

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148971.3 - phase: 0
(226 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
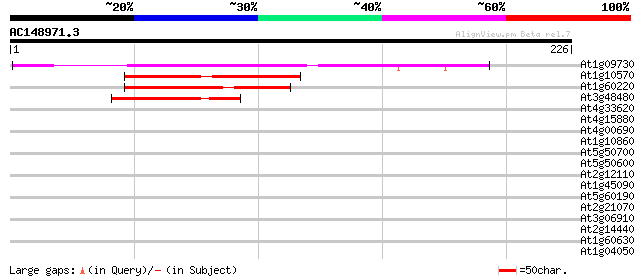
At1g09730 unknown protein 105 3e-23
At1g10570 protein-tyrosine phosphatase 2 like protein 67 1e-11
At1g60220 unknown protein 60 7e-10
At3g48480 putative protein 51 4e-07
At4g33620 putative protein 35 0.032
At4g15880 unknown protein 34 0.072
At4g00690 hypothetical protein 32 0.21
At1g10860 receptor-kinase isolog, 5' partial 32 0.21
At5g50700 11-beta-hydroxysteroid dehydrogenase-like 31 0.47
At5g50600 11-beta-hydroxysteroid dehydrogenase-like 31 0.47
At2g12110 hypothetical protein 29 2.3
At1g45090 hypothetical protein 29 2.3
At5g60190 unknown protein 28 3.0
At2g21070 unknown protein 28 3.9
At3g06910 unknown protein 28 5.1
At2g14440 putative receptor-like protein kinase 28 5.1
At1g60630 receptor kinase like protein 27 6.7
At1g04050 hypothetical protein 27 8.8
>At1g09730 unknown protein
Length = 865
Score = 105 bits (261), Expect = 3e-23
Identities = 67/195 (34%), Positives = 99/195 (50%), Gaps = 36/195 (18%)
Query: 2 ADDFSSKFLQLRFISLEVSSLQMNLTNSVELFIDFCFSLPPFFFFQLPQQDNFYDCGLFL 61
+DD SS+F+ LRF+SLE LPQQ+N +DCGLFL
Sbjct: 485 SDDISSRFMNLRFVSLE-----------------------------LPQQENSFDCGLFL 515
Query: 62 LYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQNLIYDIFENGSLKAP 121
L+++E FL EAP+ F+PFKI S FL NWFP EASL+R+ IQ LI+++ EN S +
Sbjct: 516 LHYLELFLAEAPLNFSPFKIYNASNFLYLNWFPPAEASLKRTLIQKLIFELLENRSREV- 574
Query: 122 PIDCRGKGPLSELPGVIEHKVEADSSGASCYPGI-WHGNLSNGSTETDIQFRPV--SPVR 178
+ E P + + + C P I +G+++ + I+ + S +R
Sbjct: 575 ---SNEQNQSCESPVAVNDDMGIEVLSERCSPLIDCNGDMTQTQDDQGIEMTLLERSSMR 631
Query: 179 AASCSRDPGIVFKDL 193
+ D G+V +DL
Sbjct: 632 HIQAANDSGMVLRDL 646
>At1g10570 protein-tyrosine phosphatase 2 like protein
Length = 571
Score = 66.6 bits (161), Expect = 1e-11
Identities = 31/71 (43%), Positives = 45/71 (62%), Gaps = 4/71 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
Q+PQQ N +DCGLFLL+F+ RF+EEAP + + K ++ WF +EAS R I
Sbjct: 502 QVPQQKNDFDCGLFLLFFIRRFIEEAPQRLT----LQDLKMIHKKWFKPEEASALRIKIW 557
Query: 107 NLIYDIFENGS 117
N++ D+F G+
Sbjct: 558 NILVDLFRKGN 568
>At1g60220 unknown protein
Length = 604
Score = 60.5 bits (145), Expect = 7e-10
Identities = 29/67 (43%), Positives = 42/67 (62%), Gaps = 4/67 (5%)
Query: 47 QLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWFPSDEASLRRSHIQ 106
Q+PQQ N +DCG F+L+F++RF+EEAP + + F K WF DEAS R I+
Sbjct: 535 QVPQQKNDFDCGPFVLFFIKRFIEEAPQRLKRKDLGMFDK----KWFRPDEASALRIKIR 590
Query: 107 NLIYDIF 113
N + ++F
Sbjct: 591 NTLIELF 597
>At3g48480 putative protein
Length = 169
Score = 51.2 bits (121), Expect = 4e-07
Identities = 22/52 (42%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query: 42 PFFFFQLPQQDNFYDCGLFLLYFVERFLEEAPIKFNPFKITKFSKFLNSNWF 93
PF+ +PQQ N +CG F+LY++ RF+E+AP FN + FL +WF
Sbjct: 102 PFYVPMVPQQTNDVECGSFVLYYIHRFIEDAPENFN---VEDMPYFLKEDWF 150
>At4g33620 putative protein
Length = 710
Score = 35.0 bits (79), Expect = 0.032
Identities = 25/88 (28%), Positives = 43/88 (48%), Gaps = 5/88 (5%)
Query: 91 NWFPSDEASLRRSHIQNLIYDIFENGSLKAPPIDCRGKGPLSELPGVIEHKVEADSSGAS 150
NWFP+ EASL+R +I L+Y++ + P + + + P + + + E+++
Sbjct: 408 NWFPAKEASLKRRNILELLYNLHKGHDPSILPANSKSEPPHCGVSNRNDQETESENVIEC 467
Query: 151 CYPGIWHGNL-SNGSTETDI-QFRPVSP 176
C W + ST TDI Q + SP
Sbjct: 468 CN---WIKPFDGSSSTVTDISQTKTCSP 492
>At4g15880 unknown protein
Length = 489
Score = 33.9 bits (76), Expect = 0.072
Identities = 13/25 (52%), Positives = 18/25 (72%)
Query: 44 FFFQLPQQDNFYDCGLFLLYFVERF 68
F LPQQ N YDCG+F+L +++ F
Sbjct: 435 FVEDLPQQKNGYDCGMFMLKYIDFF 459
>At4g00690 hypothetical protein
Length = 233
Score = 32.3 bits (72), Expect = 0.21
Identities = 11/29 (37%), Positives = 20/29 (68%)
Query: 49 PQQDNFYDCGLFLLYFVERFLEEAPIKFN 77
PQQ N YDCG+F+L +++ + ++F+
Sbjct: 177 PQQQNGYDCGMFMLKYIDFYSRGLSLQFS 205
>At1g10860 receptor-kinase isolog, 5' partial
Length = 604
Score = 32.3 bits (72), Expect = 0.21
Identities = 28/102 (27%), Positives = 38/102 (36%), Gaps = 4/102 (3%)
Query: 108 LIYDIFENGSLKAPPIDCRGKGPLSELPGVIEHKVEADSSGASCY----PGIWHGNLSNG 163
L+YD F NGSL R G L K+ D + A Y PG+ HGNL +
Sbjct: 369 LVYDYFPNGSLFTLIHGTRASGSGKPLHWTSCLKIAEDLASALLYIHQNPGLTHGNLKSS 428
Query: 164 STETDIQFRPVSPVRAASCSRDPGIVFKDLQAAVVPPHFDCR 205
+ F S DP V + ++ +CR
Sbjct: 429 NVLLGPDFESCLTDYGLSTLHDPDSVEETSAVSLFYKAPECR 470
>At5g50700 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 31.2 bits (69), Expect = 0.47
Identities = 13/27 (48%), Positives = 16/27 (59%)
Query: 24 MNLTNSVELFIDFCFSLPPFFFFQLPQ 50
+NLT F CF LPPF+FF+ Q
Sbjct: 8 LNLTAPFFTFFGLCFFLPPFYFFKFLQ 34
>At5g50600 11-beta-hydroxysteroid dehydrogenase-like
Length = 349
Score = 31.2 bits (69), Expect = 0.47
Identities = 13/27 (48%), Positives = 16/27 (59%)
Query: 24 MNLTNSVELFIDFCFSLPPFFFFQLPQ 50
+NLT F CF LPPF+FF+ Q
Sbjct: 8 LNLTAPFFTFFGLCFFLPPFYFFKFLQ 34
>At2g12110 hypothetical protein
Length = 550
Score = 28.9 bits (63), Expect = 2.3
Identities = 12/46 (26%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query: 33 FIDFCFSLPPFFFFQ---LPQQDNFYDCGLFLLYFVERFLEEAPIK 75
++D + + PF + + LPQ DCG + + F+E ++ P++
Sbjct: 268 YLDKPYPVTPFTYIRNQRLPQNPTTGDCGPYAMKFIELYMLNTPVE 313
>At1g45090 hypothetical protein
Length = 1210
Score = 28.9 bits (63), Expect = 2.3
Identities = 12/46 (26%), Positives = 25/46 (54%), Gaps = 3/46 (6%)
Query: 33 FIDFCFSLPPFFFFQ---LPQQDNFYDCGLFLLYFVERFLEEAPIK 75
++D + + PF + + LPQ DCG + + F+E ++ P++
Sbjct: 1135 YLDKPYPVTPFTYIRNQRLPQNPTTGDCGPYAMKFIELYMLNTPVE 1180
>At5g60190 unknown protein
Length = 226
Score = 28.5 bits (62), Expect = 3.0
Identities = 11/14 (78%), Positives = 12/14 (85%)
Query: 49 PQQDNFYDCGLFLL 62
PQQ N YDCG+FLL
Sbjct: 158 PQQKNGYDCGVFLL 171
>At2g21070 unknown protein
Length = 374
Score = 28.1 bits (61), Expect = 3.9
Identities = 23/99 (23%), Positives = 41/99 (41%), Gaps = 26/99 (26%)
Query: 35 DFCFSLPPFFFFQLPQQDNFYDCGL-----------------FLLYFVERFLEEAPIKFN 77
DFC S PPFF + F + GL FV R ++++ +
Sbjct: 235 DFCMSNPPFF-------ETFEEAGLNPKTSCGGTPEEMVCNGGEQAFVSRIIKDSAVLRQ 287
Query: 78 PFKITKFSKFLNSNWFPSDEASLRRSHIQNLIYDIFENG 116
F+ +F N W+ S ++++++ LI ++E G
Sbjct: 288 RFRTYRFVNMFNIRWYTSMLG--KKANLKLLISKLWEVG 324
>At3g06910 unknown protein
Length = 502
Score = 27.7 bits (60), Expect = 5.1
Identities = 9/23 (39%), Positives = 16/23 (69%)
Query: 44 FFFQLPQQDNFYDCGLFLLYFVE 66
F LP Q N +DCG+F++ +++
Sbjct: 448 FVQDLPMQRNGFDCGMFMVKYID 470
>At2g14440 putative receptor-like protein kinase
Length = 886
Score = 27.7 bits (60), Expect = 5.1
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 5/68 (7%)
Query: 108 LIYDIFENGSLKAPPIDCRGKGPLSELPG----VIEHKVEADSSGASCYPGIWHGNLSNG 163
LIY+ ENG+LK RG GP+ PG IE + + C P + H ++ +
Sbjct: 651 LIYEFMENGNLKEHLSGKRG-GPVLNWPGRLKIAIESALGIEYLHIGCKPPMVHRDVKST 709
Query: 164 STETDIQF 171
+ ++F
Sbjct: 710 NILLGLRF 717
>At1g60630 receptor kinase like protein
Length = 652
Score = 27.3 bits (59), Expect = 6.7
Identities = 26/104 (25%), Positives = 39/104 (37%), Gaps = 4/104 (3%)
Query: 108 LIYDIFENGSLKAPPIDCRGKGPLSELPGVIEHKVEADSSGASCY----PGIWHGNLSNG 163
L+YD F NGSL + + G L K+ D + Y PG+ HGNL +
Sbjct: 421 LVYDYFPNGSLFSLIHGSKVSGSGKPLHWTSCLKIAEDLAMGLVYIHQNPGLTHGNLKSS 480
Query: 164 STETDIQFRPVSPVRAASCSRDPGIVFKDLQAAVVPPHFDCRQM 207
+ F S DP + A++ +CR +
Sbjct: 481 NVLLGPDFESCLTDYGLSDLHDPYSIEDTSAASLFYKAPECRDL 524
>At1g04050 hypothetical protein
Length = 468
Score = 26.9 bits (58), Expect = 8.8
Identities = 20/66 (30%), Positives = 30/66 (45%), Gaps = 11/66 (16%)
Query: 139 EHKVEADSSGASCYPGIWHGNLSNGSTETDIQFRPVSPVRAASCSRDPGIVFKDLQAAVV 198
E ++E D A C G+++GN+S DI+ + D GI F D + +
Sbjct: 391 EERLEGDK--ALCLDGMFYGNISRFLNHRDIE-------AMEELAWDYGIDFNDNDSLMK 441
Query: 199 PPHFDC 204
P FDC
Sbjct: 442 P--FDC 445
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.141 0.439
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,432,935
Number of Sequences: 26719
Number of extensions: 236162
Number of successful extensions: 615
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 600
Number of HSP's gapped (non-prelim): 19
length of query: 226
length of database: 11,318,596
effective HSP length: 96
effective length of query: 130
effective length of database: 8,753,572
effective search space: 1137964360
effective search space used: 1137964360
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148971.3


