

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148969.3 + phase: 2 /pseudo
(595 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
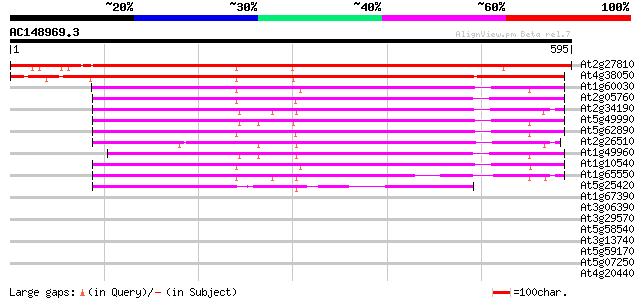
Score E
Sequences producing significant alignments: (bits) Value
At2g27810 putative membrane transporter 735 0.0
At4g38050 unknown protein 550 e-157
At1g60030 unknown protein 304 1e-82
At2g05760 putative membrane transporter 300 2e-81
At2g34190 putative membrane transporter 299 3e-81
At5g49990 permease 297 1e-80
At5g62890 permease 1 - like protein 290 2e-78
At2g26510 putative membrane transporter 288 6e-78
At1g49960 permease, putative 277 1e-74
At1g10540 putative permease 273 2e-73
At1g65550 hypothetical protein 229 3e-60
At5g25420 permease 1-like protein 179 5e-45
At1g67390 hypothetical protein 33 0.59
At3g06390 unknown protein 32 1.3
At3g29570 hypothetical protein 31 1.7
At5g58540 unknown protein 30 2.9
At3g13740 Unknown protein (MMM17.15) 30 2.9
At5g59170 cell wall protein 30 3.8
At5g07250 membrane protein 30 3.8
At4g20440 putative snRNP protein 30 5.0
>At2g27810 putative membrane transporter
Length = 721
Score = 735 bits (1897), Expect = 0.0
Identities = 403/695 (57%), Positives = 472/695 (66%), Gaps = 103/695 (14%)
Query: 1 WAQKTGFKPVFSAETKAGQSLS-------------RQPDLEAS--------PVVSTPSQA 39
WA+KTGF+P FS ET A S S QPDLEA PV + +
Sbjct: 29 WAKKTGFRPKFSGETTATDSSSGQLSLPVRAKQQETQPDLEAGQTRLRPPPPVSAAVTNG 88
Query: 40 VNDVPHGDKVPPSP------------------SDGVPTNN----------------ARVE 65
D +K PP P SDGV + R+E
Sbjct: 89 ETDKDKKEKPPPPPPGSVAVPVKDQPVKRRRDSDGVVGRSNGPDGANGSGDPVRRPGRIE 148
Query: 66 ERTTRLPVMVDHDDLVLRRRPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIA 125
E LP +D DDLV R + Y L D+P LV + YG+QHYLS++GSLIL PLVI
Sbjct: 149 ETVEVLPQSMD-DDLVARNLH--MKYGLRDTPGLVPIGFYGLQHYLSMLGSLILVPLVIV 205
Query: 126 PAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQE 185
PAMG SH+E A +V TVL VSG+TTLLHT FGSRLPLIQGPSFV+LAP LAIINSPEFQ
Sbjct: 206 PAMGGSHEEVANVVSTVLFVSGITTLLHTSFGSRLPLIQGPSFVFLAPALAIINSPEFQG 265
Query: 186 LN-ENKFKHIMKELQGAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL----- 239
LN N FKHIM+ELQGAIIIGSAFQ +LGY+GLMSL++R +NPVVV+ T+AAVGL
Sbjct: 266 LNGNNNFKHIMRELQGAIIIGSAFQAVLGYSGLMSLILRLVNPVVVAPTVAAVGLSFYSY 325
Query: 240 -FL*LARVLRSVQYRY*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENG 298
F + + L + ++ F LYLRKISV H IF IYAVPL LA+TW AFLLTE G
Sbjct: 326 GFPLVGKCLEIGVVQILLVIIFALYLRKISVLSHRIFLIYAVPLSLAITWAAAFLLTETG 385
Query: 299 --------------------------RMKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNW 332
RMK+C+V+TS ++S PWFRFPYPLQWG P+FNW
Sbjct: 386 AYTYKGCDPNVPVSNVVSTHCRKYMTRMKYCRVDTSHALSSAPWFRFPYPLQWGVPLFNW 445
Query: 333 KMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMG 392
KMA VMCVVS+I+SVDSVG+YH SSLL AS PPT GV+SR IGLEGF+S+LAGLWGTG G
Sbjct: 446 KMAFVMCVVSVIASVDSVGSYHASSLLVASRPPTRGVVSRAIGLEGFTSVLAGLWGTGTG 505
Query: 393 STTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAM 452
STTLTENVHTIA TKMGSRR V+LGAC+L++ SL GKVGGF+ASIP+ MVA LLC MWAM
Sbjct: 506 STTLTENVHTIAVTKMGSRRVVELGACVLVIFSLVGKVGGFLASIPQVMVASLLCFMWAM 565
Query: 453 LTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVT 512
TALGLSNLRY+E GSSRNIIIVGLSLFFSLS+PAYFQQY SP SN SVPSY+QPYIV+
Sbjct: 566 FTALGLSNLRYSEAGSSRNIIIVGLSLFFSLSVPAYFQQYGISPNSNLSVPSYYQPYIVS 625
Query: 513 SHGPFRSKYE------------ELNYVLNMIFSLHMVIAFLVALILDNTVPGSKQERELY 560
SHGPF+S+Y+ ++NYV+N + S+ MVIAF++A+ILDNTVPGSKQER +Y
Sbjct: 626 SHGPFKSQYKGDLQFSYLLVYLQMNYVMNTLLSMSMVIAFIMAVILDNTVPGSKQERGVY 685
Query: 561 GWSKPNDAREDPFIVSEYGLPARVGRCFRWVKWVG 595
WS A +P + +Y LP RVGR FRWVKWVG
Sbjct: 686 VWSDSETATREPALAKDYELPFRVGRFFRWVKWVG 720
>At4g38050 unknown protein
Length = 709
Score = 550 bits (1418), Expect = e-157
Identities = 301/661 (45%), Positives = 406/661 (60%), Gaps = 83/661 (12%)
Query: 1 WAQKTGFKPVFSAETKAGQSLSRQPDLEASPVVSTPS---QAVNDVPHGDKVPPSPSDGV 57
WA+KTGF +S ET S S + S P Q V H ++ P +
Sbjct: 54 WAKKTGFVSDYSGET----STSTRTKFGESSDFDLPKGRDQVVTGSSHKTEIDPI----L 105
Query: 58 PTNNARVEERTTRLPVMVDHDDLVLRR--------------------------------- 84
N +E T PV + ++ L R
Sbjct: 106 GRNRPEIEHVTGSEPVSREEEERRLNRNEATPETENEGGKINKDLENGFYYPGGGGESSE 165
Query: 85 -----RPSPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMV 139
+P + + L D+P V L YG+QHYLS++GSL+ PLVI PAM S +TA+++
Sbjct: 166 DGQWPKPILMKFGLRDNPGFVPLIYYGLQHYLSLVGSLVFIPLVIVPAMDGSDKDTASVI 225
Query: 140 CTVLLVSGVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHIMKELQ 199
T+LL++GVTT+LH FG+RLPL+QG SFVYLAPVL +INS EF+ L E+KF+ M+ELQ
Sbjct: 226 STMLLLTGVTTILHCYFGTRLPLVQGSSFVYLAPVLVVINSEEFRNLTEHKFRDTMRELQ 285
Query: 200 GAIIIGSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYR 253
GAII+GS FQ +LG++GLMSLL+RFINPVVV+ T+AAVGL F +
Sbjct: 286 GAIIVGSLFQCILGFSGLMSLLLRFINPVVVAPTVAAVGLAFFSYGFPQAGTCVEISVPL 345
Query: 254 Y*CLLFFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGR-------------- 299
LL F LYLR +S+FGH +F+IYAVPL + WT+AF LT G
Sbjct: 346 ILLLLIFTLYLRGVSLFGHRLFRIYAVPLSALLIWTYAFFLTVGGAYDYRGCNADIPSSN 405
Query: 300 ------------MKHCQVNTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSV 347
MKHC+ + S+ + W R PYP QWG P F+ + +I+M VSL++SV
Sbjct: 406 ILIDECKKHVYTMKHCRTDASNAWRTASWVRIPYPFQWGFPNFHMRTSIIMIFVSLVASV 465
Query: 348 DSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTK 407
DSVGTYH++S++ + PT G++SRGI LEGF SLLAG+WG+G GSTTLTEN+HTI TK
Sbjct: 466 DSVGTYHSASMIVNAKRPTRGIVSRGIALEGFCSLLAGIWGSGTGSTTLTENIHTINITK 525
Query: 408 MGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETG 467
+ SRR + +GA LIVLS GK+G +ASIP+A+ A +LC +WA+ +LGLSNLRYT+T
Sbjct: 526 VASRRALVIGAMFLIVLSFLGKLGAILASIPQALAASVLCFIWALTVSLGLSNLRYTQTA 585
Query: 468 SSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYV 527
S RNI IVG+SLF LSIPAYFQQY+ P S+ +PSY+ P+ S GPF++ E+L++
Sbjct: 586 SFRNITIVGVSLFLGLSIPAYFQQYQ--PLSSLILPSYYIPFGAASSGPFQTGIEQLDFA 643
Query: 528 LNMIFSLHMVIAFLVALILDNTVPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRC 587
+N + SL+MV+ FL+A ILDNTVPGSK+ER +Y W++ D + DP + ++Y LP + +
Sbjct: 644 MNAVLSLNMVVTFLLAFILDNTVPGSKEERGVYVWTRAEDMQMDPEMRADYSLPRKFAQI 703
Query: 588 F 588
F
Sbjct: 704 F 704
>At1g60030 unknown protein
Length = 538
Score = 304 bits (778), Expect = 1e-82
Identities = 180/522 (34%), Positives = 274/522 (52%), Gaps = 36/522 (6%)
Query: 87 SPLNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVS 146
S ++Y +T P + G QHYL ++G+ +L P + P MG ++E A MV T+L VS
Sbjct: 30 SSISYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTYLVPQMGGGNEEKAKMVQTLLFVS 89
Query: 147 GVTTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQEL--NENKFKHIMKELQGAIII 204
G+ TLL + FG+RLP + G S+ Y+ L+II + + ++ + KFK IM+ +QGA+I+
Sbjct: 90 GLNTLLQSFFGTRLPAVIGGSYTYVPTTLSIILAGRYSDILDPQEKFKRIMRGIQGALIV 149
Query: 205 GSAFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLL 258
S Q ++G++GL +VR ++P+ +A G F LA+ + LL
Sbjct: 150 ASILQIVVGFSGLWRNVVRLLSPLSAVPLVALAGFGLYEHGFPLLAKCIEIGLPEIILLL 209
Query: 259 FFVLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNT--------SDT 310
F Y+ + +F +AV + + W +A LLT G K+ VNT S
Sbjct: 210 LFSQYIPHLIRGERQVFHRFAVIFSVVIVWIYAHLLTVGGAYKNTGVNTQTSCRTDRSGL 269
Query: 311 MTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVL 370
++ PW R PYP QWG P F+ A M VS +S ++S GTY S A++ PP P VL
Sbjct: 270 ISGSPWIRVPYPFQWGPPTFHAGEAFAMMAVSFVSLIESTGTYIVVSRFASATPPPPSVL 329
Query: 371 SRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKV 430
SRG+G +G LL GL+G G G++ EN +A T++GSRR VQ+ A +I S+ GK
Sbjct: 330 SRGVGWQGVGVLLCGLFGAGNGASVSVENAGLLALTRVGSRRVVQISAGFMIFFSILGKF 389
Query: 431 GGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQ 490
G ASIP +VA L C+ +A + A GLS L++ S R I+G S+F LSIP YF
Sbjct: 390 GAIFASIPAPVVAALHCLFFAYVGAGGLSLLQFCNLNSFRTKFILGFSVFMGLSIPQYFN 449
Query: 491 QYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV 550
QY + V +GP + N ++N+ FS +A ++A LD T+
Sbjct: 450 QYTA----------------VNKYGPVHTHARWFNDMINVPFSSKAFVAGILAFFLDVTM 493
Query: 551 ----PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
++++R ++ W + + D Y LP + + F
Sbjct: 494 SSKDSATRKDRGMFWWDRFMSFKSDTRSEEFYSLPFNLNKYF 535
>At2g05760 putative membrane transporter
Length = 520
Score = 300 bits (768), Expect = 2e-81
Identities = 177/519 (34%), Positives = 277/519 (53%), Gaps = 35/519 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P + Q+Y+ ++G+ P ++ PAMG S + A ++ T+L V+G+
Sbjct: 15 LEYCIDSNPPWPETVLLAFQNYILMLGTSAFIPALLVPAMGGSDGDRARVIQTLLFVAGI 74
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGS 206
TLL +FG+RLP + G S Y+ P+ IIN Q+++ + +F H M+ +QGA+I+ S
Sbjct: 75 KTLLQALFGTRLPAVVGGSLAYVVPIAYIINDSSLQKISNDHERFIHTMRAIQGALIVAS 134
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
+ Q +LGY+ + L RF +P+ ++ + VGL F L + ++
Sbjct: 135 SIQIILGYSQVWGLFSRFFSPLGMAPVVGLVGLGMFQRGFPQLGNCIEIGLPMLLLVIGL 194
Query: 261 VLYLRKISVFGH-HIFQIYAVPLGLAVTWTFAFLLTENGRMK--------HCQVNTSDTM 311
YL+ + F IF+ + + + + + W +A +LT +G + C+ + ++ +
Sbjct: 195 TQYLKHVRPFKDVPIFERFPILICVTIVWIYAVILTASGAYRGKPSLTQHSCRTDKANLI 254
Query: 312 TSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLS 371
++ PWF+FPYPLQWG P F+ + M L+S V+S G Y +S LA + PP VLS
Sbjct: 255 STAPWFKFPYPLQWGPPTFSVGHSFAMMSAVLVSMVESTGAYIAASRLAIATPPPAYVLS 314
Query: 372 RGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVG 431
RGIG +G LL GL+GTG GST L ENV + T++GSRR VQ+ A +IV S GK G
Sbjct: 315 RGIGWQGIGVLLDGLFGTGTGSTVLVENVGLLGLTRVGSRRVVQVSAGFMIVFSTLGKFG 374
Query: 432 GFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQ 491
ASIP + A L CI++ ++ A+GLS L++T S RN++I GLSLF +SIP +F Q
Sbjct: 375 AVFASIPVPIYAALHCILFGLVAAVGLSFLQFTNMNSMRNLMITGLSLFLGISIPQFFAQ 434
Query: 492 YESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT-- 549
Y + +G + N LN +F + ++A+ +DNT
Sbjct: 435 Y----------------WDARHYGLVHTNAGWFNAFLNTLFMSPATVGLIIAVFMDNTME 478
Query: 550 VPGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
V SK++R + W K R D Y LP + R F
Sbjct: 479 VERSKKDRGMPWWVKFRTFRGDNRNEEFYTLPFNLNRFF 517
>At2g34190 putative membrane transporter
Length = 524
Score = 299 bits (766), Expect = 3e-81
Identities = 171/522 (32%), Positives = 282/522 (53%), Gaps = 42/522 (8%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P G +HY+ +G+ ++ P ++ P MG + +V T+L + GV
Sbjct: 20 LEYCIDSNPPWGEAIALGFEHYILALGTAVMIPSILVPMMGGDDGDKVRVVQTLLFLQGV 79
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNENKFKHI--MKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ P+++II+ + + + + + M+ +QGAII+ S
Sbjct: 80 NTLLQTLFGTRLPTVIGGSYAFMVPIISIIHDSSLTRIEDPQLRFLSTMRAVQGAIIVAS 139
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVLY 263
+ Q +LG++ + ++ RF +P+ + IA G L V V+ L+ FV++
Sbjct: 140 SVQIILGFSQMWAICSRFFSPIGMVPVIALTGFGLFNRGFPVVGNCVEIGLPMLILFVIF 199
Query: 264 LRKISVFGHHIFQI---YAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMT 312
+ + F F + +A+ + L + W +A +LT +G KH C+ + S+ ++
Sbjct: 200 SQYLKNFQFRQFPVVERFALIIALIIVWAYAHVLTASGAYKHRPHQTQLNCRTDMSNLIS 259
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
S PW + PYPLQWG P F+ A M L+S ++S G + ++ LA++ PP P VLSR
Sbjct: 260 SAPWIKIPYPLQWGAPSFDAGHAFAMMAAVLVSLIESTGAFKAAARLASATPPPPHVLSR 319
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G LL GL+GT GS+ EN+ + T++GSRR +Q+ A +I S+ GK G
Sbjct: 320 GIGWQGIGILLNGLFGTLSGSSVSVENIGLLGSTRVGSRRVIQISAGFMIFFSMLGKFGA 379
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP + A + C+++ ++ ++GLS L++T S RN+ IVG+SLF LSIP YF+ +
Sbjct: 380 LFASIPFTIFAAVYCVLFGLVASVGLSFLQFTNMNSLRNLFIVGVSLFLGLSIPEYFRDF 439
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ HGP + N LN IF ++A +VA+ LDNT+
Sbjct: 440 S----------------MKALHGPAHTNAGWFNDFLNTIFLSSPMVALMVAVFLDNTLDY 483
Query: 551 PGSKQERELYGWSK----PNDAREDPFIVSEYGLPARVGRCF 588
+ ++R L W+K D+R + F Y LP + R F
Sbjct: 484 KETARDRGLPWWAKFRTFKGDSRNEEF----YTLPFNLNRFF 521
>At5g49990 permease
Length = 528
Score = 297 bits (760), Expect = 1e-80
Identities = 168/520 (32%), Positives = 283/520 (54%), Gaps = 36/520 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
++Y +T P + G QHYL ++G+ +L P + P MG ++E A ++ T+L V+G+
Sbjct: 22 ISYCITSPPPWPEAVLLGFQHYLVMLGTTVLIPSALVPQMGGRNEEKAKLIQTILFVAGL 81
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ ++I+ S F ++ + +FK I++ QGA+I+ S
Sbjct: 82 NTLLQTVFGTRLPAVIGASYTFVPVTISIMLSGRFNDVADPVERFKRIIRATQGALIVAS 141
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL- 262
Q +LG++GL +VRF++P+ + + VG L V + ++ L+ +L
Sbjct: 142 TLQIILGFSGLWRNVVRFLSPLSAAPLVGLVGYGLYELGFPGVAKCIEIGLPGLIILILI 201
Query: 263 --YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGR--------MKHCQVNTSDTMT 312
Y+ + G H+F +AV +A+ W +AF LT G + C+ + + ++
Sbjct: 202 SQYMPHVIKGGKHVFARFAVIFSVAIVWLYAFFLTLGGAYNGVGTDTQRSCRTDRAGLIS 261
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
+ PW R P+P QWG P+F+ A M + S ++ V+S G + S A++ P P V+SR
Sbjct: 262 AAPWIRVPWPFQWGAPLFDAGEAFAMMMASFVALVESTGAFIAVSRYASATMPPPSVISR 321
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
G+G +G + L++GL+GTG+GS+ EN +A TK+GSRR VQ+ A +I S+ GK G
Sbjct: 322 GVGWQGVAILISGLFGTGIGSSVSVENAGLLALTKIGSRRVVQISAGFMIFFSILGKFGA 381
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP ++A L C+ +A + A GLS L++ S R + I+G S+F LSIP YF ++
Sbjct: 382 VFASIPSPIIAALYCLFFAYVGAGGLSLLQFCNLNSFRTLFILGFSIFLGLSIPQYFNEH 441
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ + +GP + N ++N+ FS + VA +LD T+
Sbjct: 442 TA----------------IKGYGPVHTGARWFNDMVNVPFSSKAFVGGCVAYLLDTTLHK 485
Query: 551 -PGS-KQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
GS +++R + W + + DP Y LP + + F
Sbjct: 486 KDGSIRKDRGKHWWDRFWTFKNDPRTEEFYALPFNLNKYF 525
>At5g62890 permease 1 - like protein
Length = 532
Score = 290 bits (741), Expect = 2e-78
Identities = 168/520 (32%), Positives = 272/520 (52%), Gaps = 36/520 (6%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
++Y +T P + G QHYL ++G+ +L P + P MG ++E A ++ T+L V+G+
Sbjct: 26 ISYCITSPPPWPEAILLGFQHYLVMLGTTVLIPTALVPQMGGGYEEKAKVIQTILFVAGI 85
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TLL T+FG+RLP + G S+ ++ ++II S F + + ++F+ IM+ QGA+I+ S
Sbjct: 86 NTLLQTLFGTRLPAVVGASYTFVPTTISIILSGRFSDTSNPIDRFERIMRATQGALIVAS 145
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
Q +LG++GL +VRF++P+ + VG F +A+ + L+F
Sbjct: 146 TLQMILGFSGLWRNVVRFLSPISAVPLVGLVGFGLYEFGFPGVAKCIEIGLPELLILVFV 205
Query: 261 VLYLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMK--------HCQVNTSDTMT 312
YL + G ++F +AV + + W +A LLT G C+ + + +
Sbjct: 206 SQYLPHVIKSGKNVFDRFAVIFAVVIVWIYAHLLTVGGAYNGAAPTTQTSCRTDRAGIIG 265
Query: 313 SPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSR 372
+ PW R P+P QWG P F+ A M + S ++ V+S G + S A++ P +LSR
Sbjct: 266 AAPWIRVPWPFQWGAPSFDAGEAFAMMMASFVALVESTGAFVAVSRYASATMLPPSILSR 325
Query: 373 GIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGG 432
GIG +G + L++GL+GTG GS+ EN +A T++GSRR VQ+ A +I S+ GK G
Sbjct: 326 GIGWQGVAILISGLFGTGAGSSVSVENAGLLALTRVGSRRVVQIAAGFMIFFSILGKFGA 385
Query: 433 FIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
ASIP ++A L C+ +A + A GLS L++ S R I+G S+F LSIP YF +Y
Sbjct: 386 VFASIPAPIIAALYCLFFAYVGAGGLSFLQFCNLNSFRTKFILGFSVFLGLSIPQYFNEY 445
Query: 493 ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTV-- 550
+ + +GP + N ++N+ FS +A VA LDNT+
Sbjct: 446 TA----------------IKGYGPVHTGARWFNDMVNVPFSSEPFVAGSVAFFLDNTLHK 489
Query: 551 --PGSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
+++R + W K + D Y LP + + F
Sbjct: 490 KDSSIRKDRGKHWWDKFRSFKGDTRSEEFYSLPFNLNKYF 529
>At2g26510 putative membrane transporter
Length = 551
Score = 288 bits (737), Expect = 6e-78
Identities = 176/520 (33%), Positives = 279/520 (52%), Gaps = 45/520 (8%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
L Y + +P+ V QHY+ ++G+ +L + MG + A ++ T+L +SG+
Sbjct: 44 LQYCIHSNPSWHETVVLAFQHYIVMLGTTVLIANTLVSPMGGDPGDKARVIQTILFMSGI 103
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAII---NSPEFQELNENKFKHIMKELQGAIIIG 205
TLL T+ G+RLP + G SF Y+ PVL+II N+ +F + +F+H M+ +QG++II
Sbjct: 104 NTLLQTLIGTRLPTVMGVSFAYVLPVLSIIRDYNNGQFDS-EKQRFRHTMRTVQGSLIIS 162
Query: 206 SAFQTLLGYTGLMSLLVRFINPVVVSSTIAAV--GLFL*LARVLRS-VQYRY*CLLFFVL 262
S ++GY L+R +P++V ++ V GLFL +L + V+ L+ ++
Sbjct: 163 SFVNIIIGYGQAWGNLIRIFSPIIVVPVVSVVSLGLFLRGFPLLANCVEIGLPMLILLII 222
Query: 263 ---YLRKISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTM 311
YL+ I + YA+ + LA+ W FA +LT +G + C+ + + M
Sbjct: 223 TQQYLKHAFSRISMILERYALLVCLAIIWAFAAILTVSGAYNNVSTATKQSCRTDRAFLM 282
Query: 312 TSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLS 371
+S PW R PYP QWGTP+F M ++++S +S G + +S LA + P V+S
Sbjct: 283 SSAPWIRIPYPFQWGTPIFKASHVFGMFGAAIVASAESTGVFFAASRLAGATAPPAHVVS 342
Query: 372 RGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVG 431
R IGL+G LL G++G+ G+T ENV + T++GSRR VQ+ +I S+FGK G
Sbjct: 343 RSIGLQGIGVLLEGIFGSITGNTASVENVGLLGLTRIGSRRVVQVSTFFMIFFSIFGKFG 402
Query: 432 GFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQ 491
F ASIP + AG+ CI+ ++ A+G+S +++T+T S RN+ ++G+SLF SLSI YF
Sbjct: 403 AFFASIPLPIFAGVYCILLGIVVAVGISFIQFTDTNSMRNMYVIGVSLFLSLSIAQYFLA 462
Query: 492 YESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNTVP 551
S +GP R+ N +LN IF+ ++A ++A ILDNT+
Sbjct: 463 NTSR----------------AGYGPVRTAGGWFNDILNTIFASAPLVATILATILDNTLE 506
Query: 552 GSKQERELYG--WSKP-----NDAREDPFIVSEYGLPARV 584
+ G W KP D R D F Y +P R+
Sbjct: 507 ARHASDDARGIPWWKPFQHRNGDGRNDEF----YSMPLRI 542
>At1g49960 permease, putative
Length = 526
Score = 277 bits (708), Expect = 1e-74
Identities = 163/505 (32%), Positives = 259/505 (51%), Gaps = 36/505 (7%)
Query: 104 VYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGVTTLLHTIFGSRLPLI 163
V G QHY+ ++G+ ++ P ++ P MG E A ++ TVL VSG+ TLL ++FGSRLP++
Sbjct: 35 VLGFQHYIVMLGTTVIIPSILVPLMGGGDVEKAEVINTVLFVSGINTLLQSLFGSRLPVV 94
Query: 164 QGPSFVYLAPVLAIINSPEFQELNEN--KFKHIMKELQGAIIIGSAFQTLLGYTGLMSLL 221
G S+ YL P L I S F +F+ M+ +QGA+II S ++G+ GL +L
Sbjct: 95 MGASYAYLIPALYITFSYRFTYYLHPHLRFEETMRAIQGALIIASISHMIMGFFGLWRIL 154
Query: 222 VRFINPVVVSSTIAAVGLFL*---LARVLRSVQYRY*CLLFFVL---YLRKISVFGHHIF 275
VRF++P+ + + G+ L ++ R ++ L+ ++ YL + I
Sbjct: 155 VRFLSPLSAAPLVILTGVGLLAFAFPQLARCIEIGLPALIILIILSQYLPHLFKCKRSIC 214
Query: 276 QIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMTSPPWFRFPYPLQWGT 327
+ +AV +A+ W +A +LT G C+ + S +++ PW R PYPLQWG
Sbjct: 215 EQFAVLFTIAIVWAYAEILTAAGAYDKRPDNTQLSCRTDRSGLISASPWVRIPYPLQWGR 274
Query: 328 PVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEGFSSLLAGLW 387
P F+ A M + ++ V++ G++ +S ++ P VLSRGIG +G LL GL+
Sbjct: 275 PSFHGSDAFAMMAATYVAIVETTGSFIAASRFGSATHIPPSVLSRGIGWQGIGVLLNGLF 334
Query: 388 GTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIPEAMVAGLLC 447
GT GST L EN + TK+GSRR VQ+ A +I S+FGK G +ASIP + A L C
Sbjct: 335 GTATGSTALVENTGLLGLTKVGSRRVVQISAGFMIFFSIFGKFGAVLASIPLPIFAALYC 394
Query: 448 IMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQ 507
+++A + + GL L++ S RN I+G S+F LS+ YF +Y
Sbjct: 395 VLFAYVASAGLGLLQFCNLNSFRNKFILGFSIFIGLSVAQYFTEY--------------- 439
Query: 508 PYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT----VPGSKQERELYGWS 563
++ GP ++ N ++ +IFS + + A +LD T +++ + W
Sbjct: 440 -LFISGRGPVHTRTSAFNVIMQVIFSSAATVGIMAAFLLDCTHSYGHASVRRDSGRHWWE 498
Query: 564 KPNDAREDPFIVSEYGLPARVGRCF 588
K D Y LP + R F
Sbjct: 499 KFRVYHTDTRTEEFYALPYNLNRFF 523
>At1g10540 putative permease
Length = 539
Score = 273 bits (699), Expect = 2e-73
Identities = 163/523 (31%), Positives = 271/523 (51%), Gaps = 39/523 (7%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
+ Y LT P + G QHYL ++G+ +L P ++ + A +++ ++ T+L VSG+
Sbjct: 30 ITYCLTSPPPWPETILLGFQHYLVMLGTTVLIPTMLVSKIDARNEDKVKLIQTLLFVSGI 89
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TL + FG+RLP + G S+ Y+ ++I+ + + ++ + +F+ IM+ +QGA+II S
Sbjct: 90 NTLFQSFFGTRLPAVIGASYSYVPTTMSIVLAARYNDIMDPQKRFEQIMRGIQGALIIAS 149
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGL------FL*LARVLRSVQYRY*CLLFF 260
L+G++GL + RF++P+ +A G F LA+ + L+ F
Sbjct: 150 FLHILVGFSGLWRNVTRFLSPLSAVPLVAFSGFGLYEQGFPMLAKCIEIGLPEIILLVIF 209
Query: 261 VLYLRKI--SVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKHCQVNT--------SDT 310
Y+ + + F +AV + + W +A++LT G + ++NT +
Sbjct: 210 SQYIPHLMQGETCSNFFHRFAVIFSVVIVWLYAYILTIGGAYSNTEINTQISCRTDRAGI 269
Query: 311 MTSPPWFRFPYPLQWG-TPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGV 369
+++ PW R P+P+QWG P FN M S +S V+S GTY S A++ P P V
Sbjct: 270 ISASPWIRVPHPIQWGGAPTFNAGDIFAMMAASFVSLVESTGTYIAVSRYASATPIPPSV 329
Query: 370 LSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGK 429
LSRGIG +GF LL GL+G G ++ EN +A T++GSRR +Q+ A +I S+ GK
Sbjct: 330 LSRGIGWQGFGILLCGLFGAGNATSVSVENAGLLAVTRVGSRRVIQVAAGFMIFFSILGK 389
Query: 430 VGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYF 489
G ASIP +VA L C+ ++ + A GLS +++ S R I+G S+F LSIP YF
Sbjct: 390 FGAIFASIPAPIVAALYCLFFSYVGAGGLSLIQFCNLNSFRTKFILGFSIFMGLSIPQYF 449
Query: 490 QQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHMVIAFLVALILDNT 549
QY + + ++GP R+ N ++N+ FS ++ ++A LD T
Sbjct: 450 YQYTT----------------LETYGPVRTSATWFNNIINVPFSSKAFVSGILAFFLDTT 493
Query: 550 VP----GSKQERELYGWSKPNDAREDPFIVSEYGLPARVGRCF 588
+P +K++R L W + + D Y LP + + F
Sbjct: 494 LPPKDKTTKKDRGLVWWKRFKSFQSDNRSEEFYSLPLNLSKYF 536
>At1g65550 hypothetical protein
Length = 515
Score = 229 bits (584), Expect = 3e-60
Identities = 161/533 (30%), Positives = 252/533 (47%), Gaps = 83/533 (15%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
+ Y + P + V G QHYL +G +L P V+ P MG + E ++ T+L VSG+
Sbjct: 30 IQYCVNSPPPWLEAVVLGFQHYLLSLGITVLIPSVLVPLMGGGYAEKVKVIQTLLFVSGL 89
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TTL + FG+RLP+I S+ Y+ P+ +II S F + +F M+ +QGA+II
Sbjct: 90 TTLFQSFFGTRLPVIAVASYAYIIPITSIIYSTRFTYYTDPFERFVRTMRSIQGALIITG 149
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVS--STIAAVGL----FL*LARVLRSVQYRY*CLLFF 260
FQ L+ G+ +VRF++P+ ++ +T +GL F LAR + L+F
Sbjct: 150 CFQVLICILGVWRNIVRFLSPLSIAPLATFTGLGLYHIGFPLLARCVEVGLPGLILLIFV 209
Query: 261 VLYLRKISVFGHHIFQI-------YAVPLGLAVTWTFAFLLTENGRMKH--------CQV 305
YL + + + Y + L + + W FA LLT +G H C+
Sbjct: 210 TQYLPRFLKMKKGVMILDGSRCDRYGMILCIPLVWLFAQLLTSSGVYDHKSHTTQTSCRT 269
Query: 306 NTSDTMTSPPWFRFPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPP 365
+ + +T+ PW PYP QWG+P F+ + M S ++ +S G ++ S+ ++ P
Sbjct: 270 DRTGLITNTPWIYIPYPFQWGSPTFDITDSFAMMAASFVTLFESTGLFYASARYGSATPI 329
Query: 366 TPGVLSRGIGLEGFSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLS 425
P V+SRG G LL G+ G G TT TENV +A TK+GSRR +Q+ A +I S
Sbjct: 330 PPSVVSRGTCWLGVGVLLNGMLGGITGITTSTENVGLLAMTKIGSRRVIQISAAFMIFFS 389
Query: 426 LFGKVGGFIASIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSI 485
+F V GLS L++ S I+G S F ++SI
Sbjct: 390 IFASV--------------------------GLSYLQFCNLNSFNIKFILGFSFFMAISI 423
Query: 486 PAYFQQYESSPESNFSVPSYFQPYIVTSHGPFRSKYEE--LNYVLNMIFSLHMVIAFLVA 543
P YF++Y +G +RS + L ++ +IF H +A ++A
Sbjct: 424 PQYFREY--------------------YNGGWRSDHHSNWLEDMIRVIFMSHTTVAAIIA 463
Query: 544 LILDNTV----PGSKQERELYGWSKPN----DAREDPFIVSEYGLPARVGRCF 588
++LD T+ +K++ + W K D R D F YGLP R+ + F
Sbjct: 464 IVLDCTLCRDSDEAKKDCGMKWWDKFRLYNLDVRNDEF----YGLPCRLNKFF 512
>At5g25420 permease 1-like protein
Length = 419
Score = 179 bits (453), Expect = 5e-45
Identities = 118/414 (28%), Positives = 193/414 (46%), Gaps = 72/414 (17%)
Query: 89 LNYELTDSPALVFLAVYGIQHYLSIIGSLILTPLVIAPAMGASHDETAAMVCTVLLVSGV 148
+ Y + P + V G QHYL +G +L P ++ P MG E ++ T+L VSG+
Sbjct: 42 IQYCVNSPPPWLEAVVLGFQHYLLSLGITVLIPSLLVPLMGGGDAEKVKVIQTLLFVSGL 101
Query: 149 TTLLHTIFGSRLPLIQGPSFVYLAPVLAIINSPEFQELNE--NKFKHIMKELQGAIIIGS 206
TTL + FG+RLP+I S+ Y+ P+ +II S F + +F M+ +QGA+II
Sbjct: 102 TTLFQSFFGTRLPVIASASYAYIIPITSIIYSTRFTYYTDPFERFVRTMRSIQGALIITG 161
Query: 207 AFQTLLGYTGLMSLLVRFINPVVVSSTIAAVGLFL*LARVLRSVQYRY*CLLFFVLYLRK 266
FQ L+ + G+ +VRF++P+ ++ + GL L Y + F L +
Sbjct: 162 CFQVLVCFLGVWRNIVRFLSPLSIAPLVTFTGLGL----------YH----IGFPLVKKG 207
Query: 267 ISVFGHHIFQIYAVPLGLAVTWTFAFLLTENGRMKH--------CQVNTSDTMTSPPWFR 318
++ + Y + L + V W FA LLT +G H C+ + + +T+ P
Sbjct: 208 PMIWDGNRCDRYGMMLCIPVVWLFAQLLTSSGVYDHKPQTTQTSCRTDRTGLITNTP--- 264
Query: 319 FPYPLQWGTPVFNWKMAIVMCVVSLISSVDSVGTYHTSSLLAASGPPTPGVLSRGIGLEG 378
P F+ + M S ++ +S G ++ S+
Sbjct: 265 --------CPTFDITDSFAMMAASFVTLFESTGLFYASARYG------------------ 298
Query: 379 FSSLLAGLWGTGMGSTTLTENVHTIAGTKMGSRRPVQLGACLLIVLSLFGKVGGFIASIP 438
+NV +A TK+GSRR +Q+ A ++ S+FGK G F ASIP
Sbjct: 299 -------------------KNVGLLAMTKVGSRRVIQISAAFMLFFSIFGKFGAFFASIP 339
Query: 439 EAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQY 492
++A L CI+ +++ GLS L++ S I+G S F ++SIP YF++Y
Sbjct: 340 LPIMASLYCIVLCFVSSAGLSFLQFCNLNSFNTKFILGFSFFMAISIPQYFREY 393
>At1g67390 hypothetical protein
Length = 479
Score = 32.7 bits (73), Expect = 0.59
Identities = 29/122 (23%), Positives = 53/122 (42%), Gaps = 23/122 (18%)
Query: 456 LGLSNLRYTETGSSRNIIIVGLSLFFSLSIPAYFQQYESSPESNFSVPSYFQP------- 508
L + ++ Y +N +I L F+ + + E P N+++ S+ Q
Sbjct: 269 LDIFSIFYFAFAKEQNFVIKAPRLLFN----NWTKDLEKVPNMNYNITSHAQEKKNIGHE 324
Query: 509 YIVTSHGPFRSKYEELNYVLNMIFS--LHMVIAFLVA----------LILDNTVPGSKQE 556
++V+ + K + L V+++ S +HM+ LVA L DN VP SK++
Sbjct: 325 FVVSGDASYLQKLKSLKVVVDVTNSREVHMLRDILVAWNGVLEELHILFKDNNVPNSKED 384
Query: 557 RE 558
E
Sbjct: 385 SE 386
>At3g06390 unknown protein
Length = 199
Score = 31.6 bits (70), Expect = 1.3
Identities = 22/82 (26%), Positives = 39/82 (46%), Gaps = 6/82 (7%)
Query: 69 TRLPVMVDHDDLVLRR-----RPSPLNYELTDSPALV-FLAVYGIQHYLSIIGSLILTPL 122
T L VMV D + + P+P++ E DSPA + F+ + + ++I +L+ L
Sbjct: 51 TALIVMVTSDQTEMTQLPGVSSPAPVSAEFNDSPAFIYFVVALVVASFYALISTLVSISL 110
Query: 123 VIAPAMGASHDETAAMVCTVLL 144
++ P A A + V+L
Sbjct: 111 LLKPEFTAQFSIYLASLDMVML 132
>At3g29570 hypothetical protein
Length = 327
Score = 31.2 bits (69), Expect = 1.7
Identities = 28/88 (31%), Positives = 38/88 (42%), Gaps = 11/88 (12%)
Query: 6 GFKPVFSAETKAGQSLSRQPD-LEASPVVSTPSQAVNDVPHGDKVPPSPSDGVPTNNARV 64
GFKP SA+ + G L QPD ++++PV+ P D PSPS + T
Sbjct: 57 GFKPSPSAKLQTGFKLFHQPDTIQSAPVIPPP-----DGQDQLGFKPSPSAKLQTGVKLF 111
Query: 65 EERTT-----RLPVMVDHDDLVLRRRPS 87
+ T LP D D L + PS
Sbjct: 112 HQHATIQSAPVLPPPDDQDQLGFKPSPS 139
Score = 30.0 bits (66), Expect = 3.8
Identities = 20/56 (35%), Positives = 30/56 (52%), Gaps = 6/56 (10%)
Query: 6 GFKPVFSAETKAGQSLSRQPD-LEASPVVSTPSQAVNDVPHGDKVPPSPSDGVPTN 60
GFKP SA+ + G L QPD ++++PV+ P D H P+PS + T+
Sbjct: 247 GFKPSPSAKLQTGVKLFHQPDTIQSAPVLPPP-----DGQHQLGFNPNPSAKLQTD 297
>At5g58540 unknown protein
Length = 484
Score = 30.4 bits (67), Expect = 2.9
Identities = 20/57 (35%), Positives = 25/57 (43%), Gaps = 7/57 (12%)
Query: 23 RQPDLEASPVVSTPSQAVNDV------PHGDKVPPSPSDGVPTNNARVEERTTRLPV 73
+ P L+ASP S S + D P G++ P P GVPT T LPV
Sbjct: 64 KDPALDASPP-SPESAILKDPLLPPPPPEGNETPSPPRSGVPTQTPETPPAITPLPV 119
>At3g13740 Unknown protein (MMM17.15)
Length = 247
Score = 30.4 bits (67), Expect = 2.9
Identities = 29/118 (24%), Positives = 52/118 (43%), Gaps = 19/118 (16%)
Query: 10 VFSAETKAGQSLSRQPDLEASPVVSTPSQAVNDVPHGDKV----PPSPSDGVPTNNARVE 65
+ SA + +L QP L +P + V +P+G+ P+PS G+ + A +
Sbjct: 3 ILSASSTVRAALDTQPRLPYNP---NAPRKVKKIPNGNSFLPPPSPAPSPGISISVADLL 59
Query: 66 ERTTRLPVMVDHDDLVLRRR---PSPLNYE----LTDSPALVFLA-----VYGIQHYL 111
+R + + VD DD + PSP E + + +L F+ +Y +H+L
Sbjct: 60 KRPSSKEITVDVDDTYMGYETWSPSPPKLEKPRSVFNPASLAFIGDSIYELYARRHFL 117
>At5g59170 cell wall protein
Length = 288
Score = 30.0 bits (66), Expect = 3.8
Identities = 12/40 (30%), Positives = 20/40 (50%)
Query: 287 TWTFAFLLTENGRMKHCQVNTSDTMTSPPWFRFPYPLQWG 326
T+ A L+T+ C +++ T + P WF P +WG
Sbjct: 7 TFALAVLVTQGSIFSLCLASSASTNSVPNWFHHWPPFKWG 46
>At5g07250 membrane protein
Length = 346
Score = 30.0 bits (66), Expect = 3.8
Identities = 28/130 (21%), Positives = 55/130 (41%), Gaps = 24/130 (18%)
Query: 425 SLFGKVGGFIA------SIPEAMVAGLLCIMWAMLTALGLSNLRYTETGSSRNIIIVGLS 478
+LFG +G ++ +I +A LL +++ +L L + L + + + + G
Sbjct: 203 ALFGLLGSMLSELFTNWTIYSNKIAALLTLLFVILINLAIGILPHVDNFAHVGGFVTGFL 262
Query: 479 LFFSLSIPAYFQQY--ESSPESNFSVPSYFQPYIVTSHGPFRSKYEELNYVLNMIFSLHM 536
L F L F+ E P+ P R KY+ Y+L ++ + +
Sbjct: 263 LGFILLARPQFKWLAREHMPQGT----------------PLRYKYKTYQYLLWLLSLVLL 306
Query: 537 VIAFLVALIL 546
+ F+VAL++
Sbjct: 307 IAGFVVALLM 316
>At4g20440 putative snRNP protein
Length = 257
Score = 29.6 bits (65), Expect = 5.0
Identities = 12/31 (38%), Positives = 17/31 (54%)
Query: 21 LSRQPDLEASPVVSTPSQAVNDVPHGDKVPP 51
+SR P L A P++ P Q + P G + PP
Sbjct: 147 ISRPPQLSAPPIIRPPGQMLPPPPFGGQGPP 177
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,204,503
Number of Sequences: 26719
Number of extensions: 570795
Number of successful extensions: 1679
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1605
Number of HSP's gapped (non-prelim): 28
length of query: 595
length of database: 11,318,596
effective HSP length: 105
effective length of query: 490
effective length of database: 8,513,101
effective search space: 4171419490
effective search space used: 4171419490
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148969.3


