

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148970.11 + phase: 0
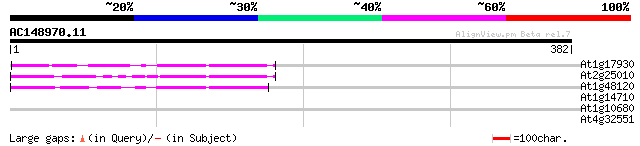
(382 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g17930 unknown protein 63 2e-10
At2g25010 unknown protein 57 2e-08
At1g48120 serine/threonine phosphatase PP7, putative 54 1e-07
At1g14710 unknown protein 31 1.3
At1g10680 putative P-glycoprotein-2 emb|CAA71277 30 2.9
At4g32551 Leunig protein 28 8.4
>At1g17930 unknown protein
Length = 478
Score = 63.2 bits (152), Expect = 2e-10
Identities = 49/181 (27%), Positives = 85/181 (46%), Gaps = 28/181 (15%)
Query: 2 PMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEYG 61
P GEMT+TLD+V+ + L ++G+ + G K + + + + LG K+ E
Sbjct: 84 PCGEMTITLDEVSLILGLAVDGKPVV-GVKEKDEDPSQVCLRLLG-------KLPKGELS 135
Query: 62 GY-ISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
G ++ L++ + A + +E+E Y R YL+Y+V +F
Sbjct: 136 GNRVTAKWLKESFAECPKGATM-------KEIE-------YHTRAYLIYIVGSTIFATTD 181
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
+I + YL D + Y+WGA LA+LY ++ +A + ++GG +TLL C
Sbjct: 182 PSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQIGNASQRSQSIIGGCLTLLQ----C 236
Query: 181 W 181
W
Sbjct: 237 W 237
>At2g25010 unknown protein
Length = 509
Score = 57.0 bits (136), Expect = 2e-08
Identities = 52/181 (28%), Positives = 83/181 (45%), Gaps = 23/181 (12%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P+GEMT+TLD+VA + L I+G + K G + M G + N+E
Sbjct: 92 LPLGEMTITLDEVALVLGLEIDGDPIVGSK-----VGDEVAMDMCGRLLGKLPSAANKE- 145
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYCVRCYLLYLVRCLLFGDRS 120
++ R++ ++L R +E PE+ + V+ + R YLLYL+ +F
Sbjct: 146 ---VNCSRVK---LNWLKR----TFSECPEDA-SFDVVKCH-TRAYLLYLIGSTIFATTD 193
Query: 121 NKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTLDDVC 180
++ + YL D + Y+WGA LA LY L +A + G +TLL C
Sbjct: 194 GDKVSVKYLPLFED-FDQAGRYAWGAAALACLYRALGNASLKSQSNICGCLTLLQ----C 248
Query: 181 W 181
W
Sbjct: 249 W 249
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 54.3 bits (129), Expect = 1e-07
Identities = 51/177 (28%), Positives = 79/177 (43%), Gaps = 25/177 (14%)
Query: 1 MPMGEMTVTLDDVACLTHLPIEGRMLDHGKKMPKHEGAALLMTYLGVAQHEAEKICNQEY 60
+P GE+TVTL DV L L ++G + K + A L LG + +
Sbjct: 101 LPAGEITVTLQDVNILLGLRVDGPAVTGSTK---YNWADLCEDLLGHRPGPKDL-----H 152
Query: 61 GGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERVRTYC-VRCYLLYLVRCLLFGDR 119
G ++S LR+ + + DP+EV C R ++L L+ L+GD+
Sbjct: 153 GSHVSLAWLRENFRNL---------PADPDEVT------LKCHTRAFVLALMSGFLYGDK 197
Query: 120 SNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGELADACRPGHRVLGGSVTLLTL 176
S + L +L + D + + SWG+ TLA LY EL A + + G + LL L
Sbjct: 198 SKHDVALTFLPLLRD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLLQL 253
>At1g14710 unknown protein
Length = 598
Score = 30.8 bits (68), Expect = 1.3
Identities = 20/77 (25%), Positives = 38/77 (48%), Gaps = 5/77 (6%)
Query: 304 IIAEQWQRYEARNSPDTYDMVSGAVAYADAQLGQEEVMSMTPQQSFEAITHMREQIAPIL 363
II Q +A + Y+ V G++ + +L +V++M Q F + + + I
Sbjct: 46 IIDSLCQHLQAVGDHNEYESVIGSIHHR--RLAWSQVLTM---QQFFPVADVSYNLQQIA 100
Query: 364 TRRRAQRPRRRHHHQDQ 380
+R+ Q P +RH++ DQ
Sbjct: 101 WKRQQQMPPQRHYNSDQ 117
>At1g10680 putative P-glycoprotein-2 emb|CAA71277
Length = 1227
Score = 29.6 bits (65), Expect = 2.9
Identities = 34/120 (28%), Positives = 56/120 (46%), Gaps = 21/120 (17%)
Query: 40 LLMTY-LGVAQHEAEKICNQEYGGYISYPRLRDFYTSYLGRANVLAGTEDPEEVEELERV 98
+L TY L ++ H +EKI Q YGG +S +YL +AN+LAG E + + V
Sbjct: 810 VLATYPLIISGHISEKIFMQGYGGNLS--------KAYL-KANMLAG----ESISNIRTV 856
Query: 99 RTYCV--RCYLLYLVRCLLFGDRSNKRIELIYLTTMADGYAGMRNYSWGAMTLAYLYGEL 156
+C + LY L +RS +R ++ + Y + + + + LA YG +
Sbjct: 857 VAFCAEEKVLDLYSKELLEPSERSFRRGQMAGIL-----YGVSQFFIFSSYGLALWYGSI 911
>At4g32551 Leunig protein
Length = 931
Score = 28.1 bits (61), Expect = 8.4
Identities = 13/49 (26%), Positives = 24/49 (48%)
Query: 334 QLGQEEVMSMTPQQSFEAITHMREQIAPILTRRRAQRPRRRHHHQDQDQ 382
Q Q +S QQ + M++ + +++ Q+ ++ HHHQ Q Q
Sbjct: 92 QQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQ 140
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,004,009
Number of Sequences: 26719
Number of extensions: 385290
Number of successful extensions: 769
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 757
Number of HSP's gapped (non-prelim): 9
length of query: 382
length of database: 11,318,596
effective HSP length: 101
effective length of query: 281
effective length of database: 8,619,977
effective search space: 2422213537
effective search space used: 2422213537
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148970.11


