

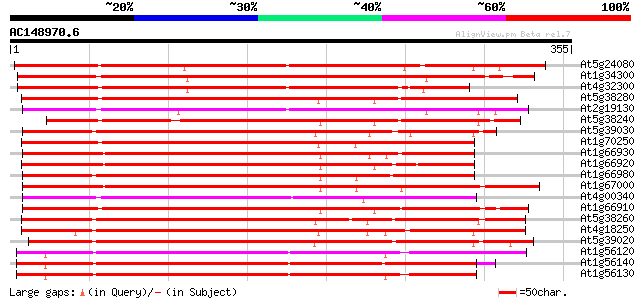
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.6 - phase: 0
(355 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g24080 receptor-like protein kinase 268 3e-72
At1g34300 putative protein 259 2e-69
At4g32300 S-receptor kinase -like protein 255 3e-68
At5g38280 receptor serine/threonine kinase PR5K 245 2e-65
At2g19130 putative receptor-like protein kinase 243 2e-64
At5g38240 receptor serine/threonine protein kinase -like 242 3e-64
At5g39030 receptor protein kinase - like protein 241 3e-64
At1g70250 hypothetical protein 241 4e-64
At1g66930 receptor serine/threonine kinase PR5K, putative 238 3e-63
At1g66920 receptor serine/threonine kinase PR5K, putative 238 4e-63
At1g66980 putative kinase 238 5e-63
At1g67000 hypothetical protein 237 6e-63
At4g00340 receptor-like protein kinase 237 8e-63
At1g66910 receptor serine/threonine kinase PR5K, putative 236 1e-62
At5g38260 receptor serine/threonine protein kinase - like 235 2e-62
At4g18250 receptor serine/threonine kinase-like protein 234 5e-62
At5g39020 receptor protein kinase - like protein 231 5e-61
At1g56120 receptor-like protein kinase, putative 230 1e-60
At1g56140 receptor-like protein kinase, putative 228 5e-60
At1g56130 receptor-like protein kinase, putative 224 6e-59
>At5g24080 receptor-like protein kinase
Length = 872
Score = 268 bits (685), Expect = 3e-72
Identities = 160/355 (45%), Positives = 214/355 (60%), Gaps = 23/355 (6%)
Query: 4 EKPIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMA 63
+ P+ FT + L+ T+N+S LLGSGGFGTVYKG T+VAVK L + E +F+
Sbjct: 515 DSPVSFTYRDLQNCTNNFSQLLGSGGFGTVYKGTVAGETLVAVKRLDRALSHG-EREFIT 573
Query: 64 EVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEK---NALGYEKLHEI 120
EV TIG +HH NLV+L G+C E + LVYEYM NGSLD+++ + N L + EI
Sbjct: 574 EVNTIGSMHHMNLVRLCGYCSEDSHRLLVYEYMINGSLDKWIFSSEQTANLLDWRTRFEI 633
Query: 121 AIGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGG 180
A+ TA+GIAY HE C +RI+H DIKP NILLD NF PKV+DFGLAK+ RE++H+ +T
Sbjct: 634 AVATAQGIAYFHEQCRNRIIHCDIKPENILLDDNFCPKVSDFGLAKMMGREHSHV-VTMI 692
Query: 181 RGTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFD 240
RGT GY APE PIT K DVYS+GMLL EIVG RRNLD+ ++P W +K+
Sbjct: 693 RGTRGYLAPEWVSNRPITVKADVYSYGMLLLEIVGGRRNLDMSYDAEDFFYPGWAYKELT 752
Query: 241 AGLLEEAM--IVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGS---LEI 295
G +A+ + G+ E+ + + +KVA WC+Q +RP M +VVK+LEG+ + +
Sbjct: 753 NGTSLKAVDKRLQGVAEEEEVV--KALKVAFWCIQDEVSMRPSMGEVVKLLEGTSDEINL 810
Query: 296 PKTFNPFQHLIDE----------TKFTTH-SDQESNTYTTSVSSVMVSDSNIVCA 339
P LI+E +F S NT TTS S S S+ C+
Sbjct: 811 PPMPQTILELIEEGLEDVYRAMRREFNNQLSSLTVNTITTSQSYRSSSRSHATCS 865
>At1g34300 putative protein
Length = 829
Score = 259 bits (661), Expect = 2e-69
Identities = 146/331 (44%), Positives = 199/331 (60%), Gaps = 14/331 (4%)
Query: 6 PIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
P++FT ++L+ T ++ LG+GGFGTVY+G+ N T+VAVK L G E+QF EV
Sbjct: 471 PVQFTYKELQRCTKSFKEKLGAGGFGTVYRGVLTNRTVVAVKQLEGIEQG--EKQFRMEV 528
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA--LGYEKLHEIAIG 123
TI HH NLV+L GFC + LVYE+M NGSLD +L +A L +E IA+G
Sbjct: 529 ATISSTHHLNLVRLIGFCSQGRHRLLVYEFMRNGSLDNFLFTTDSAKFLTWEYRFNIALG 588
Query: 124 TARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGT 183
TA+GI YLHE C IVH DIKP NIL+D NF KV+DFGLAKL N ++ M+ RGT
Sbjct: 589 TAKGITYLHEECRDCIVHCDIKPENILVDDNFAAKVSDFGLAKLLNPKDNRYNMSSVRGT 648
Query: 184 PGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGL 243
GY APE PIT K DVYS+GM+L E+V +RN D+ + + F IW +++F+ G
Sbjct: 649 RGYLAPEWLANLPITSKSDVYSYGMVLLELVSGKRNFDVSEKTNHKKFSIWAYEEFEKGN 708
Query: 244 LEEAMIVCGIEEKNREIAE--RMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNP 301
+ + E++ ++ + RMVK + WC+Q + RP M VV+MLEG EI P
Sbjct: 709 TKAILDTRLSEDQTVDMEQVMRMVKTSFWCIQEQPLQRPTMGKVVQMLEGITEIKNPLCP 768
Query: 302 FQHLIDETKFTTHSDQESNTYTTSVSSVMVS 332
I E F+ N+ +TS +S+ V+
Sbjct: 769 --KTISEVSFS------GNSMSTSHASMFVA 791
>At4g32300 S-receptor kinase -like protein
Length = 821
Score = 255 bits (651), Expect = 3e-68
Identities = 143/293 (48%), Positives = 191/293 (64%), Gaps = 13/293 (4%)
Query: 6 PIRFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEV 65
PIRF + L+ AT+N+S LG GGFG+VY+G +G+ +AVK L G K ++F AEV
Sbjct: 480 PIRFAYKDLQSATNNFSVKLGQGGFGSVYEGTLPDGSRLAVKKLEGIGQGK--KEFRAEV 537
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA---LGYEKLHEIAI 122
IG IHH +LV+L GFC E L YE++ GSL+R++ +K+ L ++ IA+
Sbjct: 538 SIIGSIHHLHLVRLRGFCAEGAHRLLAYEFLSKGSLERWIFRKKDGDVLLDWDTRFNIAL 597
Query: 123 GTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRG 182
GTA+G+AYLHE C+ RIVH DIKP NILLD NFN KV+DFGLAKL RE +H+ T RG
Sbjct: 598 GTAKGLAYLHEDCDARIVHCDIKPENILLDDNFNAKVSDFGLAKLMTREQSHV-FTTMRG 656
Query: 183 TPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAG 242
T GY APE + I+ K DVYS+GM+L E++G R+N D T + FP + +KK + G
Sbjct: 657 TRGYLAPEWITNYAISEKSDVYSYGMVLLELIGGRKNYDPSETSEKCHFPSFAFKKMEEG 716
Query: 243 LLEEAMIVCGIEEKNREI----AERMVKVALWCVQYRQQLRPMMSDVVKMLEG 291
L + IV G + KN ++ +R +K ALWC+Q Q RP MS VV+MLEG
Sbjct: 717 KLMD--IVDG-KMKNVDVTDERVQRAMKTALWCIQEDMQTRPSMSKVVQMLEG 766
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 245 bits (626), Expect = 2e-65
Identities = 132/322 (40%), Positives = 208/322 (63%), Gaps = 11/322 (3%)
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKG-IFNNGTMVAVKVLRGSSDKKIEEQFMAEVG 66
R++ +++ T++++++LG GGFGTVYKG + ++G VAVK+L+ S E+F+ EV
Sbjct: 320 RYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKILKVSEGNG--EEFINEVA 377
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYL-LHEKNALGYEKLHEIAIGTA 125
++ R H N+V L GFC+EKN A++YE+M NGSLD+Y+ + + +E+L+++A+G +
Sbjct: 378 SMSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISANMSTKMEWERLYDVAVGIS 437
Query: 126 RGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPG 185
RG+ YLH C RIVH+DIKP NIL+D N PK++DFGLAKLC + + I+M RGT G
Sbjct: 438 RGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAKLCKNKESIISMLHMRGTFG 497
Query: 186 YAAPELWMP--FPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQE---WFPIWVWKKFD 240
Y APE++ ++HK DVYS+GM++ E++G + ++ + S +FP WV+K F+
Sbjct: 498 YIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEYSGSNNGSMYFPEWVYKDFE 557
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE-IPKTF 299
G + + I ++ +IA+++V VALWC+Q RP M V++MLEG+LE +
Sbjct: 558 KGEITR-IFGDSITDEEEKIAKKLVLVALWCIQMNPSDRPPMIKVIEMLEGNLEALQVPP 616
Query: 300 NPFQHLIDETKFTTHSDQESNT 321
NP +ET T D + +
Sbjct: 617 NPLLFSPEETVPDTLEDSDDTS 638
>At2g19130 putative receptor-like protein kinase
Length = 828
Score = 243 bits (619), Expect = 2e-64
Identities = 147/338 (43%), Positives = 202/338 (59%), Gaps = 21/338 (6%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
F+ ++L+ AT N+S+ LG GGFG+V+KG + + +AVK L G S E+QF EV TI
Sbjct: 483 FSYRELQNATKNFSDKLGGGGFGSVFKGALPDSSDIAVKRLEGISQG--EKQFRTEVVTI 540
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYL----LHEKNALGYEKLHEIAIGT 124
G I H NLV+L GFC E + LVY+YM NGSLD +L + EK LG++ +IA+GT
Sbjct: 541 GTIQHVNLVRLRGFCSEGSKKLLVYDYMPNGSLDSHLFLNQVEEKIVLGWKLRFQIALGT 600
Query: 125 ARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
ARG+AYLH+ C I+H DIKP NILLD F PKVADFGLAKL R+ + + +T RGT
Sbjct: 601 ARGLAYLHDECRDCIIHCDIKPENILLDSQFCPKVADFGLAKLVGRDFSRV-LTTMRGTR 659
Query: 185 GYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDAGLL 244
GY APE IT K DVYS+GM+LFE+V RRN + E +FP W
Sbjct: 660 GYLAPEWISGVAITAKADVYSYGMMLFELVSGRRNTEQSENEKVRFFPSWAATILTKDGD 719
Query: 245 EEAMIVCGIEEKNREIAE--RMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI--PKTFN 300
+++ +E +I E R KVA WC+Q + RP MS VV++LEG LE+ P
Sbjct: 720 IRSLVDPRLEGDAVDIEEVTRACKVACWCIQDEESHRPAMSQVVQILEGVLEVNPPPFPR 779
Query: 301 PFQHLI----------DETKFTTHSDQESNTYTTSVSS 328
Q L+ + + ++H+ +++ +++S SS
Sbjct: 780 SIQALVVSDEDVVFFTESSSSSSHNSSQNHKHSSSSSS 817
>At5g38240 receptor serine/threonine protein kinase -like
Length = 588
Score = 242 bits (617), Expect = 3e-64
Identities = 132/311 (42%), Positives = 196/311 (62%), Gaps = 21/311 (6%)
Query: 24 LLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIHHFNLVKLFGFC 83
++G GGFGTVYKG +G VAVK+L+ S+ E F+ EV +I + H N+V L GFC
Sbjct: 286 VVGRGGFGTVYKGNLRDGRKVAVKILKDSNGNC--EDFINEVASISQTSHVNIVSLLGFC 343
Query: 84 FEKNLIALVYEYMGNGSLDRYLLHEKNALGYEKLHEIAIGTARGIAYLHELCEHRIVHYD 143
FEK+ A+VYE++ NGSLD + + L L+ IA+G ARGI YLH C+ RIVH+D
Sbjct: 344 FEKSKRAIVYEFLENGSLD-----QSSNLDVSTLYGIALGVARGIEYLHFGCKKRIVHFD 398
Query: 144 IKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPELWMPF--PITHKC 201
IKP N+LLD N PKVADFGLAKLC ++ + +++ RGT GY APEL+ ++HK
Sbjct: 399 IKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRVYGNVSHKS 458
Query: 202 DVYSFGMLLFEIVGRRRNLDIKNTESQE---WFPIWVWKKFDAGLLEEAMIVCGIEEKNR 258
DVYS+GML+ E+ G R ++N +S +FP W++K + G + ++ G+ +
Sbjct: 459 DVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLENGDYVK-LLADGLTREEE 517
Query: 259 EIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI------PKTFNPFQHLIDETKFT 312
+IA++M+ V LWC+Q+R RP M+ VV M+EG+L+ P P Q+ + + +
Sbjct: 518 DIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPKPLLHMPMQN--NNAESS 575
Query: 313 THSDQESNTYT 323
S+++S+ Y+
Sbjct: 576 QPSEEDSSIYS 586
>At5g39030 receptor protein kinase - like protein
Length = 806
Score = 241 bits (616), Expect = 3e-64
Identities = 137/311 (44%), Positives = 197/311 (63%), Gaps = 18/311 (5%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
+T +L+ T ++S ++G GGFGTVY G +NG VAVKVL+ K E F+ EV ++
Sbjct: 488 YTYAELKKITKSFSYIIGKGGFGTVYGGNLSNGRKVAVKVLKDL--KGSAEDFINEVASM 545
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNAL-GYEKLHEIAIGTARG 127
+ H N+V L GFCFE + A+VYE++ NGSLD+++ K+ L+ IA+G ARG
Sbjct: 546 SQTSHVNIVSLLGFCFEGSKRAIVYEFLENGSLDQFMSRNKSLTQDVTTLYGIALGIARG 605
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
+ YLH C+ RIVH+DIKP NILLDGN PKV+DFGLAKLC + + +++ RGT GY
Sbjct: 606 LEYLHYGCKTRIVHFDIKPQNILLDGNLCPKVSDFGLAKLCEKRESVLSLMDTRGTIGYI 665
Query: 188 APELW--MPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTE---SQEWFPIWVWKKFDAG 242
APE++ M ++HK DVYSFGML+ +++G R ++ + S +FP W++K + G
Sbjct: 666 APEVFSRMYGRVSHKSDVYSFGMLVIDMIGARSKEIVETVDSAASSTYFPDWIYKDLEDG 725
Query: 243 LLEEAMIVCG--IEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGS---LEIPK 297
E + G I ++ +EIA++M+ V LWC+Q RP M+ VV+M+EGS LEIP
Sbjct: 726 ---EQTWIFGDEITKEEKEIAKKMIVVGLWCIQPCPSDRPSMNRVVEMMEGSLDALEIPP 782
Query: 298 TFNPFQHLIDE 308
P H+ E
Sbjct: 783 --KPSMHISTE 791
>At1g70250 hypothetical protein
Length = 676
Score = 241 bits (615), Expect = 4e-64
Identities = 127/294 (43%), Positives = 191/294 (64%), Gaps = 9/294 (3%)
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTM-VAVKVLRGSSDKKIEEQFMAEVG 66
RF+ Q++ T ++ N+LG GGFGTVYKG +G+ VAVK+L+ S++ E F+ E+
Sbjct: 325 RFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDVAVKILKESNEDG--EDFINEIA 382
Query: 67 TIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTA 125
++ R H N+V L GFC+E A++YE M NGSLD+++ +A + ++ L+ IA+G +
Sbjct: 383 SMSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFISKNMSAKMEWKTLYNIAVGVS 442
Query: 126 RGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPG 185
G+ YLH C RIVH+DIKP NIL+DG+ PK++DFGLAKLC + I+M RGT G
Sbjct: 443 HGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISDFGLAKLCKNNESIISMLHARGTIG 502
Query: 186 YAAPELWMP--FPITHKCDVYSFGMLLFEIVGRR---RNLDIKNTESQEWFPIWVWKKFD 240
Y APE++ ++HK DVYS+GM++ E++G R R + ++ + +FP W++K +
Sbjct: 503 YIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNIGRAQNAGSSNTSMYFPDWIYKDLE 562
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
G + + EE++ +I ++MV V LWC+Q RP MS VV+MLEGSLE
Sbjct: 563 KGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNPYDRPPMSKVVEMLEGSLE 616
>At1g66930 receptor serine/threonine kinase PR5K, putative
Length = 876
Score = 238 bits (608), Expect = 3e-63
Identities = 129/295 (43%), Positives = 189/295 (63%), Gaps = 12/295 (4%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
+T Q++ T +++ ++G GGFG VY+G +G MVAVKVL+ S E+ F+ EV ++
Sbjct: 538 YTYAQVKRMTKSFAEVVGRGGFGIVYRGTLCDGRMVAVKVLKESKGNNSED-FINEVSSM 596
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTARG 127
+ H N+V L GFC E + A++YE++ NGSLD+++ + + L L+ IA+G ARG
Sbjct: 597 SQTSHVNIVSLLGFCSEGSRRAIIYEFLENGSLDKFISEKTSVILDLTALYGIALGVARG 656
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
+ YLH C+ RIVH+DIKP N+LLD N +PKV+DFGLAKLC ++ + +++ RGT GY
Sbjct: 657 LEYLHYGCKTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCEKKESVMSLMDTRGTIGYI 716
Query: 188 APELWMPF--PITHKCDVYSFGMLLFEIVGRRRNLDIKNTE---SQEWFPIWVWK---KF 239
APE+ ++HK DVYS+GML+FE++G R+ S +FP W++K K
Sbjct: 717 APEMISRVYGSVSHKSDVYSYGMLVFEMIGARKKERFGQNSANGSSMYFPEWIYKDLEKA 776
Query: 240 DAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
D G LE I GI + EIA++M V LWC+Q RP M+ VV+M+EGSL+
Sbjct: 777 DNGDLEH--IEIGISSEEEEIAKKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLD 829
>At1g66920 receptor serine/threonine kinase PR5K, putative
Length = 609
Score = 238 bits (607), Expect = 4e-63
Identities = 122/293 (41%), Positives = 195/293 (65%), Gaps = 10/293 (3%)
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGT 67
+++ +Q++ T++++ ++G GGFG VY+G ++G MVAVKVL+ E+ F+ EV +
Sbjct: 288 QYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGED-FINEVAS 346
Query: 68 IGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTAR 126
+ + H N+V L GFC E A++YE+M NGSLD+++ +K++ + + +L+ IA+G AR
Sbjct: 347 MSQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSSTMDWRELYGIALGVAR 406
Query: 127 GIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGY 186
G+ YLH C RIVH+DIKP N+LLD N +PKV+DFGLAKLC R+ + +++ RGT GY
Sbjct: 407 GLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGY 466
Query: 187 AAPELWMPF--PITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQE---WFPIWVWKKFDA 241
APE++ ++HK DVYS+GML+ +I+G R ++T S +FP W++K +
Sbjct: 467 IAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYKDLEK 526
Query: 242 GLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
G + ++ E++ EIA++M V LWC+Q RP M+ VV+M+EG+L+
Sbjct: 527 G--DNGRLIVNRSEED-EIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLD 576
>At1g66980 putative kinase
Length = 1109
Score = 238 bits (606), Expect = 5e-63
Identities = 127/293 (43%), Positives = 188/293 (63%), Gaps = 10/293 (3%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
+T Q++ T +++ ++G GGFG VYKG ++G +VAVKVL+ + K E F+ EV T+
Sbjct: 786 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDT--KGNGEDFINEVATM 843
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTARG 127
R H N+V L GFC E + A++YE++ NGSLD+++L + + + + L+ IA+G A G
Sbjct: 844 SRTSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGKTSVNMDWTALYRIALGVAHG 903
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
+ YLH C+ RIVH+DIKP N+LLD +F PKV+DFGLAKLC ++ + ++M RGT GY
Sbjct: 904 LEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIGYI 963
Query: 188 APELWMPF--PITHKCDVYSFGMLLFEIVGRRR----NLDIKNTESQEWFPIWVWKKFDA 241
APE+ ++HK DVYS+GML+ EI+G R N + S +FP WV++ ++
Sbjct: 964 APEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDLES 1023
Query: 242 GLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
I GI + E+A++M V LWC+Q RP M+ VV+M+EGSLE
Sbjct: 1024 -CKSGRHIEDGINSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLE 1075
>At1g67000 hypothetical protein
Length = 717
Score = 237 bits (605), Expect = 6e-63
Identities = 130/342 (38%), Positives = 213/342 (62%), Gaps = 18/342 (5%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
+T +++ T +++ ++G GGFG VY G ++ +MVAVKVL+ S E+ F+ EV ++
Sbjct: 371 YTYAEVKKMTKSFTEVVGRGGFGIVYSGTLSDSSMVAVKVLKDSKGTDGED-FINEVASM 429
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTARG 127
+ H N+V L GFC E + A++YE++GNGSLD+++ + + L + L+ IA+G ARG
Sbjct: 430 SQTSHVNIVSLLGFCCEGSRRAIIYEFLGNGSLDKFISDKSSVNLDLKTLYGIALGVARG 489
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
+ YLH C+ RIVH+DIKP N+LLD N PKV+DFGLAKLC ++ + +++ RGT GY
Sbjct: 490 LEYLHYGCKTRIVHFDIKPQNVLLDDNLCPKVSDFGLAKLCEKKESILSLLDTRGTIGYI 549
Query: 188 APELWMPF--PITHKCDVYSFGMLLFEIVGRRR----NLDIKNTESQEWFPIWVWKKFDA 241
APE+ ++HK DVYS+GML+ E++G R+ + + ++ S +FP W++K +
Sbjct: 550 APEMISRLYGSVSHKSDVYSYGMLVLEMIGARKKERFDQNSRSDGSSIYFPEWIYKDLEK 609
Query: 242 GLLEE-------AMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLE 294
+++ +I GI + EIA +M V LWC+Q RP M+ VV+M+EGSL+
Sbjct: 610 ANIKDIEKTENGGLIENGISSEEEEIARKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLD 669
Query: 295 IPKTFNPFQHLIDETKFTTHSDQESNT-YTTSVSSVMVSDSN 335
+ P + ++ + ++ SD N+ ++S S ++V +N
Sbjct: 670 ALEV--PPRPVLQQISASSVSDSFWNSEESSSASDILVFSTN 709
>At4g00340 receptor-like protein kinase
Length = 797
Score = 237 bits (604), Expect = 8e-63
Identities = 139/297 (46%), Positives = 180/297 (59%), Gaps = 13/297 (4%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIF-NNGTMVAVKVLRGSSDKKIEEQFMAEVGT 67
F+ ++L+ AT+ +S+ +G GGFG V+KG + T VAVK L E +F AEV T
Sbjct: 451 FSFKELQSATNGFSDKVGHGGFGAVFKGTLPGSSTFVAVKRLERPGSG--ESEFRAEVCT 508
Query: 68 IGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEK-NALGYEKLHEIAIGTAR 126
IG I H NLV+L GFC E LVY+YM GSL YL L +E IA+GTA+
Sbjct: 509 IGNIQHVNLVRLRGFCSENLHRLLVYDYMPQGSLSSYLSRTSPKLLSWETRFRIALGTAK 568
Query: 127 GIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGY 186
GIAYLHE C I+H DIKP NILLD ++N KV+DFGLAKL R+ + + T RGT GY
Sbjct: 569 GIAYLHEGCRDCIIHCDIKPENILLDSDYNAKVSDFGLAKLLGRDFSRVLAT-MRGTWGY 627
Query: 187 AAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDI-------KNTESQEW-FPIWVWKK 238
APE PIT K DVYSFGM L E++G RRN+ + K TE ++W FP W ++
Sbjct: 628 VAPEWISGLPITTKADVYSFGMTLLELIGGRRNVIVNSDTLGEKETEPEKWFFPPWAARE 687
Query: 239 FDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI 295
G ++ + E N E RM VA+WC+Q +++RP M VVKMLEG +E+
Sbjct: 688 IIQGNVDSVVDSRLNGEYNTEEVTRMATVAIWCIQDNEEIRPAMGTVVKMLEGVVEV 744
>At1g66910 receptor serine/threonine kinase PR5K, putative
Length = 655
Score = 236 bits (602), Expect = 1e-62
Identities = 129/326 (39%), Positives = 206/326 (62%), Gaps = 13/326 (3%)
Query: 9 FTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTI 68
++ Q+ T +++ ++G GGFGTVY+G +G VAVKVL+ S E F+ EV ++
Sbjct: 327 YSYAQVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLKESQGNG--EDFINEVASM 384
Query: 69 GRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTARG 127
+ H N+V L GFC E A++YE+M NGSLD+++ +K++ + + +L+ IA+G ARG
Sbjct: 385 SQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFISSKKSSTMDWRELYGIALGVARG 444
Query: 128 IAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYA 187
+ YLH C RIVH+DIKP N+LLD N +PKV+DFGLAKLC R+ + +++ RGT GY
Sbjct: 445 LEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGYI 504
Query: 188 APELWMPF--PITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQE---WFPIWVWKKFDAG 242
APE++ ++HK DVYS+GML+ +I+G R ++T S +FP W+++ +
Sbjct: 505 APEVFSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRDLEKA 564
Query: 243 LLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNPF 302
++ I I + EIA++M V LWC+Q RP M+ VV+M+EG+L+ + P
Sbjct: 565 HNGKS-IETAISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEV--PP 621
Query: 303 QHLIDETKFTTHSDQESNTYTTSVSS 328
+ ++ + T + QES+T++ +S+
Sbjct: 622 RPVLQQ--IPTATLQESSTFSEDISA 645
>At5g38260 receptor serine/threonine protein kinase - like
Length = 638
Score = 235 bits (600), Expect = 2e-62
Identities = 137/330 (41%), Positives = 202/330 (60%), Gaps = 18/330 (5%)
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGT 67
+++ ++R T +S+ LG GGFGTVY G +G VAVK+L+ K E F+ EV +
Sbjct: 310 QYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILKDF--KSNGEDFINEVAS 367
Query: 68 IGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGTAR 126
+ + H N+V L GFC+E + A+VYE++ NGSLD++L +K+ L L+ IA+G AR
Sbjct: 368 MSQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEKKSLNLDVSTLYRIALGVAR 427
Query: 127 GIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGY 186
G+ YLH C+ RIVH+DIKP NILLD F PKV+DFGLAKLC + + +++ RGT GY
Sbjct: 428 GLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKLCEKRESILSLLDARGTIGY 487
Query: 187 AAPELW--MPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNT----ESQEWFPIWVWKKFD 240
APE++ M ++HK DVYS+GML+ E++G +N +I+ T S +FP W++K +
Sbjct: 488 IAPEVFSGMYGRVSHKSDVYSYGMLVLEMIG-AKNKEIEETAASNSSSAYFPDWIYKNLE 546
Query: 241 AGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI----P 296
G + I +++E+A++M V LWC+Q RP M+ +V+M+EGSL++ P
Sbjct: 547 NG-EDTWKFGDEISREDKEVAKKMTLVGLWCIQPSPLNRPPMNRIVEMMEGSLDVLEVPP 605
Query: 297 KTFNPFQHLIDETKFTTHSDQESNTYTTSV 326
K P H E S E N+ T V
Sbjct: 606 K---PSIHYSAEPLPQLSSFSEENSIYTEV 632
>At4g18250 receptor serine/threonine kinase-like protein
Length = 687
Score = 234 bits (597), Expect = 5e-62
Identities = 130/335 (38%), Positives = 211/335 (62%), Gaps = 21/335 (6%)
Query: 8 RFTSQQLRIATDNYSNLLGSGGFGTVYKGIFNN--GTMVAVKVLRGSSDKKIEEQFMAEV 65
R++ ++++ T+++ +++G GGFGTVYKG + G +A+K+L+ S K E+F+ E+
Sbjct: 342 RYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASGRDIALKILKES--KGNGEEFINEL 399
Query: 66 GTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIAIGT 124
++ R H N+V LFGFC+E + A++YE+M NGSLD+++ + + ++ L+ IA+G
Sbjct: 400 VSMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDKFISENMSTKIEWKTLYNIAVGV 459
Query: 125 ARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTP 184
ARG+ YLH C +IVH+DIKP NIL+D + PK++DFGLAKLC ++ + I+M RGT
Sbjct: 460 ARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPKISDFGLAKLCKKKESIISMLDARGTV 519
Query: 185 GYAAPELWMP--FPITHKCDVYSFGMLLFEIVGRRRNLDIKNT---ESQEWFPIWVW--- 236
GY APE++ ++HK DVYS+GM++ E++G + +++ + +S +FP WV+
Sbjct: 520 GYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGATKREEVETSATDKSSMYFPDWVYEDL 579
Query: 237 -KKFDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGS--- 292
+K LLE+ +I EE+ +I +RM V LWC+Q RP M VV+MLEGS
Sbjct: 580 ERKETMRLLEDHIIE---EEEEEKIVKRMTLVGLWCIQTNPSDRPPMRKVVEMLEGSRLE 636
Query: 293 -LEIPKTFNPFQHLIDETKFTTHSDQESNTYTTSV 326
L++P H++ + + + S Q S T S+
Sbjct: 637 ALQVPPKPLLNLHVVTDWETSEDSQQTSRLSTQSL 671
>At5g39020 receptor protein kinase - like protein
Length = 813
Score = 231 bits (589), Expect = 5e-61
Identities = 133/330 (40%), Positives = 206/330 (62%), Gaps = 17/330 (5%)
Query: 13 QLRIATDNYSNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFMAEVGTIGRIH 72
+L+ T ++S+ +G GGFGTVY+G +NG VAVKVL+ K + F+ EV ++ +
Sbjct: 490 ELKKITKSFSHTVGKGGFGTVYRGNLSNGRTVAVKVLKDL--KGNGDDFINEVTSMSQTS 547
Query: 73 HFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNAL-GYEKLHEIAIGTARGIAYL 131
H N+V L GFC+E + A++ E++ +GSLD+++ K+ L+ IA+G ARG+ YL
Sbjct: 548 HVNIVSLLGFCYEGSKRAIISEFLEHGSLDQFISRNKSLTPNVTTLYGIALGIARGLEYL 607
Query: 132 HELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGRGTPGYAAPEL 191
H C+ RIVH+DIKP NILLD NF PKVADFGLAKLC + + +++ RGT GY APE+
Sbjct: 608 HYGCKTRIVHFDIKPQNILLDDNFCPKVADFGLAKLCEKRESILSLIDTRGTIGYIAPEV 667
Query: 192 --WMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTE-SQEWFPIWVWKKFDAGLLEEAM 248
M I+HK DVYS+GML+ +++G R ++ S +FP W++K + G ++
Sbjct: 668 VSRMYGGISHKSDVYSYGMLVLDMIGARNKVETTTCNGSTAYFPDWIYKDLENG--DQTW 725
Query: 249 IVCG-IEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGS---LEIPKTFNPFQH 304
I+ I E++ +I ++M+ V+LWC++ RP M+ VV+M+EGS LE+P P +H
Sbjct: 726 IIGDEINEEDNKIVKKMILVSLWCIRPCPSDRPPMNKVVEMIEGSLDALELPP--KPSRH 783
Query: 305 LIDETKFTTHS---DQESNTYTTSVSSVMV 331
+ E + S QE+ T ++ S ++
Sbjct: 784 ISTELVLESSSLSDGQEAEKQTQTLDSTII 813
>At1g56120 receptor-like protein kinase, putative
Length = 1045
Score = 230 bits (586), Expect = 1e-60
Identities = 144/330 (43%), Positives = 190/330 (56%), Gaps = 14/330 (4%)
Query: 5 KPIRFTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
KP FT +L+ AT ++ SN LG GGFG VYKG N+G VAVK L S ++ + QF+
Sbjct: 692 KPYTFTYSELKNATQDFDLSNKLGEGGFGAVYKGNLNDGREVAVKQLSIGS-RQGKGQFV 750
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIA 121
AE+ I + H NLVKL+G CFE + LVYEY+ NGSLD+ L +K+ L + +EI
Sbjct: 751 AEIIAISSVLHRNLVKLYGCCFEGDHRLLVYEYLPNGSLDQALFGDKSLHLDWSTRYEIC 810
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+G ARG+ YLHE RI+H D+K NILLD PKV+DFGLAKL + + THI+ T
Sbjct: 811 LGVARGLVYLHEEASVRIIHRDVKASNILLDSELVPKVSDFGLAKLYDDKKTHIS-TRVA 869
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVW----K 237
GT GY APE M +T K DVY+FG++ E+V R+N D E +++ W W K
Sbjct: 870 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRKNSDENLEEGKKYLLEWAWNLHEK 929
Query: 238 KFDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPK 297
D L+++ + E N E +RM+ +AL C Q LRP MS VV ML G E+
Sbjct: 930 NRDVELIDDE-----LSEYNMEEVKRMIGIALLCTQSSYALRPPMSRVVAMLSGDAEVND 984
Query: 298 TFNPFQHLIDETKFTTHSDQESNTYTTSVS 327
+ +L D T T S SN T S
Sbjct: 985 ATSKPGYLTDCTFDDTTSSSFSNFQTKDTS 1014
>At1g56140 receptor-like protein kinase, putative
Length = 2083
Score = 228 bits (580), Expect = 5e-60
Identities = 137/306 (44%), Positives = 180/306 (58%), Gaps = 6/306 (1%)
Query: 5 KPIRFTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
KP FT +L+ AT ++ SN LG GGFG VYKG N+G VAVK+L S ++ + QF+
Sbjct: 1727 KPYTFTYSELKSATQDFDPSNKLGEGGFGPVYKGKLNDGREVAVKLLSVGS-RQGKGQFV 1785
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIA 121
AE+ I + H NLVKL+G C+E LVYEY+ NGSLD+ L EK L + +EI
Sbjct: 1786 AEIVAISAVQHRNLVKLYGCCYEGEHRLLVYEYLPNGSLDQALFGEKTLHLDWSTRYEIC 1845
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+G ARG+ YLHE RIVH D+K NILLD PKV+DFGLAKL + + THI+ T
Sbjct: 1846 LGVARGLVYLHEEARLRIVHRDVKASNILLDSKLVPKVSDFGLAKLYDDKKTHIS-TRVA 1904
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDA 241
GT GY APE M +T K DVY+FG++ E+V R N D + + + W W +
Sbjct: 1905 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRPNSDENLEDEKRYLLEWAWNLHEK 1964
Query: 242 GLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEIPKTFNP 301
G E +I + E N E +RM+ +AL C Q LRP MS VV ML G +E+ +
Sbjct: 1965 G-REVELIDHQLTEFNMEEGKRMIGIALLCTQTSHALRPPMSRVVAMLSGDVEVSDVTSK 2023
Query: 302 FQHLID 307
+L D
Sbjct: 2024 PGYLTD 2029
Score = 214 bits (546), Expect = 4e-56
Identities = 125/294 (42%), Positives = 179/294 (60%), Gaps = 6/294 (2%)
Query: 5 KPIRFTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
+P F+ +LR AT ++ SN LG GGFG V+KG N+G +AVK L +S ++ + QF+
Sbjct: 657 RPYTFSYSELRTATQDFDPSNKLGEGGFGPVFKGKLNDGREIAVKQLSVAS-RQGKGQFV 715
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIA 121
AE+ TI + H NLVKL+G C E N LVYEY+ N SLD+ L EK+ LG+ + EI
Sbjct: 716 AEIATISAVQHRNLVKLYGCCIEGNQRMLVYEYLSNKSLDQALFEEKSLQLGWSQRFEIC 775
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+G A+G+AY+HE RIVH D+K NILLD + PK++DFGLAKL + + THI+ T
Sbjct: 776 LGVAKGLAYMHEESNPRIVHRDVKASNILLDSDLVPKLSDFGLAKLYDDKKTHIS-TRVA 834
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVWKKFDA 241
GT GY +PE M +T K DV++FG++ EIV R N + + +++ W W
Sbjct: 835 GTIGYLSPEYVMLGHLTEKTDVFAFGIVALEIVSGRPNSSPELDDDKQYLLEWAW-SLHQ 893
Query: 242 GLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI 295
+ ++ + E ++E +R++ VA C Q +RP MS VV ML G +EI
Sbjct: 894 EQRDMEVVDPDLTEFDKEEVKRVIGVAFLCTQTDHAIRPTMSRVVGMLTGDVEI 947
>At1g56130 receptor-like protein kinase, putative
Length = 1032
Score = 224 bits (571), Expect = 6e-59
Identities = 135/298 (45%), Positives = 180/298 (60%), Gaps = 14/298 (4%)
Query: 5 KPIRFTSQQLRIATDNY--SNLLGSGGFGTVYKGIFNNGTMVAVKVLRGSSDKKIEEQFM 62
KP FT +L+ AT ++ SN LG GGFG VYKG N+G +VAVK+L S ++ + QF+
Sbjct: 678 KPYIFTYSELKSATQDFDPSNKLGEGGFGPVYKGNLNDGRVVAVKLLSVGS-RQGKGQFV 736
Query: 63 AEVGTIGRIHHFNLVKLFGFCFEKNLIALVYEYMGNGSLDRYLLHEKNA-LGYEKLHEIA 121
AE+ I + H NLVKL+G CFE LVYEY+ NGSLD+ L +K L + +EI
Sbjct: 737 AEIVAISSVLHRNLVKLYGCCFEGEHRMLVYEYLPNGSLDQALFGDKTLHLDWSTRYEIC 796
Query: 122 IGTARGIAYLHELCEHRIVHYDIKPGNILLDGNFNPKVADFGLAKLCNRENTHITMTGGR 181
+G ARG+ YLHE RIVH D+K NILLD P+++DFGLAKL + + THI+ T
Sbjct: 797 LGVARGLVYLHEEASVRIVHRDVKASNILLDSRLVPQISDFGLAKLYDDKKTHIS-TRVA 855
Query: 182 GTPGYAAPELWMPFPITHKCDVYSFGMLLFEIVGRRRNLDIKNTESQEWFPIWVW----K 237
GT GY APE M +T K DVY+FG++ E+V R N D E +++ W W K
Sbjct: 856 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRPNSDENLEEEKKYLLEWAWNLHEK 915
Query: 238 KFDAGLLEEAMIVCGIEEKNREIAERMVKVALWCVQYRQQLRPMMSDVVKMLEGSLEI 295
D L+++ + + N E A+RM+ +AL C Q LRP MS VV ML G +EI
Sbjct: 916 SRDIELIDDK-----LTDFNMEEAKRMIGIALLCTQTSHALRPPMSRVVAMLSGDVEI 968
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.138 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,218,851
Number of Sequences: 26719
Number of extensions: 363662
Number of successful extensions: 4275
Number of sequences better than 10.0: 994
Number of HSP's better than 10.0 without gapping: 838
Number of HSP's successfully gapped in prelim test: 156
Number of HSP's that attempted gapping in prelim test: 911
Number of HSP's gapped (non-prelim): 1097
length of query: 355
length of database: 11,318,596
effective HSP length: 100
effective length of query: 255
effective length of database: 8,646,696
effective search space: 2204907480
effective search space used: 2204907480
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148970.6


