

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.5 + phase: 0
(402 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
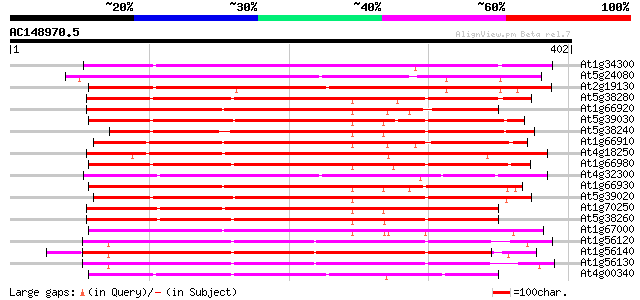
Score E
Sequences producing significant alignments: (bits) Value
At1g34300 putative protein 258 5e-69
At5g24080 receptor-like protein kinase 257 9e-69
At2g19130 putative receptor-like protein kinase 253 1e-67
At5g38280 receptor serine/threonine kinase PR5K 250 1e-66
At1g66920 receptor serine/threonine kinase PR5K, putative 249 2e-66
At5g39030 receptor protein kinase - like protein 247 7e-66
At5g38240 receptor serine/threonine protein kinase -like 247 9e-66
At1g66910 receptor serine/threonine kinase PR5K, putative 246 2e-65
At4g18250 receptor serine/threonine kinase-like protein 244 6e-65
At1g66980 putative kinase 243 1e-64
At4g32300 S-receptor kinase -like protein 242 3e-64
At1g66930 receptor serine/threonine kinase PR5K, putative 240 9e-64
At5g39020 receptor protein kinase - like protein 239 3e-63
At1g70250 hypothetical protein 237 7e-63
At5g38260 receptor serine/threonine protein kinase - like 235 4e-62
At1g67000 hypothetical protein 235 4e-62
At1g56120 receptor-like protein kinase, putative 235 4e-62
At1g56140 receptor-like protein kinase, putative 226 2e-59
At1g56130 receptor-like protein kinase, putative 222 3e-58
At4g00340 receptor-like protein kinase 221 4e-58
>At1g34300 putative protein
Length = 829
Score = 258 bits (658), Expect = 5e-69
Identities = 142/338 (42%), Positives = 202/338 (59%), Gaps = 5/338 (1%)
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVI 113
PV+FT ++L T+ + LG+G FG V++G L+N VAVK L ++ G E+QF+ EV
Sbjct: 471 PVQFTYKELQRCTKSFKEKLGAGGFGTVYRGVLTNRTVVAVKQLEGIEQG-EKQFRMEVA 529
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGT 173
TI T+H+NLV+L GFC R LVYE++ NGSLD ++F + + ++ IA+GT
Sbjct: 530 TISSTHHLNLVRLIGFCSQGRHRLLVYEFMRNGSLDNFLFTTDSAKFLTWEYRFNIALGT 589
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
AKGI YLHEEC+ I+H DIKPEN+L+D K++DFGLAKL + + N + RGTR
Sbjct: 590 AKGITYLHEECRDCIVHCDIKPENILVDDNFAAKVSDFGLAKLLNPKDNRYNMSSVRGTR 649
Query: 234 GYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFE--NN 291
GY APE P+T K DVYS+G++L E+V +R+FD S + + F W +E FE N
Sbjct: 650 GYLAPEWLANLPITSKSDVYSYGMVLLELVSGKRNFDVSEKTNHKKFSIWAYEEFEKGNT 709
Query: 292 ELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFPF 351
+ ++ L E + D E RM+K + WC+Q P RP M VV+MLEG +I +P P
Sbjct: 710 KAILDTRLSEDQTVDMEQVMRMVKTSFWCIQEQPLQRPTMGKVVQMLEGITEIKNPLCP- 768
Query: 352 HNLVPAKENSTQEGSTADSDTTTSSWRTESSRESGFKT 389
+ S ST+ + +S T SS S ++
Sbjct: 769 -KTISEVSFSGNSMSTSHASMFVASGPTRSSSFSATRS 805
>At5g24080 receptor-like protein kinase
Length = 872
Score = 257 bits (656), Expect = 9e-69
Identities = 154/363 (42%), Positives = 213/363 (58%), Gaps = 28/363 (7%)
Query: 41 TMERIFSN--INREKPVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLN 98
T++R N I + PV FT L T +S +LGSG FG V+KG ++ VAVK L+
Sbjct: 502 TLKRAAKNSLILCDSPVSFTYRDLQNCTNNFSQLLGSGGFGTVYKGTVAGETLVAVKRLD 561
Query: 99 CLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGS-KN 157
E +F EV TIG +H+NLV+L G+C R LVYEY+ NGSLDK+IF S +
Sbjct: 562 RALSHGEREFITEVNTIGSMHHMNLVRLCGYCSEDSHRLLVYEYMINGSLDKWIFSSEQT 621
Query: 158 RNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLR 217
N D++ +IA+ TA+GIAY HE+C++RIIH DIKPEN+LLD PK++DFGLAK+
Sbjct: 622 ANLLDWRTRFEIAVATAQGIAYFHEQCRNRIIHCDIKPENILLDDNFCPKVSDFGLAKMM 681
Query: 218 SRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQ 277
RE + + T RGTRGY APE P+T K DVYS+G+LL EIVG RR+ D SY
Sbjct: 682 GREHS-HVVTMIRGTRGYLAPEWVSNRPITVKADVYSYGMLLLEIVGGRRNLDMSYDAED 740
Query: 278 QWFPRWTWEMFENNELVVMLALCEIEEKDSEIAE-----RMLKVALWCVQYSPNDRPLMS 332
++P W ++ EL +L ++++ +AE + LKVA WC+Q + RP M
Sbjct: 741 FFYPGWAYK-----ELTNGTSLKAVDKRLQGVAEEEEVVKALKVAFWCIQDEVSMRPSMG 795
Query: 333 TVVKMLEGEID-ISSPPFP-------------FHNLVPAKENSTQEGSTADSDTTTSSWR 378
VVK+LEG D I+ PP P + + + N+ T ++ TT+ S+R
Sbjct: 796 EVVKLLEGTSDEINLPPMPQTILELIEEGLEDVYRAMRREFNNQLSSLTVNTITTSQSYR 855
Query: 379 TES 381
+ S
Sbjct: 856 SSS 858
>At2g19130 putative receptor-like protein kinase
Length = 828
Score = 253 bits (646), Expect = 1e-67
Identities = 144/340 (42%), Positives = 208/340 (60%), Gaps = 10/340 (2%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
F+ +L T+ +S LG G FG VFKG L + ++AVK L + G E+QF+ EV+TIG
Sbjct: 483 FSYRELQNATKNFSDKLGGGGFGSVFKGALPDSSDIAVKRLEGISQG-EKQFRTEVVTIG 541
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF--DFQKLHKIAIGTA 174
H+NLV+L GFC K+ LVY+Y+ NGSLD ++F ++ ++ +IA+GTA
Sbjct: 542 TIQHVNLVRLRGFCSEGSKKLLVYDYMPNGSLDSHLFLNQVEEKIVLGWKLRFQIALGTA 601
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+AYLH+EC+ IIH DIKPEN+LLD + PK+ADFGLAKL R+ + L T RGTRG
Sbjct: 602 RGLAYLHDECRDCIIHCDIKPENILLDSQFCPKVADFGLAKLVGRDFSRVLTT-MRGTRG 660
Query: 235 YAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNELV 294
Y APE +T K DVYS+G++LFE+V RR+ + S +E ++FP W + + +
Sbjct: 661 YLAPEWISGVAITAKADVYSYGMMLFELVSGRRNTEQSENEKVRFFPSWAATILTKDGDI 720
Query: 295 VMLALCEIEEKDSEIAE--RMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP-- 350
L +E +I E R KVA WC+Q + RP MS VV++LEG ++++ PPFP
Sbjct: 721 RSLVDPRLEGDAVDIEEVTRACKVACWCIQDEESHRPAMSQVVQILEGVLEVNPPPFPRS 780
Query: 351 FHNLVPAKENST--QEGSTADSDTTTSSWRTESSRESGFK 388
LV + E+ E S++ S ++ + + SS S K
Sbjct: 781 IQALVVSDEDVVFFTESSSSSSHNSSQNHKHSSSSSSSKK 820
>At5g38280 receptor serine/threonine kinase PR5K
Length = 665
Score = 250 bits (638), Expect = 1e-66
Identities = 129/326 (39%), Positives = 213/326 (64%), Gaps = 13/326 (3%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSN-GENVAVKVLNCLDMGMEEQFKAEVIT 114
R++ ++ ++T ++ +LG G FG V+KG+L++ G +VAVK+L + G E+F EV +
Sbjct: 320 RYSYTRVKKMTNSFAHVLGKGGFGTVYKGKLADSGRDVAVKILKVSE-GNGEEFINEVAS 378
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTA 174
+ RT H+N+V L GFC+ ++KRA++YE++ NGSLDKYI + + ++++L+ +A+G +
Sbjct: 379 MSRTSHVNIVSLLGFCYEKNKRAIIYEFMPNGSLDKYISANMSTK-MEWERLYDVAVGIS 437
Query: 175 KGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRG 234
+G+ YLH C RI+H+DIKP+N+L+D L PKI+DFGLAKL + +I H RGT G
Sbjct: 438 RGLEYLHNRCVTRIVHFDIKPQNILMDENLCPKISDFGLAKLCKNKESIISMLHMRGTFG 497
Query: 235 YAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSYSES---QQWFPRWTWEMFE 289
Y APEM+ V++K DVYS+G+++ E++G + YS S +FP W ++ FE
Sbjct: 498 YIAPEMFSKNFGAVSHKSDVYSYGMVVLEMIGAKNIEKVEYSGSNNGSMYFPEWVYKDFE 557
Query: 290 NNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPP 348
E+ + I +++ +IA++++ VALWC+Q +P+DRP M V++MLEG ++ + PP
Sbjct: 558 KGEITRIFG-DSITDEEEKIAKKLVLVALWCIQMNPSDRPPMIKVIEMLEGNLEALQVPP 616
Query: 349 FPFHNLVPAKENSTQEGSTADSDTTT 374
P L+ + E + + DT+T
Sbjct: 617 NP---LLFSPEETVPDTLEDSDDTST 639
>At1g66920 receptor serine/threonine kinase PR5K, putative
Length = 609
Score = 249 bits (635), Expect = 2e-66
Identities = 133/304 (43%), Positives = 197/304 (64%), Gaps = 16/304 (5%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
+++ E++ IT ++ ++G G FG+V++G LS+G VAVKVL L E F EV ++
Sbjct: 288 QYSYEQVKRITNSFAEVVGRGGFGIVYRGTLSDGRMVAVKVLKDLKGNNGEDFINEVASM 347
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAK 175
+T H+N+V L GFC KRA++YE++ENGSLDK+I SK + D+++L+ IA+G A+
Sbjct: 348 SQTSHVNIVTLLGFCSEGYKRAIIYEFMENGSLDKFI-SSKKSSTMDWRELYGIALGVAR 406
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
G+ YLH C+ RI+H+DIKP+NVLLD L PK++DFGLAKL R+ +I RGT GY
Sbjct: 407 GLEYLHHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGY 466
Query: 236 AAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHF---DSSYSESQQWFPRWTW---EM 287
APE++ V++K DVYS+G+L+ +I+G R D++ S S +FP W + E
Sbjct: 467 IAPEVFSRVYGSVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYKDLEK 526
Query: 288 FENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISS 346
+N L+V ++ EIA++M V LWC+Q P DRP M+ VV+M+EG +D +
Sbjct: 527 GDNGRLIVN------RSEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEV 580
Query: 347 PPFP 350
PP P
Sbjct: 581 PPRP 584
>At5g39030 receptor protein kinase - like protein
Length = 806
Score = 247 bits (631), Expect = 7e-66
Identities = 139/321 (43%), Positives = 206/321 (63%), Gaps = 15/321 (4%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
+T +L +IT+ +S I+G G FG V+ G LSNG VAVKVL L G E F EV ++
Sbjct: 488 YTYAELKKITKSFSYIIGKGGFGTVYGGNLSNGRKVAVKVLKDLK-GSAEDFINEVASMS 546
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKG 176
+T H+N+V L GFCF KRA+VYE++ENGSLD+++ +K+ D L+ IA+G A+G
Sbjct: 547 QTSHVNIVSLLGFCFEGSKRAIVYEFLENGSLDQFMSRNKSLTQ-DVTTLYGIALGIARG 605
Query: 177 IAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYA 236
+ YLH CK RI+H+DIKP+N+LLD L PK++DFGLAKL + ++ RGT GY
Sbjct: 606 LEYLHYGCKTRIVHFDIKPQNILLDGNLCPKVSDFGLAKLCEKRESVLSLMDTRGTIGYI 665
Query: 237 APEMWKPY--PVTYKCDVYSFGILLFEIVGRR-----RHFDSSYSESQQWFPRWTWEMFE 289
APE++ V++K DVYSFG+L+ +++G R DS+ S + +FP W ++ E
Sbjct: 666 APEVFSRMYGRVSHKSDVYSFGMLVIDMIGARSKEIVETVDSAASST--YFPDWIYKDLE 723
Query: 290 NNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPP 348
+ E + EI +++ EIA++M+ V LWC+Q P+DRP M+ VV+M+EG +D + PP
Sbjct: 724 DGEQTWIFG-DEITKEEKEIAKKMIVVGLWCIQPCPSDRPSMNRVVEMMEGSLDALEIPP 782
Query: 349 FPFHNLVPAKENSTQEGSTAD 369
P ++ + E T+ S +D
Sbjct: 783 KPSMHI--STEVITESSSLSD 801
>At5g38240 receptor serine/threonine protein kinase -like
Length = 588
Score = 247 bits (630), Expect = 9e-66
Identities = 133/311 (42%), Positives = 200/311 (63%), Gaps = 16/311 (5%)
Query: 72 ILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCF 131
++G G FG V+KG L +G VAVK+L + G E F EV +I +T H+N+V L GFCF
Sbjct: 286 VVGRGGFGTVYKGNLRDGRKVAVKILKDSN-GNCEDFINEVASISQTSHVNIVSLLGFCF 344
Query: 132 HRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHY 191
+ KRA+VYE++ENGSLD+ ++ D L+ IA+G A+GI YLH CK RI+H+
Sbjct: 345 EKSKRAIVYEFLENGSLDQ-------SSNLDVSTLYGIALGVARGIEYLHFGCKKRIVHF 397
Query: 192 DIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPY--PVTYK 249
DIKP+NVLLD L+PK+ADFGLAKL ++ +I RGT GY APE++ V++K
Sbjct: 398 DIKPQNVLLDENLKPKVADFGLAKLCEKQESILSLLDTRGTIGYIAPELFSRVYGNVSHK 457
Query: 250 CDVYSFGILLFEIVGRR---RHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKD 306
DVYS+G+L+ E+ G R R ++ + S +FP W ++ EN + V +LA + ++
Sbjct: 458 SDVYSYGMLVLEMTGARNKERVQNADSNNSSAYFPDWIFKDLENGDYVKLLA-DGLTREE 516
Query: 307 SEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPFPFHNLVPAKENSTQEG 365
+IA++M+ V LWC+Q+ P+DRP M+ VV M+EG +D + PP P ++ P + N+ +
Sbjct: 517 EDIAKKMILVGLWCIQFRPSDRPSMNKVVGMMEGNLDSLDPPPKPLLHM-PMQNNNAESS 575
Query: 366 STADSDTTTSS 376
++ D++ S
Sbjct: 576 QPSEEDSSIYS 586
>At1g66910 receptor serine/threonine kinase PR5K, putative
Length = 655
Score = 246 bits (628), Expect = 2e-65
Identities = 137/319 (42%), Positives = 201/319 (62%), Gaps = 15/319 (4%)
Query: 61 KLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYH 120
++ IT+ ++ ++G G FG V++G L +G +VAVKVL G E F EV ++ +T H
Sbjct: 331 QVTSITKSFAEVIGKGGFGTVYRGTLYDGRSVAVKVLK-ESQGNGEDFINEVASMSQTSH 389
Query: 121 INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYL 180
+N+V L GFC KRA++YE++ENGSLDK+I SK + D+++L+ IA+G A+G+ YL
Sbjct: 390 VNIVTLLGFCSEGYKRAIIYEFMENGSLDKFI-SSKKSSTMDWRELYGIALGVARGLEYL 448
Query: 181 HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEM 240
H C+ RI+H+DIKP+NVLLD L PK++DFGLAKL R+ +I RGT GY APE+
Sbjct: 449 HHGCRTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCERKESILSLMDTRGTIGYIAPEV 508
Query: 241 WKPY--PVTYKCDVYSFGILLFEIVGRRRHF---DSSYSESQQWFPRWTWEMFE--NNEL 293
+ V++K DVYS+G+L+ +I+G R D++ S S +FP W + E +N
Sbjct: 509 FSRVYGRVSHKSDVYSYGMLVLDIIGARNKTSTEDTTSSTSSMYFPEWIYRDLEKAHNGK 568
Query: 294 VVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPFPFH 352
+ A I ++ EIA++M V LWC+Q P DRP M+ VV+M+EG +D + PP P
Sbjct: 569 SIETA---ISNEEDEIAKKMTLVGLWCIQPWPLDRPAMNRVVEMMEGNLDALEVPPRPVL 625
Query: 353 NLVPAKENSTQEGSTADSD 371
+P + QE ST D
Sbjct: 626 QQIPTA--TLQESSTFSED 642
>At4g18250 receptor serine/threonine kinase-like protein
Length = 687
Score = 244 bits (623), Expect = 6e-65
Identities = 136/341 (39%), Positives = 215/341 (62%), Gaps = 14/341 (4%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGEL--SNGENVAVKVLNCLDMGMEEQFKAEVI 113
R++ EK+ ++T + ++G G FG V+KG+L ++G ++A+K+L G E+F E++
Sbjct: 342 RYSFEKVKKMTNSFDHVIGKGGFGTVYKGKLPDASGRDIALKILK-ESKGNGEEFINELV 400
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRND-FDFQKLHKIAIG 172
++ R H+N+V L+GFC+ +RA++YE++ NGSLDK+I S+N + +++ L+ IA+G
Sbjct: 401 SMSRASHVNIVSLFGFCYEGSQRAIIYEFMPNGSLDKFI--SENMSTKIEWKTLYNIAVG 458
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
A+G+ YLH C +I+H+DIKP+N+L+D L PKI+DFGLAKL ++ +I RGT
Sbjct: 459 VARGLEYLHNSCVSKIVHFDIKPQNILIDEDLCPKISDFGLAKLCKKKESIISMLDARGT 518
Query: 233 RGYAAPEMW-KPY-PVTYKCDVYSFGILLFEIVGRRRHFD---SSYSESQQWFPRWTWEM 287
GY APEM+ K Y V++K DVYS+G+++ E++G + + S+ +S +FP W +E
Sbjct: 519 VGYIAPEMFSKNYGGVSHKSDVYSYGMVVLEMIGATKREEVETSATDKSSMYFPDWVYED 578
Query: 288 FENNELVVMLALCEIEEKDSE-IAERMLKVALWCVQYSPNDRPLMSTVVKMLEGE--IDI 344
E E + +L IEE++ E I +RM V LWC+Q +P+DRP M VV+MLEG +
Sbjct: 579 LERKETMRLLEDHIIEEEEEEKIVKRMTLVGLWCIQTNPSDRPPMRKVVEMLEGSRLEAL 638
Query: 345 SSPPFPFHNLVPAKENSTQEGSTADSDTTTSSWRTESSRES 385
PP P NL + T E S S +T S +R +
Sbjct: 639 QVPPKPLLNLHVVTDWETSEDSQQTSRLSTQSLLERKTRSN 679
>At1g66980 putative kinase
Length = 1109
Score = 243 bits (621), Expect = 1e-64
Identities = 136/324 (41%), Positives = 199/324 (60%), Gaps = 12/324 (3%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
+T ++ IT+ ++ ++G G FG+V+KG LS+G VAVKVL G E F EV T+
Sbjct: 786 YTYAQVKRITKSFAEVVGRGGFGIVYKGTLSDGRVVAVKVLKDTK-GNGEDFINEVATMS 844
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKG 176
RT H+N+V L GFC KRA++YE++ENGSLDK+I G + N D+ L++IA+G A G
Sbjct: 845 RTSHLNIVSLLGFCSEGSKRAIIYEFLENGSLDKFILGKTSVN-MDWTALYRIALGVAHG 903
Query: 177 IAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYA 236
+ YLH CK RI+H+DIKP+NVLLD PK++DFGLAKL ++ +I RGT GY
Sbjct: 904 LEYLHHSCKTRIVHFDIKPQNVLLDDSFCPKVSDFGLAKLCEKKESILSMLDTRGTIGYI 963
Query: 237 APEMWKPY--PVTYKCDVYSFGILLFEIVGRRRHFDSSY----SESQQWFPRWTWEMFEN 290
APEM V++K DVYS+G+L+ EI+G R ++ + S +FP W + E+
Sbjct: 964 APEMISRVYGNVSHKSDVYSYGMLVLEIIGARNKEKANQACASNTSSMYFPEWVYRDLES 1023
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPF 349
+ + I ++ E+A++M V LWC+Q SP DRP M+ VV+M+EG ++ + PP
Sbjct: 1024 CKSGRHIE-DGINSEEDELAKKMTLVGLWCIQPSPVDRPAMNRVVEMMEGSLEALEVPPR 1082
Query: 350 PFHNLVPAKENSTQEGSTADSDTT 373
P +P ++ E S D +
Sbjct: 1083 PVLQQIPI--SNLHESSILSEDVS 1104
>At4g32300 S-receptor kinase -like protein
Length = 821
Score = 242 bits (617), Expect = 3e-64
Identities = 135/335 (40%), Positives = 201/335 (59%), Gaps = 7/335 (2%)
Query: 54 PVRFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVI 113
P+RF + L T +S LG G FG V++G L +G +AVK L + G +E F+AEV
Sbjct: 480 PIRFAYKDLQSATNNFSVKLGQGGFGSVYEGTLPDGSRLAVKKLEGIGQGKKE-FRAEVS 538
Query: 114 TIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDF-DFQKLHKIAIG 172
IG +H++LV+L GFC R L YE++ GSL+++IF K+ + D+ IA+G
Sbjct: 539 IIGSIHHLHLVRLRGFCAEGAHRLLAYEFLSKGSLERWIFRKKDGDVLLDWDTRFNIALG 598
Query: 173 TAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGT 232
TAKG+AYLHE+C RI+H DIKPEN+LLD K++DFGLAKL +RE + + T RGT
Sbjct: 599 TAKGLAYLHEDCDARIVHCDIKPENILLDDNFNAKVSDFGLAKLMTREQS-HVFTTMRGT 657
Query: 233 RGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFENNE 292
RGY APE Y ++ K DVYS+G++L E++G R+++D S + + FP + ++ E +
Sbjct: 658 RGYLAPEWITNYAISEKSDVYSYGMVLLELIGGRKNYDPSETSEKCHFPSFAFKKMEEGK 717
Query: 293 L--VVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP 350
L +V + ++ D + +R +K ALWC+Q RP MS VV+MLEG + PP
Sbjct: 718 LMDIVDGKMKNVDVTDERV-QRAMKTALWCIQEDMQTRPSMSKVVQMLEGVFPVVQPP-S 775
Query: 351 FHNLVPAKENSTQEGSTADSDTTTSSWRTESSRES 385
+ +S + + D TTSS ++ + E+
Sbjct: 776 SSTMGSRLYSSFFKSISEDGGATTSSGPSDCNSEN 810
>At1g66930 receptor serine/threonine kinase PR5K, putative
Length = 876
Score = 240 bits (613), Expect = 9e-64
Identities = 136/325 (41%), Positives = 202/325 (61%), Gaps = 17/325 (5%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
+T ++ +T+ ++ ++G G FG+V++G L +G VAVKVL E F EV ++
Sbjct: 538 YTYAQVKRMTKSFAEVVGRGGFGIVYRGTLCDGRMVAVKVLKESKGNNSEDFINEVSSMS 597
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKG 176
+T H+N+V L GFC +RA++YE++ENGSLDK+I K D L+ IA+G A+G
Sbjct: 598 QTSHVNIVSLLGFCSEGSRRAIIYEFLENGSLDKFI-SEKTSVILDLTALYGIALGVARG 656
Query: 177 IAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYA 236
+ YLH CK RI+H+DIKP+NVLLD L PK++DFGLAKL ++ ++ RGT GY
Sbjct: 657 LEYLHYGCKTRIVHFDIKPQNVLLDDNLSPKVSDFGLAKLCEKKESVMSLMDTRGTIGYI 716
Query: 237 APEMWKPY--PVTYKCDVYSFGILLFEIVGRR---RHFDSSYSESQQWFPRWTW---EMF 288
APEM V++K DVYS+G+L+FE++G R R +S + S +FP W + E
Sbjct: 717 APEMISRVYGSVSHKSDVYSYGMLVFEMIGARKKERFGQNSANGSSMYFPEWIYKDLEKA 776
Query: 289 ENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSP 347
+N +L + I ++ EIA++M V LWC+Q SP+DRP M+ VV+M+EG +D + P
Sbjct: 777 DNGDLEHI--EIGISSEEEEIAKKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLDALEVP 834
Query: 348 PFPFHNLV---PAKENS--TQEGST 367
P P + P E+S T+E S+
Sbjct: 835 PRPVLQQIHVGPLLESSWITEESSS 859
>At5g39020 receptor protein kinase - like protein
Length = 813
Score = 239 bits (609), Expect = 3e-63
Identities = 129/321 (40%), Positives = 206/321 (63%), Gaps = 10/321 (3%)
Query: 61 KLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYH 120
+L +IT+ +S +G G FG V++G LSNG VAVKVL L G + F EV ++ +T H
Sbjct: 490 ELKKITKSFSHTVGKGGFGTVYRGNLSNGRTVAVKVLKDLK-GNGDDFINEVTSMSQTSH 548
Query: 121 INLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYL 180
+N+V L GFC+ KRA++ E++E+GSLD++I +K+ + L+ IA+G A+G+ YL
Sbjct: 549 VNIVSLLGFCYEGSKRAIISEFLEHGSLDQFISRNKSLTP-NVTTLYGIALGIARGLEYL 607
Query: 181 HEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEM 240
H CK RI+H+DIKP+N+LLD PK+ADFGLAKL + +I RGT GY APE+
Sbjct: 608 HYGCKTRIVHFDIKPQNILLDDNFCPKVADFGLAKLCEKRESILSLIDTRGTIGYIAPEV 667
Query: 241 WKPY--PVTYKCDVYSFGILLFEIVGRRRHFD-SSYSESQQWFPRWTWEMFENNELVVML 297
+++K DVYS+G+L+ +++G R + ++ + S +FP W ++ EN + ++
Sbjct: 668 VSRMYGGISHKSDVYSYGMLVLDMIGARNKVETTTCNGSTAYFPDWIYKDLENGDQTWII 727
Query: 298 ALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSPPFPFHNL-- 354
EI E+D++I ++M+ V+LWC++ P+DRP M+ VV+M+EG +D + PP P ++
Sbjct: 728 G-DEINEEDNKIVKKMILVSLWCIRPCPSDRPPMNKVVEMIEGSLDALELPPKPSRHIST 786
Query: 355 -VPAKENSTQEGSTADSDTTT 374
+ + +S +G A+ T T
Sbjct: 787 ELVLESSSLSDGQEAEKQTQT 807
>At1g70250 hypothetical protein
Length = 676
Score = 237 bits (605), Expect = 7e-63
Identities = 128/303 (42%), Positives = 197/303 (64%), Gaps = 11/303 (3%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGE-NVAVKVLNCLDMGMEEQFKAEVIT 114
RF+ ++ ++T+ + +LG G FG V+KG+L +G +VAVK+L + E+ F E+ +
Sbjct: 325 RFSYVQVKKMTKSFENVLGKGGFGTVYKGKLPDGSRDVAVKILKESNEDGED-FINEIAS 383
Query: 115 IGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRN-DFDFQKLHKIAIGT 173
+ RT H N+V L GFC+ K+A++YE + NGSLDK+I SKN + +++ L+ IA+G
Sbjct: 384 MSRTSHANIVSLLGFCYEGRKKAIIYELMPNGSLDKFI--SKNMSAKMEWKTLYNIAVGV 441
Query: 174 AKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTR 233
+ G+ YLH C RI+H+DIKP+N+L+D L PKI+DFGLAKL +I H RGT
Sbjct: 442 SHGLEYLHSHCVSRIVHFDIKPQNILIDGDLCPKISDFGLAKLCKNNESIISMLHARGTI 501
Query: 234 GYAAPEMWKPY--PVTYKCDVYSFGILLFEIVGRR---RHFDSSYSESQQWFPRWTWEMF 288
GY APE++ V++K DVYS+G+++ E++G R R ++ S + +FP W ++
Sbjct: 502 GYIAPEVFSQNFGGVSHKSDVYSYGMVVLEMIGARNIGRAQNAGSSNTSMYFPDWIYKDL 561
Query: 289 ENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID-ISSP 347
E E++ LA EE+D +I ++M+ V LWC+Q +P DRP MS VV+MLEG ++ + P
Sbjct: 562 EKGEIMSFLADQITEEEDEKIVKKMVLVGLWCIQTNPYDRPPMSKVVEMLEGSLEALQIP 621
Query: 348 PFP 350
P P
Sbjct: 622 PKP 624
>At5g38260 receptor serine/threonine protein kinase - like
Length = 638
Score = 235 bits (599), Expect = 4e-62
Identities = 125/301 (41%), Positives = 191/301 (62%), Gaps = 9/301 (2%)
Query: 56 RFTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITI 115
+++ ++ +IT+ +S LG G FG V+ G L +G VAVK+L E+ F EV ++
Sbjct: 310 QYSYAEVRKITKLFSHTLGKGGFGTVYGGNLCDGRKVAVKILKDFKSNGED-FINEVASM 368
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAK 175
+T H+N+V L GFC+ KRA+VYE++ENGSLD+++ K+ N D L++IA+G A+
Sbjct: 369 SQTSHVNIVSLLGFCYEGSKRAIVYEFLENGSLDQFLSEKKSLN-LDVSTLYRIALGVAR 427
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
G+ YLH CK RI+H+DIKP+N+LLD PK++DFGLAKL + +I RGT GY
Sbjct: 428 GLDYLHHGCKTRIVHFDIKPQNILLDDTFCPKVSDFGLAKLCEKRESILSLLDARGTIGY 487
Query: 236 AAPEMWKPY--PVTYKCDVYSFGILLFEIVGRRR---HFDSSYSESQQWFPRWTWEMFEN 290
APE++ V++K DVYS+G+L+ E++G + ++ + S +FP W ++ EN
Sbjct: 488 IAPEVFSGMYGRVSHKSDVYSYGMLVLEMIGAKNKEIEETAASNSSSAYFPDWIYKNLEN 547
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDI-SSPPF 349
E EI +D E+A++M V LWC+Q SP +RP M+ +V+M+EG +D+ PP
Sbjct: 548 GEDTWKFG-DEISREDKEVAKKMTLVGLWCIQPSPLNRPPMNRIVEMMEGSLDVLEVPPK 606
Query: 350 P 350
P
Sbjct: 607 P 607
>At1g67000 hypothetical protein
Length = 717
Score = 235 bits (599), Expect = 4e-62
Identities = 137/346 (39%), Positives = 206/346 (58%), Gaps = 21/346 (6%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKAEVITIG 116
+T ++ ++T+ ++ ++G G FG+V+ G LS+ VAVKVL E F EV ++
Sbjct: 371 YTYAEVKKMTKSFTEVVGRGGFGIVYSGTLSDSSMVAVKVLKDSKGTDGEDFINEVASMS 430
Query: 117 RTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKG 176
+T H+N+V L GFC +RA++YE++ NGSLDK+I K+ + D + L+ IA+G A+G
Sbjct: 431 QTSHVNIVSLLGFCCEGSRRAIIYEFLGNGSLDKFI-SDKSSVNLDLKTLYGIALGVARG 489
Query: 177 IAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGYA 236
+ YLH CK RI+H+DIKP+NVLLD L PK++DFGLAKL ++ +I RGT GY
Sbjct: 490 LEYLHYGCKTRIVHFDIKPQNVLLDDNLCPKVSDFGLAKLCEKKESILSLLDTRGTIGYI 549
Query: 237 APEMWKPY--PVTYKCDVYSFGILLFEIVGRRR--HFD--SSYSESQQWFPRWTWEMFEN 290
APEM V++K DVYS+G+L+ E++G R+ FD S S +FP W ++ E
Sbjct: 550 APEMISRLYGSVSHKSDVYSYGMLVLEMIGARKKERFDQNSRSDGSSIYFPEWIYKDLEK 609
Query: 291 NELVVM-------LALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEID 343
+ + L I ++ EIA +M V LWC+Q SP+DRP M+ VV+M+EG +D
Sbjct: 610 ANIKDIEKTENGGLIENGISSEEEEIARKMTLVGLWCIQSSPSDRPPMNKVVEMMEGSLD 669
Query: 344 -ISSPPFPFHNLVPAKE------NSTQEGSTADSDTTTSSWRTESS 382
+ PP P + A NS + S +D +++ + ESS
Sbjct: 670 ALEVPPRPVLQQISASSVSDSFWNSEESSSASDILVFSTNSKLESS 715
>At1g56120 receptor-like protein kinase, putative
Length = 1045
Score = 235 bits (599), Expect = 4e-62
Identities = 136/342 (39%), Positives = 197/342 (56%), Gaps = 21/342 (6%)
Query: 53 KPVRFTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKA 110
KP FT +L T+ + S LG G FG V+KG L++G VAVK L+ + QF A
Sbjct: 692 KPYTFTYSELKNATQDFDLSNKLGEGGFGAVYKGNLNDGREVAVKQLSIGSRQGKGQFVA 751
Query: 111 EVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIA 170
E+I I H NLVKLYG CF D R LVYEY+ NGSLD+ +FG K+ + D+ ++I
Sbjct: 752 EIIAISSVLHRNLVKLYGCCFEGDHRLLVYEYLPNGSLDQALFGDKSLH-LDWSTRYEIC 810
Query: 171 IGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFR 230
+G A+G+ YLHEE RIIH D+K N+LLD +L PK++DFGLAKL + ++T
Sbjct: 811 LGVARGLVYLHEEASVRIIHRDVKASNILLDSELVPKVSDFGLAKLYD-DKKTHISTRVA 869
Query: 231 GTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFEN 290
GT GY APE +T K DVY+FG++ E+V R++ D + E +++ W W + E
Sbjct: 870 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRKNSDENLEEGKKYLLEWAWNLHEK 929
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP 350
N V ++ E+ E + E +RM+ +AL C Q S RP MS VV ML G+ +++
Sbjct: 930 NRDVELID-DELSEYNMEEVKRMIGIALLCTQSSYALRPPMSRVVAMLSGDAEVN----- 983
Query: 351 FHNLVPAKENSTQEGSTAD---SDTTTSSWRTESSRESGFKT 389
+ +++ G D DTT+SS+ ++++ F T
Sbjct: 984 --------DATSKPGYLTDCTFDDTTSSSFSNFQTKDTSFST 1017
>At1g56140 receptor-like protein kinase, putative
Length = 2083
Score = 226 bits (576), Expect = 2e-59
Identities = 122/295 (41%), Positives = 179/295 (60%), Gaps = 5/295 (1%)
Query: 53 KPVRFTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKA 110
KP FT +L T+ + S LG G FG V+KG+L++G VAVK+L+ + QF A
Sbjct: 1727 KPYTFTYSELKSATQDFDPSNKLGEGGFGPVYKGKLNDGREVAVKLLSVGSRQGKGQFVA 1786
Query: 111 EVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIA 170
E++ I H NLVKLYG C+ + R LVYEY+ NGSLD+ +FG K + D+ ++I
Sbjct: 1787 EIVAISAVQHRNLVKLYGCCYEGEHRLLVYEYLPNGSLDQALFGEKTLH-LDWSTRYEIC 1845
Query: 171 IGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFR 230
+G A+G+ YLHEE + RI+H D+K N+LLD KL PK++DFGLAKL + ++T
Sbjct: 1846 LGVARGLVYLHEEARLRIVHRDVKASNILLDSKLVPKVSDFGLAKLYD-DKKTHISTRVA 1904
Query: 231 GTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFEN 290
GT GY APE +T K DVY+FG++ E+V R + D + + +++ W W + E
Sbjct: 1905 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRPNSDENLEDEKRYLLEWAWNLHEK 1964
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDIS 345
V ++ ++ E + E +RM+ +AL C Q S RP MS VV ML G++++S
Sbjct: 1965 GREVELID-HQLTEFNMEEGKRMIGIALLCTQTSHALRPPMSRVVAMLSGDVEVS 2018
Score = 208 bits (530), Expect = 4e-54
Identities = 127/355 (35%), Positives = 196/355 (54%), Gaps = 13/355 (3%)
Query: 27 IEVVVQTDSKHEFATMERIFSNINREKPVRFTPEKLDEITEKY--STILGSGAFGVVFKG 84
I +++ K + A E + ++++ +P F+ +L T+ + S LG G FG VFKG
Sbjct: 632 IAILLFIRRKRKRAADEEVLNSLHI-RPYTFSYSELRTATQDFDPSNKLGEGGFGPVFKG 690
Query: 85 ELSNGENVAVKVLNCLDMGMEEQFKAEVITIGRTYHINLVKLYGFCFHRDKRALVYEYVE 144
+L++G +AVK L+ + QF AE+ TI H NLVKLYG C ++R LVYEY+
Sbjct: 691 KLNDGREIAVKQLSVASRQGKGQFVAEIATISAVQHRNLVKLYGCCIEGNQRMLVYEYLS 750
Query: 145 NGSLDKYIFGSKNRNDFDFQKLHKIAIGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKL 204
N SLD+ +F K+ + + +I +G AKG+AY+HEE RI+H D+K N+LLD L
Sbjct: 751 NKSLDQALFEEKSL-QLGWSQRFEICLGVAKGLAYMHEESNPRIVHRDVKASNILLDSDL 809
Query: 205 EPKIADFGLAKLRSRESNIELNTHFRGTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVG 264
PK++DFGLAKL + ++T GT GY +PE +T K DV++FGI+ EIV
Sbjct: 810 VPKLSDFGLAKLYD-DKKTHISTRVAGTIGYLSPEYVMLGHLTEKTDVFAFGIVALEIVS 868
Query: 265 RRRHFDSSYSESQQWFPRWTWEMFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYS 324
R + + +Q+ W W + + + ++ ++ E D E +R++ VA C Q
Sbjct: 869 GRPNSSPELDDDKQYLLEWAWSLHQEQRDMEVVD-PDLTEFDKEEVKRVIGVAFLCTQTD 927
Query: 325 PNDRPLMSTVVKMLEGEIDISSPPFPFHNLVP--AKENSTQEGSTADSDTTTSSW 377
RP MS VV ML G+++I+ N P E + + + S +T+SSW
Sbjct: 928 HAIRPTMSRVVGMLTGDVEITEA-----NAKPGYVSERTFENAMSFMSGSTSSSW 977
>At1g56130 receptor-like protein kinase, putative
Length = 1032
Score = 222 bits (565), Expect = 3e-58
Identities = 132/343 (38%), Positives = 189/343 (54%), Gaps = 28/343 (8%)
Query: 53 KPVRFTPEKLDEITEKY--STILGSGAFGVVFKGELSNGENVAVKVLNCLDMGMEEQFKA 110
KP FT +L T+ + S LG G FG V+KG L++G VAVK+L+ + QF A
Sbjct: 678 KPYIFTYSELKSATQDFDPSNKLGEGGFGPVYKGNLNDGRVVAVKLLSVGSRQGKGQFVA 737
Query: 111 EVITIGRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIA 170
E++ I H NLVKLYG CF + R LVYEY+ NGSLD+ +FG K + D+ ++I
Sbjct: 738 EIVAISSVLHRNLVKLYGCCFEGEHRMLVYEYLPNGSLDQALFGDKTLH-LDWSTRYEIC 796
Query: 171 IGTAKGIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFR 230
+G A+G+ YLHEE RI+H D+K N+LLD +L P+I+DFGLAKL + ++T
Sbjct: 797 LGVARGLVYLHEEASVRIVHRDVKASNILLDSRLVPQISDFGLAKLYD-DKKTHISTRVA 855
Query: 231 GTRGYAAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRHFDSSYSESQQWFPRWTWEMFEN 290
GT GY APE +T K DVY+FG++ E+V R + D + E +++ W W + E
Sbjct: 856 GTIGYLAPEYAMRGHLTEKTDVYAFGVVALELVSGRPNSDENLEEEKKYLLEWAWNLHEK 915
Query: 291 NELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISSPPFP 350
+ + ++ ++ + + E A+RM+ +AL C Q S RP MS VV ML G+++I
Sbjct: 916 SRDIELID-DKLTDFNMEEAKRMIGIALLCTQTSHALRPPMSRVVAMLSGDVEI------ 968
Query: 351 FHNLVPAKENSTQEGSTADSDTTTSSWR---TESSRESGFKTK 390
G S WR T S SGF+ K
Sbjct: 969 --------------GDVTSKPGYVSDWRFDDTTGSSLSGFQIK 997
>At4g00340 receptor-like protein kinase
Length = 797
Score = 221 bits (564), Expect = 4e-58
Identities = 134/304 (44%), Positives = 178/304 (58%), Gaps = 14/304 (4%)
Query: 57 FTPEKLDEITEKYSTILGSGAFGVVFKGELSNGEN-VAVKVLNCLDMGMEEQFKAEVITI 115
F+ ++L T +S +G G FG VFKG L VAVK L G E +F+AEV TI
Sbjct: 451 FSFKELQSATNGFSDKVGHGGFGAVFKGTLPGSSTFVAVKRLERPGSG-ESEFRAEVCTI 509
Query: 116 GRTYHINLVKLYGFCFHRDKRALVYEYVENGSLDKYIFGSKNRNDFDFQKLHKIAIGTAK 175
G H+NLV+L GFC R LVY+Y+ GSL Y+ + + ++ +IA+GTAK
Sbjct: 510 GNIQHVNLVRLRGFCSENLHRLLVYDYMPQGSLSSYLSRTSPKL-LSWETRFRIALGTAK 568
Query: 176 GIAYLHEECKHRIIHYDIKPENVLLDMKLEPKIADFGLAKLRSRESNIELNTHFRGTRGY 235
GIAYLHE C+ IIH DIKPEN+LLD K++DFGLAKL R+ + L T RGT GY
Sbjct: 569 GIAYLHEGCRDCIIHCDIKPENILLDSDYNAKVSDFGLAKLLGRDFSRVLAT-MRGTWGY 627
Query: 236 AAPEMWKPYPVTYKCDVYSFGILLFEIVGRRRH-------FDSSYSESQQW-FPRW-TWE 286
APE P+T K DVYSFG+ L E++G RR+ +E ++W FP W E
Sbjct: 628 VAPEWISGLPITTKADVYSFGMTLLELIGGRRNVIVNSDTLGEKETEPEKWFFPPWAARE 687
Query: 287 MFENNELVVMLALCEIEEKDSEIAERMLKVALWCVQYSPNDRPLMSTVVKMLEGEIDISS 346
+ + N V+ + E E+ RM VA+WC+Q + RP M TVVKMLEG ++++
Sbjct: 688 IIQGNVDSVVDSRLNGEYNTEEVT-RMATVAIWCIQDNEEIRPAMGTVVKMLEGVVEVTV 746
Query: 347 PPFP 350
PP P
Sbjct: 747 PPPP 750
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,362,790
Number of Sequences: 26719
Number of extensions: 417892
Number of successful extensions: 4166
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 837
Number of HSP's successfully gapped in prelim test: 176
Number of HSP's that attempted gapping in prelim test: 1215
Number of HSP's gapped (non-prelim): 1139
length of query: 402
length of database: 11,318,596
effective HSP length: 102
effective length of query: 300
effective length of database: 8,593,258
effective search space: 2577977400
effective search space used: 2577977400
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148970.5


