

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148994.1 + phase: 0
(347 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
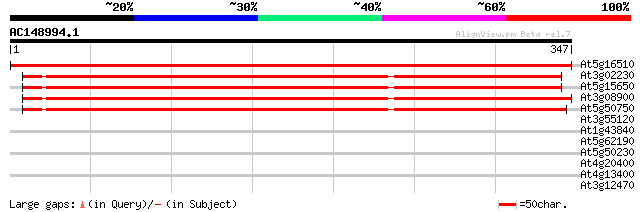
Score E
Sequences producing significant alignments: (bits) Value
At5g16510 amylogenin; reversibly glycosylatable polypeptide 535 e-152
At3g02230 reversibly glycosylated polypeptide-1 362 e-100
At5g15650 reversibly glycosylated polypeptide-2 (AtRGP) 360 e-100
At3g08900 reversibly glycosylated polypeptide-3 (RGP) 360 e-100
At5g50750 UDP-glucose:protein transglucosylase; reversibly glyco... 355 3e-98
At3g55120 chalcone isomerase 30 1.5
At1g43840 hypothetical protein 29 3.3
At5g62190 putative RNA helicase (MMI9.2) 29 4.3
At5g50230 putative protein 29 4.3
At4g20400 unknown protein 28 5.7
At4g13400 putative protein 28 5.7
At3g12470 unknown protein 28 9.7
>At5g16510 amylogenin; reversibly glycosylatable polypeptide
Length = 348
Score = 535 bits (1378), Expect = e-152
Identities = 249/347 (71%), Positives = 297/347 (84%)
Query: 1 MSQVIINDNEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADV 60
MS IN NEVDIVIGAL+++LT F+ W+ FS FHLI+VKDP LKEEL IPEGF DV
Sbjct: 1 MSLAEINKNEVDIVIGALNADLTQFLTSWRPFFSGFHLIVVKDPELKEELNIPEGFDVDV 60
Query: 61 YTNSEIERVVGSSTSIRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQH 120
Y+ +++E+VVG+S S FSGY+CRYFG+LVSKKKY+V IDDDCVPAKD G +VDAV QH
Sbjct: 61 YSKTDMEKVVGASNSTMFSGYSCRYFGYLVSKKKYIVSIDDDCVPAKDPKGFLVDAVTQH 120
Query: 121 IVNLKTPATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQAL 180
++NL+ PATP FFNTLYDP+ +GADFVRGYPFSLRSGV CA SCGLWLNLADLDAPTQAL
Sbjct: 121 VINLENPATPLFFNTLYDPYCEGADFVRGYPFSLRSGVPCAASCGLWLNLADLDAPTQAL 180
Query: 181 KPTQRNSRYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDI 240
K +RN+ YVDAV+TVP +AMLP+SGINIAFNRELVGPALVPAL LAGEGK+RWET+ED+
Sbjct: 181 KTEKRNTAYVDAVMTVPAKAMLPISGINIAFNRELVGPALVPALRLAGEGKVRWETLEDV 240
Query: 241 WCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQ 300
WCG+C+K + DHL GVK+GLPYVWRNERG+A++SL+K+WEG+KLME VPFF S+KLP+
Sbjct: 241 WCGMCLKHISDHLGYGVKTGLPYVWRNERGDAVESLRKKWEGMKLMEKSVPFFDSLKLPE 300
Query: 301 SATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
+A EDCVIE+AK+VKEQLG DP F +AADAM +WV+LW SV S+
Sbjct: 301 TALKVEDCVIELAKAVKEQLGSDDPAFTQAADAMVKWVQLWNSVNSS 347
>At3g02230 reversibly glycosylated polypeptide-1
Length = 357
Score = 362 bits (930), Expect = e-100
Identities = 169/334 (50%), Positives = 237/334 (70%), Gaps = 6/334 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ +HLIIV+D + + +PEGF ++Y ++I R
Sbjct: 20 DELDIVIPTIRN--LDFLEMWRPFLQPYHLIIVQDGDPSKTIAVPEGFDYELYNRNDINR 77
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD +G V+A+ QHI NL P
Sbjct: 78 ILGPKASCISFKDSACRCFGYMVSKKKYIFTIDDDCFVAKDPSGKAVNALEQHIKNLLCP 137
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TPFFFNTLYDP+R+GADFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 138 STPFFFNTLYDPYREGADFVRGYPFSLREGVSTAVSHGLWLNIPDYDAPTQLVKPKERNT 197
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+REL+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 198 RYVDAVMTIPKGTLFPMCGMNLAFDRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 254
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHL LGVK+GLPY++ ++ N +LKKE++G+ ED++PFFQS KL + A T +
Sbjct: 255 VICDHLGLGVKTGLPYIYHSKASNPFVNLKKEYKGIFWQEDIIPFFQSAKLTKEAVTVQQ 314
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C +E++K VKE+L +DP F K ADAM W++ W
Sbjct: 315 CYMELSKLVKEKLSPIDPYFDKLADAMVTWIEAW 348
>At5g15650 reversibly glycosylated polypeptide-2 (AtRGP)
Length = 360
Score = 360 bits (923), Expect = e-100
Identities = 166/334 (49%), Positives = 239/334 (70%), Gaps = 6/334 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ +HLIIV+D +++ +PEG+ ++Y ++I R
Sbjct: 20 DELDIVIPTIRN--LDFLEMWRPFLQPYHLIIVQDGDPSKKIHVPEGYDYELYNRNDINR 77
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD +G V+A+ QHI NL P
Sbjct: 78 ILGPKASCISFKDSACRCFGYMVSKKKYIFTIDDDCFVAKDPSGKAVNALEQHIKNLLCP 137
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
++PFFFNTLYDP+R+GADFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 138 SSPFFFNTLYDPYREGADFVRGYPFSLREGVSTAVSHGLWLNIPDYDAPTQLVKPKERNT 197
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+R+L+GPA+ L+ G+ R+ +D+W G C+K
Sbjct: 198 RYVDAVMTIPKGTLFPMCGMNLAFDRDLIGPAMYFGLMGDGQPIGRY---DDMWAGWCIK 254
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDHLSLGVK+GLPY++ ++ N +LKKE++G+ E+++PFFQ+ KL + A T +
Sbjct: 255 VICDHLSLGVKTGLPYIYHSKASNPFVNLKKEYKGIFWQEEIIPFFQNAKLSKEAVTVQQ 314
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLW 341
C IE++K VKE+L +DP F K ADAM W++ W
Sbjct: 315 CYIELSKMVKEKLSSLDPYFDKLADAMVTWIEAW 348
>At3g08900 reversibly glycosylated polypeptide-3 (RGP)
Length = 362
Score = 360 bits (923), Expect = e-100
Identities = 167/340 (49%), Positives = 237/340 (69%), Gaps = 6/340 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+E+DIVI + + F+ W+ F ++HLIIV+D + + IP GF ++Y ++I R
Sbjct: 16 DELDIVIPTIRN--LDFLEMWRPFFEQYHLIIVQDGDPSKVINIPVGFDYELYNRNDINR 73
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G S I F ACR FG++VSKKKY+ IDDDC AKD G ++A+ QHI NL +P
Sbjct: 74 ILGPKASCISFKDSACRCFGYMVSKKKYIYTIDDDCFVAKDPTGKEINALEQHIKNLLSP 133
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TP FFNTLYDP+R GADFVRGYPFS+R G A+S GLWLN+ D DAPTQ +KP ++NS
Sbjct: 134 STPHFFNTLYDPYRDGADFVRGYPFSMREGAITAVSHGLWLNIPDYDAPTQLVKPLEKNS 193
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P + P+ G+N+AF+REL+GPA+ L+ G+ R+ +D+W G CVK
Sbjct: 194 RYVDAVMTIPKGTLFPMCGMNLAFDRELIGPAMYFGLMGDGQPIGRY---DDMWAGWCVK 250
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
++CDH+ GVK+GLPY+W ++ N +LKKE+ G+ E+ +PFFQSV LP+ T+ +
Sbjct: 251 VICDHMGWGVKTGLPYIWHSKASNPFVNLKKEYNGIFWQEEAIPFFQSVTLPKECTSVQQ 310
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSVGSA 347
C +E+AK V+E+LGKVDP F+ A M W++ W+ + SA
Sbjct: 311 CYLELAKLVREKLGKVDPYFITLATGMVTWIEAWEELNSA 350
>At5g50750 UDP-glucose:protein transglucosylase; reversibly
glycosylated polypeptide
Length = 364
Score = 355 bits (910), Expect = 3e-98
Identities = 166/337 (49%), Positives = 236/337 (69%), Gaps = 6/337 (1%)
Query: 9 NEVDIVIGALHSNLTPFMNEWKSIFSRFHLIIVKDPALKEELQIPEGFSADVYTNSEIER 68
+++DIVI + S F+ +W+ +HLIIV+D ++++PEG+ ++Y ++I R
Sbjct: 16 DDLDIVIPTIRS--LDFLEQWRPFLHHYHLIIVQDGDPSIKIRVPEGYDYELYNRNDINR 73
Query: 69 VVGSSTS-IRFSGYACRYFGFLVSKKKYVVCIDDDCVPAKDDAGNVVDAVAQHIVNLKTP 127
++G + I + CR FGF+VSKKKY+ IDDDC AKD +G ++ +AQHI NL+TP
Sbjct: 74 ILGPRANCISYKDGGCRCFGFMVSKKKYIYTIDDDCFVAKDPSGKDINVIAQHIKNLETP 133
Query: 128 ATPFFFNTLYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLDAPTQALKPTQRNS 187
+TP +FNTLYDPFR G DFVRGYPFSLR GV A+S GLWLN+ D DAPTQ +KP +RN+
Sbjct: 134 STPHYFNTLYDPFRDGTDFVRGYPFSLREGVQTAISHGLWLNIPDYDAPTQLVKPRERNT 193
Query: 188 RYVDAVLTVPTRAMLPVSGINIAFNRELVGPALVPALVLAGEGKLRWETVEDIWCGLCVK 247
RYVDAV+T+P R + P+ G+N+AFNRELVGPA+ L+ G+ R+ +D+W G K
Sbjct: 194 RYVDAVMTIPKRVLYPMCGMNLAFNRELVGPAMYFGLMGEGQPISRY---DDMWAGWAAK 250
Query: 248 IVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAED 307
+VCDHL GVK+GLPY+W ++ N +LKKE +G+ ED+VPFFQ+++L + + TA
Sbjct: 251 VVCDHLGFGVKTGLPYLWHSKASNPFVNLKKEHKGLHWQEDMVPFFQNLRLSKESDTAAK 310
Query: 308 CVIEMAKSVKEQLGKVDPMFLKAADAMAEWVKLWKSV 344
C +E++ KE+L KVDP F K ADAM W++ W+ +
Sbjct: 311 CYMEISNMTKEKLTKVDPYFEKLADAMVVWIEAWEEL 347
>At3g55120 chalcone isomerase
Length = 246
Score = 30.4 bits (67), Expect = 1.5
Identities = 22/85 (25%), Positives = 37/85 (42%), Gaps = 15/85 (17%)
Query: 270 GNAIDSLKKEWEG--VKLMEDVVPFFQSV-------------KLPQSATTAEDCVIEMAK 314
GNA+ SL +W+G + + + +PFF+ + KLP + + V E
Sbjct: 67 GNAVPSLSVKWKGKTTEELTESIPFFREIVTGAFEKFIKVTMKLPLTGQQYSEKVTENCV 126
Query: 315 SVKEQLGKVDPMFLKAADAMAEWVK 339
++ +QLG KA + E K
Sbjct: 127 AIWKQLGLYTDCEAKAVEKFLEIFK 151
>At1g43840 hypothetical protein
Length = 418
Score = 29.3 bits (64), Expect = 3.3
Identities = 17/53 (32%), Positives = 26/53 (48%), Gaps = 2/53 (3%)
Query: 265 WRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAE--DCVIEMAKS 315
W+ D K +W + + +V F+Q K + TTAE D +I MAK+
Sbjct: 151 WKGNWKEKGDEAKPKWIDPDVSKGLVQFWQDPKSEKRVTTAEMLDIMIRMAKA 203
>At5g62190 putative RNA helicase (MMI9.2)
Length = 671
Score = 28.9 bits (63), Expect = 4.3
Identities = 21/78 (26%), Positives = 37/78 (46%), Gaps = 9/78 (11%)
Query: 259 SGLPYVWRNERGNAIDSLKKEWEGVKLMEDVVPFFQSVKLPQSATTAEDCVIEMAKSVKE 318
+G+ + R +++ ++KE G+K F+ + PQ A +E A+ VK+
Sbjct: 443 TGVAVTLYDSRKSSVSRIEKE-AGIK--------FEHLAAPQPDEIARSGGMEAAEKVKQ 493
Query: 319 QLGKVDPMFLKAADAMAE 336
V P FL+AA + E
Sbjct: 494 VCDSVVPAFLEAAKELLE 511
>At5g50230 putative protein
Length = 515
Score = 28.9 bits (63), Expect = 4.3
Identities = 13/51 (25%), Positives = 23/51 (44%), Gaps = 8/51 (15%)
Query: 241 WCGLCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKE--------WEGV 283
W C+ D+++ G G +VW +GN + LK++ W G+
Sbjct: 448 WSRSCISPDDDYVAAGSADGSVHVWSLSKGNIVSILKEQTSPILCCSWSGI 498
>At4g20400 unknown protein
Length = 954
Score = 28.5 bits (62), Expect = 5.7
Identities = 10/39 (25%), Positives = 22/39 (55%)
Query: 136 LYDPFRKGADFVRGYPFSLRSGVDCALSCGLWLNLADLD 174
+Y ++ +F+ +P + SG +C +C +N+A +D
Sbjct: 378 VYRAVQRSGEFILTFPKAYHSGFNCGFNCAEAVNVAPVD 416
>At4g13400 putative protein
Length = 306
Score = 28.5 bits (62), Expect = 5.7
Identities = 21/56 (37%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 30 KSIFSRFHLII-VKDPALKEELQIPEGFSADV-YTNSEIERVVGSSTSIRFSGYAC 83
++IFSR + + DPA +Q G V Y EI +G +TSI SGY C
Sbjct: 173 RAIFSRDSVEVPCPDPASGLYIQTRSGQIVKVVYGEDEIAYQIGETTSILSSGYLC 228
>At3g12470 unknown protein
Length = 220
Score = 27.7 bits (60), Expect = 9.7
Identities = 27/102 (26%), Positives = 41/102 (39%), Gaps = 9/102 (8%)
Query: 244 LCVKIVCDHLSLGVKSGLPYVWRNERGNAIDSLKKEWEG--VKLME------DVVPFFQS 295
LCV C + LG GLP V R + + W G K +E ++
Sbjct: 80 LCVGSRCLIIQLGYNYGLPKVLRTFLADPKTTFVGVWNGQDQKKLEKCRHRVEIGKLLDI 139
Query: 296 VKLPQSATTAEDCVIEMAKSVKEQLGKVDPMFLKAADAMAEW 337
+ + A C + VKE+LG+V + L A M++W
Sbjct: 140 RMFVRDSRGARMCFCSFEQIVKERLGRVG-VRLDPAICMSDW 180
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,154,608
Number of Sequences: 26719
Number of extensions: 349222
Number of successful extensions: 780
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 765
Number of HSP's gapped (non-prelim): 12
length of query: 347
length of database: 11,318,596
effective HSP length: 100
effective length of query: 247
effective length of database: 8,646,696
effective search space: 2135733912
effective search space used: 2135733912
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148994.1


