

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148965.10 + phase: 0
(636 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
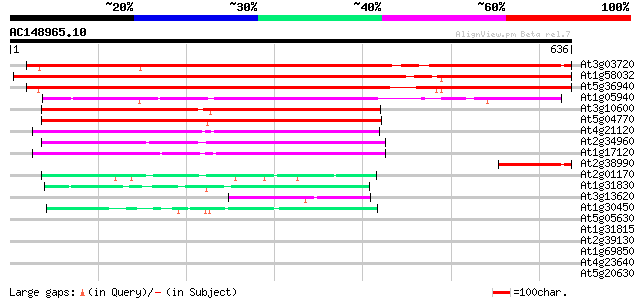
Score E
Sequences producing significant alignments: (bits) Value
At3g03720 putative cationic amino acid transporter 798 0.0
At1g58032 unknown protein 797 0.0
At5g36940 cationic amino acid transporter -like protein 761 0.0
At1g05940 unknown protein (At1g05940) 322 4e-88
At3g10600 putative amino acid transporter 287 1e-77
At5g04770 amino acid transporter-like protein 285 4e-77
At4g21120 amino acid transport protein AAT1 257 2e-68
At2g34960 amino acid transporter like protein 240 2e-63
At1g17120 putative amino acid transporter 228 8e-60
At2g38990 similar to MURA transposase of maize Mutator transposon 96 6e-20
At2g01170 putative amino acid or GABA permease 53 6e-07
At1g31830 unknown protein 47 3e-05
At3g13620 unknown protein 46 6e-05
At1g30450 cation-chloride co-transporter, putative 45 2e-04
At5g05630 unknown protein 42 0.001
At1g31815 hypothetical protein 38 0.020
At2g39130 unknown protein 36 0.075
At1g69850 nitrate transporter (NTL1) 32 0.83
At4g23640 potassium transport like protein 31 1.8
At5g20630 germin-like protein (GLP3b) 30 3.2
>At3g03720 putative cationic amino acid transporter
Length = 614
Score = 798 bits (2062), Expect = 0.0
Identities = 401/634 (63%), Positives = 485/634 (76%), Gaps = 39/634 (6%)
Query: 20 SLIRRKQVDSIHY---RGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPAL 76
SL+RRKQVDS+H G QLA+KLS VDLV IGVG TIGAGVYIL+GTVARE GPAL
Sbjct: 3 SLVRRKQVDSVHLIKNDGPHQLAKKLSAVDLVAIGVGTTIGAGVYILVGTVAREHTGPAL 62
Query: 77 VISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGAS 136
+S FIAG+AAALSA CYAELA RCPSAGSAYHY YIC+GEG+AWLVGW+L+L+YTIG S
Sbjct: 63 AVSFFIAGVAAALSACCYAELASRCPSAGSAYHYAYICLGEGIAWLVGWALVLDYTIGGS 122
Query: 137 AVARGITPNL--------------ALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
A+ARGITPNL A FFGG DNLP FLAR T+PG+GIVVDPCAA+LI++
Sbjct: 123 AIARGITPNLVFAFELYVFGFSQEASFFGGLDNLPVFLARQTIPGVGIVVDPCAALLIMI 182
Query: 183 ITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGM 242
+T+LLC GIKESSTVQ+IVT++NV ++FII+VGGYL K GWVGY+LPSGYFP+G+NG+
Sbjct: 183 VTILLCFGIKESSTVQAIVTSVNVCTLVFIIVVGGYLACKTGWVGYDLPSGYFPFGLNGI 242
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
AGSA+VFFSYIGFD+VTSTAEEVKNPQRDLP+GI AL ICC+LYML+S VIVGLVPYY
Sbjct: 243 LAGSAVVFFSYIGFDTVTSTAEEVKNPQRDLPLGIGIALLICCILYMLLSVVIVGLVPYY 302
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
LNPDTPISSAF GM+WA YI+TTGA+TAL +SLLGS+L QPR+FMAMARDGLLP FF
Sbjct: 303 SLNPDTPISSAFGDSGMQWAAYILTTGAITALCASLLGSLLAQPRIFMAMARDGLLPAFF 362
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
S+I RTQ+P+KSTI G+ AA LAFFMDV+QL+ MVSVGTL+AFT VAV VL++RYVPP
Sbjct: 363 SEISPRTQVPVKSTIAIGVLAAALAFFMDVAQLSEMVSVGTLMAFTAVAVCVLVLRYVPP 422
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKE 482
D +P+ +S T D +E R + L ++S + PL+ E
Sbjct: 423 DGVPLSSSSQTL---------SDTDESRAETENFLVDAIESS----------DSPLLGNE 463
Query: 483 VTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIV 542
+++ RRK+AAW+IAL+CIG+L ++ +ASAER P R T+ G VI L S+I
Sbjct: 464 TARDEKYFGKRRKIAAWSIALVCIGVLGLASAASAERLPSFPRFTICGVSAVILLGSLIT 523
Query: 543 LACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLI 602
L I +D+ RH FGH GGF CPFVP+LP CILINTYL+I++G TW+RV +WLLIG +I
Sbjct: 524 LGYIDEDEERHNFGHKGGFLCPFVPYLPVLCILINTYLIINIGAGTWIRVLIWLLIGSMI 583
Query: 603 YLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
Y+FYGR+HS L NA+YVP+ R +HLA
Sbjct: 584 YIFYGRSHSLLNNAVYVPTMTCT---RKTTDHLA 614
>At1g58032 unknown protein
Length = 635
Score = 797 bits (2058), Expect = 0.0
Identities = 402/643 (62%), Positives = 496/643 (76%), Gaps = 23/643 (3%)
Query: 5 VGNGEEGGGRFRGF-GSLIRRKQVDSIHYRGHP-QLARKLSVVDLVGIGVGATIGAGVYI 62
V +EGGG G+ SL+RRKQVDS + + H QLAR L+V LV IGVGATIGAGVYI
Sbjct: 5 VDTQKEGGGHSWGYVRSLVRRKQVDSANGQSHGHQLARALTVPHLVAIGVGATIGAGVYI 64
Query: 63 LIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWL 122
L+GTVARE +GP+L +S IAGIAA LSA CYAEL+ RCPSAGSAYHY+YIC+GEGVAW+
Sbjct: 65 LVGTVAREHSGPSLALSFLIAGIAAGLSAFCYAELSSRCPSAGSAYHYSYICVGEGVAWI 124
Query: 123 VGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVL 182
+GW+LILEYTIG SAVARGI+PNLAL FGG+D LP+ LARH +PGL IVVDPCAA+L+ +
Sbjct: 125 IGWALILEYTIGGSAVARGISPNLALIFGGEDGLPAILARHQIPGLDIVVDPCAAILVFV 184
Query: 183 ITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGM 242
+T LLC+GIKES+ Q IVT +NV V+LF+I+ G YLGFK GW GYELP+G+FP+GV+GM
Sbjct: 185 VTGLLCMGIKESTFAQGIVTAVNVCVLLFVIVAGSYLGFKTGWPGYELPTGFFPFGVDGM 244
Query: 243 FAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYY 302
FAGSA VFF++IGFDSV STAEEV+NPQRDLPIGI AL +CC LYM+VS VIVGL+PYY
Sbjct: 245 FAGSATVFFAFIGFDSVASTAEEVRNPQRDLPIGIGLALLLCCSLYMMVSIVIVGLIPYY 304
Query: 303 ELNPDTPISSAFSSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFF 362
++PDTPISSAF+S+ M+WAVY+IT GAV AL S+L+G++LPQPR+ MAMARDGLLP+ F
Sbjct: 305 AMDPDTPISSAFASHDMQWAVYLITLGAVMALCSALMGALLPQPRILMAMARDGLLPSIF 364
Query: 363 SDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPP 422
SDI++RTQ+P+K+T+ TGL AA LAFFMDVSQLAGMVSVGTLLAFT VA+SVLI+RYVPP
Sbjct: 365 SDINKRTQVPVKATVATGLCAATLAFFMDVSQLAGMVSVGTLLAFTMVAISVLILRYVPP 424
Query: 423 DEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASYSDNSH--LHDKSDVLLEHPLII 480
DE P+P+SL +D + G+ SD+SH L +D L++ PLI
Sbjct: 425 DEQPLPSSLQERIDSVSFICGETTSSGH-------VGTSDSSHQPLIVNNDALVDVPLI- 476
Query: 481 KEVTKEQH-------NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGV 533
K Q +E+TRR +A W+I C+G ++S +AS+ P ++R L G G
Sbjct: 477 ----KNQEALGCLVLSEETRRIVAGWSIMFTCVGAFLLSYAASSLSFPGLIRYPLCGVGG 532
Query: 534 VIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVS 593
+ L +I L+ I QDD RHTFGHSGG+ CPFVP LP CILIN YLL++LG TW RVS
Sbjct: 533 CLLLAGLIALSSIDQDDARHTFGHSGGYMCPFVPLLPIICILINMYLLVNLGSATWARVS 592
Query: 594 VWLLIGVLIYLFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
VWLLIGV++Y+FYGR +SSL NA+YV +A A+EI+R LA
Sbjct: 593 VWLLIGVIVYVFYGRKNSSLANAVYVTTAHAEEIYREHEGSLA 635
>At5g36940 cationic amino acid transporter -like protein
Length = 609
Score = 761 bits (1966), Expect = 0.0
Identities = 382/633 (60%), Positives = 475/633 (74%), Gaps = 45/633 (7%)
Query: 20 SLIRRKQVDSIH-----YRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGP 74
SL+RRKQ DS + + H QLA+ L+ L+ IGVG+TIGAGVYIL+GTVARE +GP
Sbjct: 6 SLVRRKQFDSSNGKAETHHHHQQLAKALTFPHLIAIGVGSTIGAGVYILVGTVAREHSGP 65
Query: 75 ALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIG 134
AL +S IAGI+AALSA CYAEL+ R PSAGSAYHY+YICIGEGVAWL+GW+LILEYTIG
Sbjct: 66 ALALSFLIAGISAALSAFCYAELSSRFPSAGSAYHYSYICIGEGVAWLIGWALILEYTIG 125
Query: 135 ASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKES 194
S VARGI+PNLA+ FGG+D LP+ LARH +PGL IVVDPCAAVL+ ++T L CLG+KES
Sbjct: 126 GSTVARGISPNLAMIFGGEDCLPTILARHQIPGLDIVVDPCAAVLVFIVTGLCCLGVKES 185
Query: 195 STVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYI 254
+ Q IVTT NV VM+F+I+ G YL FK GWVGYELP+GYFPYGV+GM GSA VFF+YI
Sbjct: 186 TFAQGIVTTANVFVMIFVIVAGSYLCFKTGWVGYELPTGYFPYGVDGMLTGSATVFFAYI 245
Query: 255 GFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAF 314
GFD+V S AEEVKNP+RDLP+GI +L +CC+LYM+VS VIVGLVPYY ++PDTPISSAF
Sbjct: 246 GFDTVASMAEEVKNPRRDLPLGIGISLLLCCLLYMMVSVVIVGLVPYYAMDPDTPISSAF 305
Query: 315 SSYGMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLK 374
SS+G++WA Y+I GAV AL S+L+GS+LPQPR+ MAMARDGLLP++FS +++RTQ+P+
Sbjct: 306 SSHGIQWAAYLINLGAVMALCSALMGSILPQPRILMAMARDGLLPSYFSYVNQRTQVPIN 365
Query: 375 STIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV-PPDEIPIPASLLT 433
TI TG+ AA+LAFFMDVSQLAGMVSVGTL+AFT VA+S+LI+RYV PPDE+P+P+SL
Sbjct: 366 GTITTGVCAAILAFFMDVSQLAGMVSVGTLVAFTMVAISLLIVRYVVPPDEVPLPSSL-- 423
Query: 434 SVDPLLRHSGDDIEEDRTVSPVDLASYSDNSHLHDKSDVLLEHPLIIKE----VTKEQH- 488
+NS H + + + PL+ K V KE
Sbjct: 424 ---------------------------QENSSSHVGTSIRSKQPLLGKVDDSVVDKENAP 456
Query: 489 -----NEKTRRKLAAWTIALLCIGILIVSGSASAERCPRILRVTLFGAGVVIFLCSIIVL 543
N+K RRK A W+I CIG ++S +AS+ P +LR +L G G + L +IVL
Sbjct: 457 GSWVLNKKNRRKFAGWSIMFTCIGNFLLSYAASSFLLPGLLRYSLCGVGGLFLLVGLIVL 516
Query: 544 ACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIY 603
CI QDD RH+FGHSGGF CPFVP LP CILIN YLL++LG TW+RVSVWL +GV++Y
Sbjct: 517 ICIDQDDARHSFGHSGGFICPFVPLLPIVCILINMYLLVNLGAATWVRVSVWLFLGVVVY 576
Query: 604 LFYGRTHSSLLNAIYVPSARADEIHRSQANHLA 636
+FYGR +SSL+NA+YV +A EI R+ + LA
Sbjct: 577 IFYGRRNSSLVNAVYVSTAHLQEIRRTSGHSLA 609
>At1g05940 unknown protein (At1g05940)
Length = 569
Score = 322 bits (825), Expect = 4e-88
Identities = 203/600 (33%), Positives = 315/600 (51%), Gaps = 91/600 (15%)
Query: 38 LARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAEL 97
L R+L + DL+ +GVGA+IGAGV+++ GTVAR+ AGP + IS +AG + L+ALCYAEL
Sbjct: 48 LVRRLGLFDLILLGVGASIGAGVFVVTGTVARD-AGPGVTISFLLAGASCVLNALCYAEL 106
Query: 98 ACRCPSA-GSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPN----LALFFGG 152
+ R P+ G AY Y+Y E A+LV L+L+Y IGA++++R + L LF
Sbjct: 107 SSRFPAVVGGAYMYSYSAFNEITAFLVFVQLMLDYHIGAASISRSLASYAVALLELFPAL 166
Query: 153 QDNLPSFLA--RHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVML 210
+ ++P ++ + L GL + ++ A +L+ L+TL+LC G++ESS V S++T V ++L
Sbjct: 167 KGSIPLWMGSGKELLGGL-LSLNILAPILLALLTLVLCQGVRESSAVNSVMTATKVVIVL 225
Query: 211 FIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQ 270
+I G + A W S + P G + G+ +VFFSY+GFD+V ++AEE KNPQ
Sbjct: 226 VVICAGAFEIDVANW------SPFAPNGFKAVLTGATVVFFSYVGFDAVANSAEESKNPQ 279
Query: 271 RDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGA 330
RDLPIGI +L +C LY+ V V+ G+VP+ L+ D P++ AFSS GM++ +I+ GA
Sbjct: 280 RDLPIGIMGSLLVCISLYIGVCLVLTGMVPFSLLSEDAPLAEAFSSKGMKFVSILISIGA 339
Query: 331 VTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFM 390
V L ++LL + Q R+++ + RDGLLP+ FS IH PL S I G+ A VLA
Sbjct: 340 VAGLTTTLLVGLYVQSRLYLGLGRDGLLPSIFSRIHPTLHTPLHSQIWCGIVAGVLAGIF 399
Query: 391 DVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDR 450
+V L+ ++SVGTL ++ VA V+ +R
Sbjct: 400 NVHSLSHILSVGTLTGYSVVAACVVALR-------------------------------- 427
Query: 451 TVSPVDLASYSDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILI 510
L+DK D E + R ++W ++C+ I+
Sbjct: 428 ---------------LNDKKD-----------------RESSNRWTSSWQEGVICLVIIA 455
Query: 511 VSGSASAERCPRILRVTLFGAGVVIFLCSI---IVLACIKQDDTRHTFGHSGGFACPFVP 567
SG + F A V+ L S+ +V + + + GF+CP VP
Sbjct: 456 CSGFGAGV-------FYRFSASVIFILLSVGVAVVASAVLHYRQAYALPLGSGFSCPGVP 508
Query: 568 FLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHS--SLLNAIYVPSARAD 625
+P+ CI N +L L + W+R V ++ +Y YG+ H+ S+L+ P +D
Sbjct: 509 IVPSVCIFFNIFLFAQLHYEAWIRFVVVSVLATAVYALYGQYHADPSMLDYQRAPETESD 568
>At3g10600 putative amino acid transporter
Length = 584
Score = 287 bits (734), Expect = 1e-77
Identities = 153/392 (39%), Positives = 244/392 (62%), Gaps = 13/392 (3%)
Query: 37 QLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAE 96
Q+ R L DL+G+G+G IGAGV++ G +R AGP++V+S IAG+ A LSA CY E
Sbjct: 55 QMRRTLRWYDLIGLGIGGMIGAGVFVTTGRASRLYAGPSIVVSYAIAGLCALLSAFCYTE 114
Query: 97 LACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNL 156
A P AG A+ Y I GE A++ G +LI++Y + +AV+RG T L FG +
Sbjct: 115 FAVHLPVAGGAFSYIRITFGEFPAFITGANLIMDYVLSNAAVSRGFTAYLGSAFGISTSE 174
Query: 157 PSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVG 216
F+ LP +DP A ++++ +T ++C +ESS V ++T ++++ ++F+I+
Sbjct: 175 WRFIV-SGLPNGFNEIDPIAVIVVLAVTFVICYSTRESSKVNMVLTALHIAFIVFVIV-- 231
Query: 217 GYLGFKAGWV-------GYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNP 269
+GF G V E PSG+FP+GV+G+F G+A+V+ SYIG+D+V++ AEEVK+P
Sbjct: 232 --MGFSKGDVKNLTRPDNPENPSGFFPFGVSGVFNGAAMVYLSYIGYDAVSTMAEEVKDP 289
Query: 270 QRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFS-SYGMEWAVYIITT 328
+D+P+GIS ++AI VLY L++ + L+PY ++ + P S+AFS S G EW ++
Sbjct: 290 VKDIPMGISGSVAIVIVLYCLMAISMSMLLPYDLIDAEAPYSAAFSKSEGWEWVTRVVGI 349
Query: 329 GAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAF 388
GA + +SL+ ++L Q R + R ++P +F+ +H +T P+ ++ G+F AVLA
Sbjct: 350 GASFGILTSLIVAMLGQARYMCVIGRSRVVPIWFAKVHPKTSTPVNASAFLGIFTAVLAL 409
Query: 389 FMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYV 420
F D++ L +VS+GTL F VA +V+ RYV
Sbjct: 410 FTDLNVLLNLVSIGTLFVFYMVANAVIFRRYV 441
Score = 35.4 bits (80), Expect = 0.098
Identities = 21/85 (24%), Positives = 39/85 (45%), Gaps = 6/85 (7%)
Query: 528 LFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVD 587
+ GA V + + + C+ F + P +P+ P I +N +LL L
Sbjct: 481 ILGASTVTAIAIVQIFHCVVPQARIPEF-----WGVPLMPWTPCVSIFLNIFLLGSLDAP 535
Query: 588 TWLRVSVWLLIGVLIYLFYGRTHSS 612
+++R + + VL+Y+FY H+S
Sbjct: 536 SYIRFGFFSGLVVLVYVFYS-VHAS 559
>At5g04770 amino acid transporter-like protein
Length = 583
Score = 285 bits (730), Expect = 4e-77
Identities = 148/389 (38%), Positives = 238/389 (61%), Gaps = 5/389 (1%)
Query: 37 QLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAE 96
Q+ R L DL+G+G+G +GAGV++ G +R AGP++V+S IAG+ A LSA CY E
Sbjct: 56 QMRRTLRWYDLIGLGIGGMVGAGVFVTTGRASRLDAGPSIVVSYAIAGLCALLSAFCYTE 115
Query: 97 LACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNL 156
A P AG A+ Y I GE A+ G +L+++Y + +AV+R T L FG +
Sbjct: 116 FAVHLPVAGGAFSYIRITFGEFPAFFTGANLVMDYVMSNAAVSRSFTAYLGTAFGISTSK 175
Query: 157 PSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVG 216
F+ G +DP A +++++IT+++C +ESS V I+T +++ + F+I++G
Sbjct: 176 WRFVVSGLPKGFN-EIDPVAVLVVLVITVIICCSTRESSKVNMIMTAFHIAFIFFVIVMG 234
Query: 217 GYLGFK---AGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDL 273
G + E PSG+FP+G G+F G+A+V+ SYIG+D+V++ AEEV+NP +D+
Sbjct: 235 FIKGDSKNLSSPANPEHPSGFFPFGAAGVFNGAAMVYLSYIGYDAVSTMAEEVENPVKDI 294
Query: 274 PIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAF-SSYGMEWAVYIITTGAVT 332
P+G+S ++AI VLY L++ + L+PY ++P+ P S+AF S G EW ++ GA
Sbjct: 295 PVGVSGSVAIVTVLYCLMAVSMSMLLPYDLIDPEAPFSAAFRGSNGWEWVTKVVGIGASF 354
Query: 333 ALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDV 392
+ +SLL ++L Q R + R ++P +F+ IH +T P+ ++ G+F A LA F D+
Sbjct: 355 GILTSLLVAMLGQARYMCVIGRSRVVPFWFAKIHPKTSTPVNASTFLGIFTAALALFTDL 414
Query: 393 SQLAGMVSVGTLLAFTTVAVSVLIIRYVP 421
+ L +VS+GTL F VA +++ RYVP
Sbjct: 415 NVLLNLVSIGTLFVFYMVANALIFRRYVP 443
Score = 42.0 bits (97), Expect = 0.001
Identities = 23/85 (27%), Positives = 42/85 (49%), Gaps = 6/85 (7%)
Query: 528 LFGAGVVIFLCSIIVLACIKQDDTRHTFGHSGGFACPFVPFLPAACILINTYLLIDLGVD 587
+ GA V+ + ++ C+ + + PF+P+ P I +N +LL L
Sbjct: 482 MLGASAVVAIAIVLSFQCVVPQARKPEL-----WGVPFMPWTPCVSIFLNIFLLGSLDAP 536
Query: 588 TWLRVSVWLLIGVLIYLFYGRTHSS 612
+++R + + VL+YLFYG H+S
Sbjct: 537 SYVRFGFFSGLIVLVYLFYG-VHAS 560
>At4g21120 amino acid transport protein AAT1
Length = 594
Score = 257 bits (656), Expect = 2e-68
Identities = 140/394 (35%), Positives = 232/394 (58%), Gaps = 7/394 (1%)
Query: 26 QVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGI 85
+++ + R ++ + L+ DL+ G+GA IG+G+++L G AR +GPA+V+S ++G+
Sbjct: 58 EINEMKARSGHEMKKTLTWWDLMWFGIGAVIGSGIFVLTGLEARNHSGPAVVLSYVVSGV 117
Query: 86 AAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPN 145
+A LS CY E A P AG ++ Y + +G+ +A++ ++ILEY +G +AVAR T
Sbjct: 118 SAMLSVFCYTEFAVEIPVAGGSFAYLRVELGDFMAFIAAGNIILEYVVGGAAVARSWTSY 177
Query: 146 LALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTIN 205
A + + H L +DP A + +I +L +G K SS I + I+
Sbjct: 178 FATLLNHKPEDFRIIV-HKLGEDYSHLDPIAVGVCAIICVLAVVGTKGSSRFNYIASIIH 236
Query: 206 VSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEE 265
+ V+LF+II G KA Y S + PYGV G+F +A++FF+YIGFD+V++ AEE
Sbjct: 237 MVVILFVIIAGFT---KADVKNY---SDFTPYGVRGVFKSAAVLFFAYIGFDAVSTMAEE 290
Query: 266 VKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYI 325
KNP RD+PIG+ ++ + V Y L++ + + PY +++PD P S AFS+ G +WA YI
Sbjct: 291 TKNPGRDIPIGLVGSMVVTTVCYCLMAVTLCLMQPYQQIDPDAPFSVAFSAVGWDWAKYI 350
Query: 326 ITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAV 385
+ GA+ + + LL + Q R +AR ++P + + ++ +T P+ +T+V A+
Sbjct: 351 VAFGALKGMTTVLLVGAIGQARYMTHIARAHMMPPWLAQVNAKTGTPINATVVMLAATAL 410
Query: 386 LAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRY 419
+AFF + LA ++SV TL F VAV++L+ RY
Sbjct: 411 IAFFTKLKILADLLSVSTLFIFMFVAVALLVRRY 444
Score = 38.9 bits (89), Expect = 0.009
Identities = 16/44 (36%), Positives = 29/44 (65%)
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYG 607
P VP+LP+A I IN +LL + +++R ++W I ++ Y+ +G
Sbjct: 516 PLVPWLPSASIAINIFLLGSIDTKSFVRFAIWTGILLIYYVLFG 559
>At2g34960 amino acid transporter like protein
Length = 569
Score = 240 bits (613), Expect = 2e-63
Identities = 129/392 (32%), Positives = 224/392 (56%), Gaps = 12/392 (3%)
Query: 37 QLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAE 96
++ R L+ DLV G G+ IGAG+++L G A EQAGPA+V+S ++G++A LS CY E
Sbjct: 63 EMKRCLTWWDLVWFGFGSVIGAGIFVLTGQEAHEQAGPAIVLSYVVSGLSAMLSVFCYTE 122
Query: 97 LACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNL 156
A P AG ++ Y I +G+ A++ +++LE +G +AVAR T A N
Sbjct: 123 FAVEIPVAGGSFAYLRIELGDFAAFITAGNILLESIVGTAAVARAWTSYFATLLNRSPN- 181
Query: 157 PSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVG 216
+ + L ++DP A V+I + + +++S + I + IN V+ F+II
Sbjct: 182 -ALRIKTDLSSGFNLLDPIAVVVIAASATIASISTRKTSLLNWIASAINTLVIFFVII-- 238
Query: 217 GYLGFKAGWVGYELPS--GYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLP 274
AG++ + + + P+G G+F +A+V+F+Y GFDS+ + AEE KNP RD+P
Sbjct: 239 ------AGFIHADTSNLTPFLPFGPEGVFRAAAVVYFAYGGFDSIATMAEETKNPSRDIP 292
Query: 275 IGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYIITTGAVTAL 334
IG+ +++I V+Y L++ + + Y +++P+ S AF S GM+W Y++ GA+ +
Sbjct: 293 IGLLGSMSIITVIYCLMALSLSMMQKYTDIDPNAAYSVAFQSVGMKWGKYLVALGALKGM 352
Query: 335 FSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLAFFMDVSQ 394
+ LL L Q R +AR ++P F+ +H +T P+ + ++ + +A++AFF +
Sbjct: 353 TTVLLVGALGQARYVTHIARTHMIPPIFALVHPKTGTPINANLLVAIPSALIAFFSGLDV 412
Query: 395 LAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIP 426
LA ++S+ TL FT + +++L+ RY + P
Sbjct: 413 LASLLSISTLFIFTMMPIALLVRRYYVRQDTP 444
Score = 32.7 bits (73), Expect = 0.64
Identities = 26/108 (24%), Positives = 47/108 (43%), Gaps = 9/108 (8%)
Query: 503 LLCIGILIVSGSASAERCPRILRVTLFGAGVVI---FLCSIIVLACIKQDDTRHTFGHSG 559
+ C+ ++VS ++ + + G V + FL ++ ++ + Q T +G
Sbjct: 452 ITCLLFVVVSSMGTSAYWGMQRKGSWIGYTVTVPFWFLGTLGIVFFVPQQRTPKVWG--- 508
Query: 560 GFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYG 607
P VP+LP I N +L+ LG ++R V L +L Y G
Sbjct: 509 ---VPLVPWLPCLSIATNIFLMGSLGAMAFVRFGVCTLAMLLYYFLLG 553
>At1g17120 putative amino acid transporter
Length = 590
Score = 228 bits (581), Expect = 8e-60
Identities = 122/401 (30%), Positives = 225/401 (55%), Gaps = 7/401 (1%)
Query: 26 QVDSIHYRGHPQLARKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAGI 85
++D+ + R L+ DL+ + G+ +G+GV+++ G AR AGPA+V+S I+G+
Sbjct: 69 ELDAARRESENPMRRCLTWWDLLWLSFGSVVGSGVFVITGQEARVGAGPAVVLSYAISGV 128
Query: 86 AAALSALCYAELACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPN 145
+A LS LCYAE P AG ++ Y + +G+ +A++ +++LE +GA+ + R +
Sbjct: 129 SALLSVLCYAEFGVEIPVAGGSFSYLRVELGDFIAFIAAGNILLEAMVGAAGLGRSWSSY 188
Query: 146 LALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTIN 205
LA + G ++ DP A ++++ + G K +S + I + +
Sbjct: 189 LASLVKNDSDYFRIKVDSFAKGFDLL-DPVAVAVLLVANGIAMTGTKRTSWLNLITSMVT 247
Query: 206 VSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEE 265
V +++FI++VG F +P +FPYG G+ +A+V++SY GFD V + AEE
Sbjct: 248 VCIIVFIVVVG----FTHSKTSNLVP--FFPYGAKGVVQSAAVVYWSYTGFDMVANMAEE 301
Query: 266 VKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTPISSAFSSYGMEWAVYI 325
+ P RD+PIG+ ++++ V+Y L++ + +V Y E++ + S AF+ GM+WA Y+
Sbjct: 302 TEKPSRDIPIGLVGSMSMITVVYCLMALALTMMVKYTEIDANAAYSVAFAQIGMKWAKYL 361
Query: 326 ITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAV 385
+ A+ + +SLL L Q R +AR ++P +F+ +H +T P+ +T++ + +++
Sbjct: 362 VGICALKGMTTSLLVGSLGQARYTTQIARSHMIPPWFALVHPKTGTPIYATLLVTILSSI 421
Query: 386 LAFFMDVSQLAGMVSVGTLLAFTTVAVSVLIIRYVPPDEIP 426
++FF + L+ + S TL F VAV++L+ RY D P
Sbjct: 422 ISFFTSLEVLSSVFSFATLFIFMLVAVALLVRRYYVKDVTP 462
Score = 36.6 bits (83), Expect = 0.044
Identities = 17/44 (38%), Positives = 27/44 (60%)
Query: 564 PFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYG 607
P VP+LP+ I +N +L+ LG +LR + ++ +L YLF G
Sbjct: 526 PLVPWLPSFSIAMNLFLIGSLGYVAFLRFIICTMVMLLYYLFVG 569
>At2g38990 similar to MURA transposase of maize Mutator transposon
Length = 142
Score = 95.9 bits (237), Expect = 6e-20
Identities = 46/82 (56%), Positives = 56/82 (68%), Gaps = 3/82 (3%)
Query: 555 FGHSGGFACPFVPFLPAACILINTYLLIDLGVDTWLRVSVWLLIGVLIYLFYGRTHSSLL 614
FGH GGF C FVP+LP CILINTYL+I++G TW+RV V LLIG +I LF GR+H L
Sbjct: 64 FGHKGGFLCSFVPYLPVLCILINTYLIINIGAGTWIRVLVSLLIGSMICLFCGRSHCLLN 123
Query: 615 NAIYVPSARADEIHRSQANHLA 636
N +YVP+ R +HLA
Sbjct: 124 NVVYVPTTTCT---RKTTDHLA 142
>At2g01170 putative amino acid or GABA permease
Length = 516
Score = 52.8 bits (125), Expect = 6e-07
Identities = 88/420 (20%), Positives = 168/420 (39%), Gaps = 57/420 (13%)
Query: 37 QLARKLSVVDLVGIGVGA-TIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYA 95
+L R LSV I ++ G+ T R LV F+AG L A
Sbjct: 28 ELKRDLSVFSNFAISFSIISVLTGITTTYNTGLRFGGTVTLVYGWFLAGSFTMCVGLSMA 87
Query: 96 ELACRCPSAGSAYHYTYICIGEG----VAWLVGW-SLILEYTIGAS---AVARGITPNLA 147
E+ P++G Y+++ + G +W+ GW +++ ++ + AS ++A+ I +
Sbjct: 88 EICSSYPTSGGLYYWSAMLAGPRWAPLASWMTGWFNIVGQWAVTASVDFSLAQLIQVIVL 147
Query: 148 LFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINVS 207
L GG++ G VV ++ + LL L I S + + N+
Sbjct: 148 LSTGGRNG-------GGYKGSDFVVIGIHGGILFIHALLNSLPISVLSFIGQLAALWNLL 200
Query: 208 VMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYI------------- 254
+L ++I+ V E + F + G I ++YI
Sbjct: 201 GVLVLMIL-------IPLVSTERATTKFVFTNFNTDNGLGITSYAYIFVLGLLMSQYTIT 253
Query: 255 GFDSVTSTAEEVKNPQRDLPIGISTALAICCVL---YMLVSAVIVGLVPYYELNPDTPIS 311
G+D+ EE + ++ P GI +A+ I + Y+L + V +P L +T S
Sbjct: 254 GYDASAHMTEETVDADKNGPRGIISAIGISILFGWGYILGISYAVTDIP--SLLSETNNS 311
Query: 312 SAFSSYGMEWAVY----------IITTGAV-TALFSSLLGSVLPQPRVFMAMARDGLLP- 359
++ + + + I+ G V A+F + SV R+ A +RDG +P
Sbjct: 312 GGYAIAEIFYLAFKNRFGSGTGGIVCLGVVAVAVFFCGMSSVTSNSRMAYAFSRDGAMPM 371
Query: 360 -TFFSDIHRRTQIPLKSTIVTGL--FAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLI 416
+ ++ R ++P+ + ++ L F L + MVS+ T+ + A+ +++
Sbjct: 372 SPLWHKVNSR-EVPINAVWLSALISFCMALTSLGSIVAFQAMVSIATIGLYIAYAIPIIL 430
>At1g31830 unknown protein
Length = 495
Score = 47.0 bits (110), Expect = 3e-05
Identities = 80/383 (20%), Positives = 154/383 (39%), Gaps = 48/383 (12%)
Query: 40 RKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAG--IAAALSALCYAEL 97
RK+S++ LV + + + G + + +V AGP L + F+ I + AL AE+
Sbjct: 47 RKVSMLPLVFL-IFYEVSGGPFGVEDSV--NAAGPLLALLGFVIFPFIWSIPEALITAEM 103
Query: 98 ACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNLP 157
P G + +G + GW L + + + P L L +
Sbjct: 104 GTMYPENGGYVVWVSSALGPFWGFQQGWMKWL-----SGVIDNALYPVLFLDY------- 151
Query: 158 SFLARHTLPGLGIVVDPCAAVLI--VLITLLLCLGIKESSTVQSIVTTINVSVMLFIIIV 215
+ +P LG + A++L+ +L+T L G+ +IV + V + +F I+
Sbjct: 152 ---LKSGVPALGSGLPRVASILVLTILLTYLNYRGL-------TIVGWVAVLMGVFSILP 201
Query: 216 GGYLGF-------KAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKN 268
+G + W+ +L + + +N +F ++ +DS+++ A EV+N
Sbjct: 202 FAVMGLISIPQLEPSRWLVMDLGNVNWNLYLNTLF-------WNLNYWDSISTLAGEVEN 254
Query: 269 PQRDLPIGISTALAICCVLYMLVSAVIVGLVPY-YELNPDTPISSAFSSYGMEWAVYIIT 327
P LP + + + Y+ +G +P E D S + G W + +
Sbjct: 255 PNHTLPKALFYGVILVACSYIFPLLAGIGAIPLEREKWTDGYFSDVAKALGGAWLRWWVQ 314
Query: 328 TGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRRTQIPLKSTIVTGLFAAVLA 387
A T+ + + + MA G+LP FF+ R PL + + +L+
Sbjct: 315 AAAATSNMGMFIAEMSSDSFQLLGMAERGMLPEFFAK-RSRYGTPLLGILFSASGVVLLS 373
Query: 388 F--FMDVSQLAGMV-SVGTLLAF 407
+ F ++ ++ VG +L F
Sbjct: 374 WLSFQEIVAAENLLYCVGMILEF 396
>At3g13620 unknown protein
Length = 478
Score = 46.2 bits (108), Expect = 6e-05
Identities = 42/168 (25%), Positives = 74/168 (44%), Gaps = 10/168 (5%)
Query: 249 VFFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDT 308
+F++ +D+V++ A EV PQ+ P+ + A+ CV Y++ + G V + +
Sbjct: 216 LFWNLNFWDNVSTLAGEVDEPQKTFPLALLIAVIFTCVAYLIPLFAVTGAVSVDQSRWEN 275
Query: 309 PI-SSAFSSYGMEWAVYIITTGAVTA---LFSSLLGSVLPQPRVFMAMARDGLLPTFFSD 364
+ A +W I GAV + LF + L S Q MA G LP FF
Sbjct: 276 GFHAEAAEMIAGKWLKIWIEIGAVLSSIGLFEAQLSSSAYQ---LEGMAELGFLPKFFGV 332
Query: 365 IHRRTQIPLKSTIVTGLFAAVLAF--FMDVSQLAG-MVSVGTLLAFTT 409
+ P +++ L + L++ F D+ A + ++G L F +
Sbjct: 333 RSKWFNTPWVGILISALMSLGLSYMNFTDIISSANFLYTLGMFLEFAS 380
>At1g30450 cation-chloride co-transporter, putative
Length = 1321
Score = 44.7 bits (104), Expect = 2e-04
Identities = 85/409 (20%), Positives = 147/409 (35%), Gaps = 87/409 (21%)
Query: 42 LSVVDLVGIGV-GATIGAGVYILIGTVAREQAGPALVISLFIAG-IAAALSALCYAELAC 99
L+ + L I GA G G Y LIG + G ++ + F+ +A AL L E
Sbjct: 180 LTTISLSAIATNGAMKGGGPYYLIGRALGPEVGISIGLCFFLGNAVAGALYVLGAVETFL 239
Query: 100 RCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGI-TPNLALFFGGQDNLPS 158
+ A + T + +AV+ I +PN
Sbjct: 240 KAFPAAGIFRETIT------------------KVNGTAVSESIQSPN------------- 268
Query: 159 FLARHTLPGLGIVVDPCAAVLIVLITLLLCL----GIKESSTVQSIVTTINVSVMLFIII 214
H L GIVV T+LLC G+K + V + V + +F I
Sbjct: 269 ---SHDLQVYGIVV-----------TILLCFIVFGGVKMINRVAPAFL-VPVLLSIFCIF 313
Query: 215 VGGYLG----------------FKAGW---------VGYELPSGYFPYGVNGMFAGSAIV 249
+G +L FK W G P+G + N + +
Sbjct: 314 IGIFLAKTDDPDNGITGLRLKSFKDNWGSAYQMTNDAGIPDPTGGTYWSFNELVG---LF 370
Query: 250 FFSYIGFDSVTSTAEEVKNPQRDLPIGISTALAICCVLYMLVSAVIVGLVPYYELNPDTP 309
F + G + ++ + +K+ Q+ +P+G A LY L+S + G V D
Sbjct: 371 FPAVTGIMAGSNRSASLKDTQKSIPVGTLAATLTTTSLY-LISVLFFGAVA----TRDKL 425
Query: 310 ISSAFSSYGMEWAV-YIITTGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHRR 368
++ + + W I+ G + + + L S+ PR+ A+A D +LP
Sbjct: 426 LTDRLLTATIAWPFPAIVHVGIILSTLGAALQSLTGAPRLLAAIANDDILPILNYFKVAD 485
Query: 369 TQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTLLAFTTVAVSVLII 417
T P +T+ T ++ + V++ LL ++ V +S ++
Sbjct: 486 TSEPHIATLFTAFICIGCVVIGNLDLITPTVTMFYLLCYSGVNLSCFLL 534
>At5g05630 unknown protein
Length = 490
Score = 41.6 bits (96), Expect = 0.001
Identities = 72/328 (21%), Positives = 130/328 (38%), Gaps = 26/328 (7%)
Query: 40 RKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAG--IAAALSALCYAEL 97
+K++V+ LV + + + G + + +V + AGP L I FI I + AL AE+
Sbjct: 51 KKITVLPLVFL-IFYEVSGGPFGIEDSV--KAAGPLLAIVGFIVFPFIWSIPEALITAEM 107
Query: 98 ACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNLP 157
P G + + +G + GW L + + + P L L +
Sbjct: 108 GTMFPENGGYVVWVTLAMGPYWGFQQGWVKWL-----SGVIDNALYPILFLDY------- 155
Query: 158 SFLARHTLPGLGIVVDPCAAVLIVLITLLLCLGIKESSTVQSIVTTINV-SVMLFIIIVG 216
+ +P LG + AA+L++ + L L + S V + V S++ F+++
Sbjct: 156 ---LKSGIPILGSGIPRVAAILVLTVALTY-LNYRGLSIVGVAAVLLGVFSILPFVVM-- 209
Query: 217 GYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKNPQRDLPIG 276
++ L GVN + + F++ +DSV++ EV+NP + LP
Sbjct: 210 SFMSIPKLKPSRWLVVSKKMKGVNWSLYLNTL-FWNLNYWDSVSTLTGEVENPSKTLPRA 268
Query: 277 ISTALAICCVLYMLVSAVIVGLVPY-YELNPDTPISSAFSSYGMEWAVYIITTGAVTALF 335
+ AL + Y+ G + +L D + G W + I A T+
Sbjct: 269 LFYALLLVVFSYIFPVLTGTGAIALDQKLWTDGYFADIGKVIGGVWLGWWIQAAAATSNM 328
Query: 336 SSLLGSVLPQPRVFMAMARDGLLPTFFS 363
L + + MA G+LP F+
Sbjct: 329 GMFLAEMSSDSFQLLGMAERGMLPEVFA 356
>At1g31815 hypothetical protein
Length = 482
Score = 37.7 bits (86), Expect = 0.020
Identities = 72/340 (21%), Positives = 128/340 (37%), Gaps = 44/340 (12%)
Query: 40 RKLSVVDLVGIGVGATIGAGVYILIGTVAREQAGPALVISLFIAG--IAAALSALCYAEL 97
+K+S++ LV + + + G + G+V AGP L + F+ I AL AE+
Sbjct: 34 QKVSMLPLVFL-IFYEVSGGPFGAEGSV--NAAGPLLALLGFVIFPFIWCIPEALITAEM 90
Query: 98 ACRCPSAGSAYHYTYICIGEGVAWLVGWSLILEYTIGASAVARGITPNLALFFGGQDNLP 157
+ P G + +G + VGW L + + P L L +
Sbjct: 91 STMFPINGGFVVWVSSALGTFWGFQVGWMKWL-----CGVIDNALYPVLFLDY------- 138
Query: 158 SFLARHTLPGLGIVVDPCAAVLIV--LITLLLCLGIKE---SSTVQSIVTTINVSVMLFI 212
+ +P L + A++LI+ L+T L G+ ++ + + + +VM +
Sbjct: 139 ---LKSAVPALATGLPRVASILILTLLLTYLNYRGLTIVGWTAVFMGVFSMLPFAVMSLV 195
Query: 213 IIV----GGYLGFKAGWVGYELPSGYFPYGVNGMFAGSAIVFFSYIGFDSVTSTAEEVKN 268
I +L G V + L + +N +DSV++ A EV N
Sbjct: 196 SIPQLEPSRWLVMDLGNVNWNLYLNTLLWNLNY--------------WDSVSTLAGEVAN 241
Query: 269 PQRDLPIGISTALAICCVLYMLVSAVIVGLVPY-YELNPDTPISSAFSSYGMEWAVYIIT 327
P++ LP + + + L G +P EL D ++ + G W +
Sbjct: 242 PKKTLPKALCYGVIFVALSNFLPLLSGTGAIPLDRELWTDGYLAEVAKAIGGGWLQLWVQ 301
Query: 328 TGAVTALFSSLLGSVLPQPRVFMAMARDGLLPTFFSDIHR 367
A T+ L + + MA G+LP F+ R
Sbjct: 302 AAAATSNMGMFLAEMSSDSFQLLGMAELGILPEIFAQRSR 341
>At2g39130 unknown protein
Length = 550
Score = 35.8 bits (81), Expect = 0.075
Identities = 84/363 (23%), Positives = 147/363 (40%), Gaps = 69/363 (19%)
Query: 53 GATIGAGVYILIGTVAREQAGPALVISLFIAGIAAALSALCYAELACRCPSAGSAYHYTY 112
G + GV IL A ++ G ++ LF+ G+ + + + L C + S TY
Sbjct: 169 GLNVLCGVGILSTPYAAKEGGWLGLMILFVYGLLSFYTGI----LLRYCLDSESDLE-TY 223
Query: 113 ICIGEG------------VAWLVGWSLILEYTIGASAVARGITPNLALFFGG--QDNLPS 158
IG+ V +L ++ +EY I S + PN AL GG D
Sbjct: 224 PDIGQAAFGTTGRIFVSIVLYLELYACCVEYIILESDNLSSLYPNAALSIGGFQLDARHL 283
Query: 159 FLARHTLP--------GLGIVVDPCAAVLIVLITLLLCL---GIKESSTVQSIVTTINVS 207
F TL L ++ A +I + ++LCL G+ + + S TT+N+S
Sbjct: 284 FALLTTLAVLPTVWLRDLSVLSYISAGGVIASVLVVLCLFWIGLVDEVGIHSKGTTLNLS 343
Query: 208 VM-----LFIIIVGGYLGFKAGWVGYELPSGY-----FPYGV-NGMFAGSAIVFFSYIGF 256
+ L+ G+ F + PS Y +G+ M+AG A++ ++ G
Sbjct: 344 TLPVAIGLYGYCYSGHAVFPNIYTSMAKPSQYPAVLLTCFGICTLMYAGVAVMGYTMFG- 402
Query: 257 DSVTSTAEEVKNPQRDLPIGISTALAICCVLY-------MLVSAVIVGLVPYYELNPDTP 309
++ ++ N +DL I+T +A+ + + +S V + L EL P
Sbjct: 403 --ESTQSQFTLNLPQDL---IATKIAVWTTVVNPFTKYALTISPVAMSL---EELIPSRH 454
Query: 310 ISSAFSSYGMEWAVYIITT---------GAVTALFSSLLG---SVLPQPRVFMAMARDGL 357
I S + + G+ + T G V +L SLL +++ P F+++ R +
Sbjct: 455 IRSHWYAIGIRTLLVFSTLLVGLAIPFFGLVMSLIGSLLTMLVTLILPPACFLSIVRRKV 514
Query: 358 LPT 360
PT
Sbjct: 515 TPT 517
>At1g69850 nitrate transporter (NTL1)
Length = 585
Score = 32.3 bits (72), Expect = 0.83
Identities = 38/159 (23%), Positives = 67/159 (41%), Gaps = 18/159 (11%)
Query: 401 VGTLLAFTTVAVSVLIIRYVPPDEIPIPASLLTSVDPLLRHSGDDIEEDRTVSPVDLASY 460
V T+ F ++ + + R+ ++IP + L T + LL S + + V S
Sbjct: 218 VSTIAIFVSILIFLSGSRFYR-NKIPCGSPLTTILKVLLAASVKCCSSGSSSNAVASMSV 276
Query: 461 SDNSHLHDKSDVLLEHPLIIKEVTKEQHNEKTRRKLAAWTIALLCIGILIVSGSASAERC 520
S ++H K KEV + EK R++ A A L + +++G+A +
Sbjct: 277 SPSNHCVSKGK---------KEVESQGELEKPRQEEALPPRAQLTNSLKVLNGAADEKPV 327
Query: 521 PRILRVTLFGAGVV--------IFLCSIIVLACIKQDDT 551
R+L T+ V IF C+I++ C+ Q T
Sbjct: 328 HRLLECTVQQVEDVKIVLKMLPIFACTIMLNCCLAQLST 366
>At4g23640 potassium transport like protein
Length = 775
Score = 31.2 bits (69), Expect = 1.8
Identities = 71/316 (22%), Positives = 118/316 (36%), Gaps = 49/316 (15%)
Query: 122 LVGWSLILEYTIGASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIVVDPCAAVLIV 181
LVG S+++ TIG +TP +++ ++ +A+ +L +V+ CA
Sbjct: 148 LVGTSMVI--TIGV------LTPAISV----SSSIDGLVAKTSLKHSTVVMIACA----- 190
Query: 182 LITLLLCLGIKESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFPYGVNG 241
L+ L L + ++ V + I + +L I G Y Y+ S Y+ Y
Sbjct: 191 LLVGLFVLQHRGTNKVAFLFAPIMILWLLIIATAGVYNIVTWNPSVYKALSPYYIY---- 246
Query: 242 MFAGSAIVFFSYIGFDSVTS--------TAEEVKNPQRDLPIGISTALAICCVLYMLVSA 293
VFF G D S T E + S A CCV+Y +
Sbjct: 247 -------VFFRDTGIDGWLSLGGILLCITGTEAIFAELGQFTATSIRFAFCCVVYPCLVL 299
Query: 294 VIVGLVPYYELNPDTPISSAFSSY--GMEWAVYIITTGAVTALFSSLLGSVLPQPRVFMA 351
+G + N SS +SS W V ++ A +++ + + A
Sbjct: 300 QYMGQAAFLSKNFSALPSSFYSSIPDPFFWPVLMMAMLAAMVASQAVIFATFSIVKQCYA 359
Query: 352 MARDGLLPTFFSDIHR------RTQIPLKSTIVTGLFAAVLAFFMDVSQLAGMVSVGTL- 404
+ G P +H+ + IP + +V L AV F D +A + +
Sbjct: 360 L---GCFPR-VKIVHKPRWVLGQIYIPEINWVVMILTLAVTICFRDTRHIAFAFGLACMT 415
Query: 405 LAFTTVAVSVLIIRYV 420
LAF T + LII +V
Sbjct: 416 LAFVTTWLMPLIINFV 431
>At5g20630 germin-like protein (GLP3b)
Length = 211
Score = 30.4 bits (67), Expect = 3.2
Identities = 27/105 (25%), Positives = 46/105 (43%), Gaps = 13/105 (12%)
Query: 135 ASAVARGITPNLALFFGGQDNLPSFLARHTLPGLGIV---VDPCAAVLIVLITLLLCLGI 191
++ + +TP A + G + L LAR L G G++ P A+ ++V+I +C G
Sbjct: 64 SNIIKAAVTPAFAPAYAGINGLGVSLARLDLAGGGVIPLHTHPGASEVLVVIQGTICAGF 123
Query: 192 KESSTVQSIVTTINVSVMLFIIIVGGYLGFKAGWVGYELPSGYFP 236
S+ V L + G + F G + ++L SG P
Sbjct: 124 ISSAN----------KVYLKTLNRGDSMVFPQGLLHFQLNSGKGP 158
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,922,559
Number of Sequences: 26719
Number of extensions: 602266
Number of successful extensions: 2005
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 1946
Number of HSP's gapped (non-prelim): 41
length of query: 636
length of database: 11,318,596
effective HSP length: 105
effective length of query: 531
effective length of database: 8,513,101
effective search space: 4520456631
effective search space used: 4520456631
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148965.10


