSPNPIHVMLVSFQGQGHVNPLLRLGKLIASKGLLVTFVTTELWGKKMRQ 62
Query: 112 AGL----EFYPLGGDPKVLAEYMVKNKGFLPSGPSEIHLQRSQIRAIIHSLLPACNSRYP 167
A E P+G + E+ + ++ L + + ++ + RY
Sbjct: 63 ANKIVDGELKPVGSG-SIRFEFFDEEWAEDDDRRADFSLYIAHLESVGIREVSKLVRRYE 121
Query: 168 ESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWTPTSDFPH------------- 214
E+NEP +I NP HVAE N+P + + S + H
Sbjct: 122 EANEP--VSCLINNPFIPWVCHVAEEFNIPCAVLWVQSCACFSAYYHYQDGSVSFPTETE 179
Query: 215 PLSRVRQPIGYRLSYQIVDALI-----WLGIRD-LINEFRKKKLKLRAVTYLRGSYTFPP 268
P V+ P L + + + + G R ++ +F+ ++ L S+
Sbjct: 180 PELDVKLPCVPVLKNDEIPSFLHPSSRFTGFRQAILGQFKNLS---KSFCVLIDSFDSLE 236
Query: 269 DMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASNY-EPPKSLVDWLEE--GENPIYVG 325
Y+ S L P K GP + D++ + + ++WL+ + +Y+
Sbjct: 237 QEVIDYMSS--LCP-VKTVGPLFKVARTVTSDVSGDICKSTDKCLEWLDSRPKSSVVYIS 293
Query: 326 FGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGL------------------GNLAEL 367
FG++ + E++ I G++ G L L E
Sbjct: 294 FGTVAYLKQEQIEEI---------AHGVLKSGLSFLWVIRPPPHDLKVETHVLPQELKES 344
Query: 368 NTSKSVYLLDNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGER 425
+ ++D CP + + P A V H G +T L + P P +GDQ +
Sbjct: 345 SAKGKGMIVDWCPQEQVLSHPSVACFVTHCGWNSTMESLSSGVPVVCCPQWGDQV--TDA 402
Query: 426 VHARGVGPAPIRV-EEFTLERLV---DAIRFMLNPEVKKRAVEL-ANAMK 470
V+ V +R+ T ER+V + +L V ++A EL NA+K
Sbjct: 403 VYLIDVFKTGVRLGRGATEERVVPREEVAEKLLEATVGEKAEELRKNALK 452
>At5g18930 S-adenosyl-L-methionine decarboxylase - like protein
Length = 347
Score = 33.1 bits (74), Expect = 0.38
Identities = 17/35 (48%), Positives = 20/35 (56%)
Query: 240 IRDLINEFRKKKLKLRAVTYLRGSYTFPPDMPYGY 274
IR LI+ R L LRA Y RGS+ FP P+ Y
Sbjct: 90 IRPLIHLARNLGLTLRACRYSRGSFIFPKAQPFPY 124
>At2g22930 putative flavonol 3-O-glucosyltransferase
Length = 442
Score = 32.3 bits (72), Expect = 0.65
Identities = 25/106 (23%), Positives = 45/106 (41%), Gaps = 9/106 (8%)
Query: 386 PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEE---FT 442
P V+H G GT L +C ++PF GDQ + + + E+ F+
Sbjct: 328 PSIGCFVNHCGPGTIWECLMTDCQMVLLPFLGDQVLFTRLMTEEFKVSVEVSREKTGWFS 387
Query: 443 LERLVDAIRFMLNPE------VKKRAVELANAMKNEDGVAGAVNAF 482
E L DAI+ +++ + V+ +L + + + G V+ F
Sbjct: 388 KESLSDAIKSVMDKDSDLGKLVRSNHAKLKETLGSHGLLTGYVDKF 433
>At4g15500 indole-3-acetate beta-glucosyltransferase like protein
Length = 475
Score = 32.0 bits (71), Expect = 0.85
Identities = 46/202 (22%), Positives = 79/202 (38%), Gaps = 45/202 (22%)
Query: 306 EPPKSLVDWLEEGE--NPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLG- 362
+P ++WL+ E + +Y+ FG+L + ++ I GI+N G L
Sbjct: 261 KPDSDCIEWLDSREPSSVVYISFGTLAFLKQNQIDEIA---------HGILNSGLSCLWV 311
Query: 363 ------------NLAELNTSKSVYLLDNCPHDWLF--PRCAAVVHHGGAGTTAAGLRAEC 408
++ L + +++ C + + P A + H G +T L +
Sbjct: 312 LRPPLEGLAIEPHVLPLELEEKGKIVEWCQQEKVLAHPAVACFLSHCGWNSTMEALTSGV 371
Query: 409 PTTVVPFFGDQPF----------WGERVHARGVGPAPIRVEEFTLERLVDAIRFMLNPEV 458
P P +GDQ G R+ +RG I E ERL++A V
Sbjct: 372 PVICFPQWGDQVTNAVYMIDVFKTGLRL-SRGASDERIVPREEVAERLLEA-------TV 423
Query: 459 KKRAVEL-ANAMKNEDGVAGAV 479
++AVEL NA + ++ AV
Sbjct: 424 GEKAVELRENARRWKEEAESAV 445
>At4g36180 receptor protein kinase like protein
Length = 1136
Score = 31.6 bits (70), Expect = 1.1
Identities = 28/80 (35%), Positives = 42/80 (52%), Gaps = 15/80 (18%)
Query: 203 TMPWTPTSDFPHPLSRVRQPIGYRLSYQIVDALIWLGIR-----DLINE--FRKK----- 250
T+ T D + LSR R + ++ +Y D ++ L IR L+NE F+K+
Sbjct: 834 TIEATRQFDEENVLSRTRYGLLFKANYN--DGMV-LSIRRLPNGSLLNENLFKKEAEVLG 890
Query: 251 KLKLRAVTYLRGSYTFPPDM 270
K+K R +T LRG Y PPD+
Sbjct: 891 KVKHRNITVLRGYYAGPPDL 910
>At2g22590 putative anthocyanidin-3-glucoside rhamnosyltransferase
Length = 470
Score = 31.2 bits (69), Expect = 1.5
Identities = 33/132 (25%), Positives = 51/132 (38%), Gaps = 21/132 (15%)
Query: 346 EQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLR 405
E+T RG++ +GW V L HD + V+ H G GT +R
Sbjct: 337 ERTADRGMVWRGW--------------VEQLRTLSHDSI----GLVLTHPGWGTIIEAIR 378
Query: 406 AECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEE---FTLERLVDAIRFMLNPEVKKRA 462
P ++ F DQ + + +G R E FT E + +++R ++ E K
Sbjct: 379 FAKPMAMLVFVYDQGLNARVIEEKKIGYMIPRDETEGFFTKESVANSLRLVMVEEEGKVY 438
Query: 463 VELANAMKNEDG 474
E MK G
Sbjct: 439 RENVKEMKGVFG 450
>At3g21560 UDP-glucose:indole-3-acetate beta-D-glucosyltransferase,
putative
Length = 496
Score = 30.8 bits (68), Expect = 1.9
Identities = 39/185 (21%), Positives = 69/185 (37%), Gaps = 35/185 (18%)
Query: 306 EPPKSLVDWLEEG--ENPIYVGFGSLPLQEPEKMTRIIV---------------QALEQT 348
EP ++WL+ + +Y+ FG++ + E++ I Q L
Sbjct: 271 EPTDPCMEWLDSQPVSSVVYISFGTVAYLKQEQIDEIAYGVLNADVTFLWVIRQQELGFN 330
Query: 349 GQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLRAEC 408
++ ++ + G G + E + + V P A V H G +T + +
Sbjct: 331 KEKHVLPEEVKGKGKIVEWCSQEKVLS---------HPSVACFVTHCGWNSTMEAVSSGV 381
Query: 409 PTTVVPFFGDQ--------PFWGERVH-ARGVGPAPIRVEEFTLERLVDAIRFMLNPEVK 459
PT P +GDQ W V +RG + E ERL + + E+K
Sbjct: 382 PTVCFPQWGDQVTDAVYMIDVWKTGVRLSRGEAEERLVPREEVAERLREVTKGEKAIELK 441
Query: 460 KRAVE 464
K A++
Sbjct: 442 KNALK 446
>At1g59910 hypothetical protein
Length = 929
Score = 30.8 bits (68), Expect = 1.9
Identities = 19/67 (28%), Positives = 31/67 (45%), Gaps = 2/67 (2%)
Query: 108 FVLSAGLEFYPLGGDPKVLAEYMVKNKGFLPSGPSEIHL--QRSQIRAIIHSLLPACNSR 165
+ SAG ++ GG + +++ G PS PS+IH RS + A N+
Sbjct: 199 YATSAGSDYGGGGGGKQSQSKFQAPGGGSFPSSPSQIHSGGGRSPPLPLPPGQFTAGNAS 258
Query: 166 YPESNEP 172
+P S +P
Sbjct: 259 FPSSTQP 265
>At5g16900 receptor protein kinase -like protein
Length = 851
Score = 30.4 bits (67), Expect = 2.5
Identities = 14/43 (32%), Positives = 24/43 (55%), Gaps = 6/43 (13%)
Query: 268 PDMPYGYIWSPHLVPKPKDWGPNIDIVGFCFLDLASNYEPPKS 310
PD Y +WSP +P+ ++D+ + ++NYEPPK+
Sbjct: 204 PDDVYDRVWSPFFLPEWTQITTSLDV------NNSNNYEPPKA 240
>At1g24370 hypothetical protein
Length = 413
Score = 30.4 bits (67), Expect = 2.5
Identities = 23/93 (24%), Positives = 38/93 (40%), Gaps = 8/93 (8%)
Query: 291 IDIVGFCFLDLASNYEPPKSLVDWLEEGENPIYVGFGSLPLQEPEKMTRIIVQALEQTGQ 350
ID++ C L N+ P +++DW E LQE R+ ++ T
Sbjct: 270 IDVIKRCSSSLYKNFSPIDTMLDWPPENPFEHSKLLNYYELQEDNDWIRLYLELAVATTN 329
Query: 351 RGIINKGWGGLGNLAELNTSKSVYL-LDNCPHD 382
RG I +L +L+ K + + +D P D
Sbjct: 330 RGTIR-------DLDDLSNLKIIQVAIDTTPQD 355
>At4g32370 putative protein
Length = 503
Score = 29.6 bits (65), Expect = 4.2
Identities = 20/68 (29%), Positives = 28/68 (40%), Gaps = 8/68 (11%)
Query: 158 LLPACNSRYPESNEPFKADAIIANPPAYGHTHVAEYLNVPLHIFFTMPWTPTSDFPHPLS 217
L PA + R+ E P K + P A P H+ MP P + FP PL+
Sbjct: 424 LAPAKSPRHVELPMPTKPPTMFPKPLAPAKP--------PRHVEPPMPTKPPTMFPKPLA 475
Query: 218 RVRQPIGY 225
+ P+ Y
Sbjct: 476 PAKPPVYY 483
>At4g27560 UDP rhamnose-anthocyanidin-3-glucoside
rhamnosyltransferase - like protein
Length = 455
Score = 29.6 bits (65), Expect = 4.2
Identities = 26/88 (29%), Positives = 35/88 (39%), Gaps = 8/88 (9%)
Query: 386 PRCAAVVHHGGAGTTAAGLRAECPTTVVPFFGDQPFWGERVHARGVGPAPIRVEE---FT 442
P V H G G+ L ++C +VP GDQ + + EE F+
Sbjct: 334 PSVGCFVSHCGFGSMWESLLSDCQIVLVPQLGDQVLNTRLLSDELKVSVEVAREETGWFS 393
Query: 443 LERLVDAIRFMLNPEVKKRAVELANAMK 470
E L DAI V KR E+ N +K
Sbjct: 394 KESLFDAIN-----SVMKRDSEIGNLVK 416
>At2g36780 putative glucosyl transferase
Length = 496
Score = 29.6 bits (65), Expect = 4.2
Identities = 30/120 (25%), Positives = 50/120 (41%), Gaps = 20/120 (16%)
Query: 346 EQTGQRGIINKGWGGLGNLAELNTSKSVYLLDNCPHDWLFPRCAAVVHHGGAGTTAAGLR 405
E+ +RG++ KGW + V +L + P + H G +T G+
Sbjct: 344 ERIKERGLLIKGW-----------APQVLILSH-------PSVGGFLTHCGWNSTLEGIT 385
Query: 406 AECPTTVVPFFGDQPFWGERVHARGV-GPAPIRVEEFTLERLVDAIRFMLNPEVKKRAVE 464
+ P P FGDQ F +++ + + VEE D I +++ E K+AVE
Sbjct: 386 SGIPLITWPLFGDQ-FCNQKLVVQVLKAGVSAGVEEVMKWGEEDKIGVLVDKEGVKKAVE 444
>At2g31750 putative glucosyltransferase
Length = 456
Score = 29.6 bits (65), Expect = 4.2
Identities = 33/124 (26%), Positives = 50/124 (39%), Gaps = 24/124 (19%)
Query: 312 VDWLEEGE--NPIYVGFGSLPLQEPEKMTRIIVQALEQTGQRGIINKGWGGLGNLAELNT 369
+DWL+ + IYV FGSL + + ++M + L+QTG L + E T
Sbjct: 261 LDWLDSKPPGSVIYVSFGSLAVLKDDQMIE-VAAGLKQTGH--------NFLWVVRETET 311
Query: 370 SK--SVYLLDNCPHDWLF---PRCAAVVH--------HGGAGTTAAGLRAECPTTVVPFF 416
K S Y+ D C + P+ + H H G +T L +P +
Sbjct: 312 KKLPSNYIEDICDKGLIVNWSPQLQVLAHKSIGCFMTHCGWNSTLEALSLGVALIGMPAY 371
Query: 417 GDQP 420
DQP
Sbjct: 372 SDQP 375
>At1g27410 unknown protein (At1g27420)
Length = 422
Score = 29.6 bits (65), Expect = 4.2
Identities = 13/40 (32%), Positives = 22/40 (54%)
Query: 49 SDESCNETINEDIEPIRPQQIAMLIVGTRGDVQPFVAIGK 88
SD SC E I E I+ ++P+ + ++V + V P G+
Sbjct: 311 SDHSCYEEIGEFIKLVKPKSMKGIVVSSSSYVDPLYYFGR 350
>At5g16800 unknown protein
Length = 236
Score = 29.3 bits (64), Expect = 5.5
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Query: 46 IDPSDESCNETINEDIEPIRPQQIAMLIVGTRGDVQPFVAIGKRLQADGHRVRL 99
I+PSD E I+ DI PIR + V GD+ + A+ R + DGH L
Sbjct: 30 INPSDLERLEQIHRDIFPIRYESEFFQNVVNGGDIVSWAAV-DRSRPDGHSEEL 82
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.141 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,092,353
Number of Sequences: 26719
Number of extensions: 611357
Number of successful extensions: 1268
Number of sequences better than 10.0: 25
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 1246
Number of HSP's gapped (non-prelim): 33
length of query: 520
length of database: 11,318,596
effective HSP length: 104
effective length of query: 416
effective length of database: 8,539,820
effective search space: 3552565120
effective search space used: 3552565120
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148994.4


