

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.6 - phase: 0
(156 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
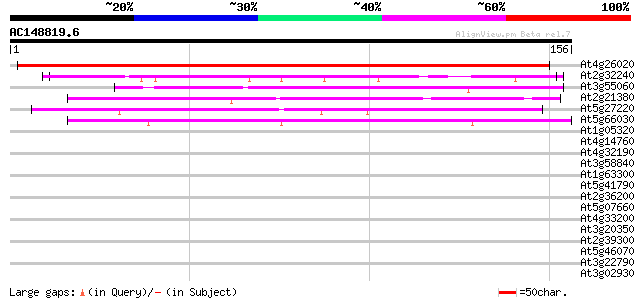
Score E
Sequences producing significant alignments: (bits) Value
At4g26020 hypothetical protein 155 1e-38
At2g32240 putative myosin heavy chain 45 2e-05
At3g55060 centromere protein - like 43 8e-05
At2g21380 putative kinesin heavy chain 42 1e-04
At5g27220 putative protein 42 2e-04
At5g66030 Golgi-localized protein GRIP 40 7e-04
At1g05320 unknown protein 39 0.001
At4g14760 centromere protein homolog 39 0.001
At4g32190 unknown protein 38 0.002
At3g58840 unknown protein 38 0.003
At1g63300 hypothetical protein 38 0.003
At5g41790 myosin heavy chain-like protein 37 0.003
At2g36200 putative kinesin-related cytokinesis protein 37 0.003
At5g07660 SMC-like protein 37 0.004
At4g33200 myosin - like protein 37 0.004
At3g20350 unknown protein 37 0.004
At2g39300 hypothetical protein 36 0.007
At5g46070 putative protein 36 0.010
At3g22790 unknown protein 36 0.010
At3g02930 unknown protein 36 0.010
>At4g26020 hypothetical protein
Length = 273
Score = 155 bits (391), Expect = 1e-38
Identities = 75/148 (50%), Positives = 111/148 (74%)
Query: 3 FSEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILA 62
F++Q HR AL+ L +++ K+ LE +++ L EKA+ + I +L D+ A+K+H+Q ++
Sbjct: 124 FADQEHRNALESLRQKHVTKVEELEYKIRSLLVEKATNDMVIDRLRQDLTANKSHIQAMS 183
Query: 63 NRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEM 122
+LD+V EVE KY EI+DLKDCL+ EQ EKND++ K+Q L+KELL+ + + ++Q++
Sbjct: 184 KKLDRVVTEVECKYELEIQDLKDCLLMEQAEKNDISNKLQSLQKELLISRTSIAEKQRDT 243
Query: 123 TSNWQVETLKQKIMKLRKENEVLKRKFS 150
TSN QVETLKQK+MKLRKENE+LKRK S
Sbjct: 244 TSNRQVETLKQKLMKLRKENEILKRKLS 271
>At2g32240 putative myosin heavy chain
Length = 1333
Score = 44.7 bits (104), Expect = 2e-05
Identities = 46/175 (26%), Positives = 84/175 (47%), Gaps = 41/175 (23%)
Query: 10 RALKLLEEEYNEKIARLEAQVKESLHEKAS----YEATISQLHGDIAAHKNHMQILANRL 65
++ K +E EK+A L+ ++KE L+EK S EA + G++AA + + + +RL
Sbjct: 243 KSTKESAKEMEEKMASLQQEIKE-LNEKMSENEKVEAALKSSAGELAAVQEELALSKSRL 301
Query: 66 --------------DQVHFEVESKYSSEIRDLKDC------------LMAEQEEKNDLNR 99
D++ E+E K +SE R ++ L A+ E+ +N
Sbjct: 302 LETEQKVSSTEALIDELTQELEQKKASESRFKEELSVLQDLDAQTKGLQAKLSEQEGINS 361
Query: 100 KM--QLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHI 152
K+ +L EKELL +K DQ++++ T +K+ ++ KE E L+ + +
Sbjct: 362 KLAEELKEKELLESLSK--DQEEKL------RTANEKLAEVLKEKEALEANVAEV 408
Score = 39.3 bits (90), Expect = 9e-04
Identities = 41/159 (25%), Positives = 72/159 (44%), Gaps = 18/159 (11%)
Query: 12 LKLLEEEYNEKIARLEAQVKESLH----------EKASYEATISQLHGDIAAHKNHMQIL 61
LK LE E A+ + KES E A++ + ++L ++A + +
Sbjct: 983 LKNLESTIEELGAKCQGLEKESGDLAEVNLKLNLELANHGSEANELQTKLSALEAEKEQT 1042
Query: 62 ANRLDQVHFEVES---KYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQ 118
AN L+ +E + +SE L+ + + EE N +N Q ++EL AK+ +
Sbjct: 1043 ANELEASKTTIEDLTKQLTSEGEKLQSQISSHTEENNQVNAMFQSTKEELQSVIAKL--E 1100
Query: 119 QQEMTSNWQVETLKQKIMKLR---KENEVLKRKFSHIEE 154
+Q + + +TL +I KLR E VL+ F +E+
Sbjct: 1101 EQLTVESSKADTLVSEIEKLRAVAAEKSVLESHFEELEK 1139
Score = 33.9 bits (76), Expect = 0.036
Identities = 34/151 (22%), Positives = 76/151 (49%), Gaps = 17/151 (11%)
Query: 13 KLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHK--NHMQILANRLDQVHF 70
KL+E E K + EAQ E LH++ S H D + K ++L + +
Sbjct: 199 KLIELEEGLKRSAEEAQKFEELHKQ-------SASHADSESQKALEFSELLKSTKESAK- 250
Query: 71 EVESKYSS---EIRDLKDCLMAEQEEKNDLNR---KMQLLEKELLLFKAKMVDQQQEMTS 124
E+E K +S EI++L + + ++ + L ++ +++EL L K+++++ +Q+++S
Sbjct: 251 EMEEKMASLQQEIKELNEKMSENEKVEAALKSSAGELAAVQEELALSKSRLLETEQKVSS 310
Query: 125 NWQ-VETLKQKIMKLRKENEVLKRKFSHIEE 154
++ L Q++ + + K + S +++
Sbjct: 311 TEALIDELTQELEQKKASESRFKEELSVLQD 341
Score = 28.1 bits (61), Expect = 2.0
Identities = 29/156 (18%), Positives = 66/156 (41%), Gaps = 21/156 (13%)
Query: 20 NEKIARLEAQVKESLHEKASYEA---TISQLH---GDIAAHKNHMQILANRLDQVHFEVE 73
+E ++ +A + ++L + E ++ +LH G AA + + + +
Sbjct: 430 DENFSKTDALLSQALSNNSELEQKLKSLEELHSEAGSAAAAATQKNLELEDVVRSSSQAA 489
Query: 74 SKYSSEIRDLKDCLMAEQEEKNDLNRKMQLL---------------EKELLLFKAKMVDQ 118
+ S+I++L+ A +++ +L +++ LL EK L A V +
Sbjct: 490 EEAKSQIKELETKFTAAEQKNAELEQQLNLLQLKSSDAERELKELSEKSSELQTAIEVAE 549
Query: 119 QQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+++ + Q++ KQK +L + S +EE
Sbjct: 550 EEKKQATTQMQEYKQKASELELSLTQSSARNSELEE 585
Score = 26.2 bits (56), Expect = 7.6
Identities = 33/159 (20%), Positives = 70/159 (43%), Gaps = 16/159 (10%)
Query: 12 LKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFE 71
+K+ +E ++ K +A +K E + S ++ + + L L++V E
Sbjct: 49 IKVEKEAFDAKDDAEKADHVPVEEQKEVIERSSSGSQRELHESQEKAKELELELERVAGE 108
Query: 72 VESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS------- 124
++ +Y SE LKD L++ +E+ + +K LE + K+V+ ++ +S
Sbjct: 109 LK-RYESENTHLKDELLSAKEKLEETEKKHGDLEVVQKKQQEKIVEGEERHSSQLKSLED 167
Query: 125 --------NWQVETLKQKIMKLRKENEVLKRKFSHIEEG 155
+ ++ +K+ L E E ++K +EEG
Sbjct: 168 ALQSHDAKDKELTEVKEAFDALGIELESSRKKLIELEEG 206
>At3g55060 centromere protein - like
Length = 896
Score = 42.7 bits (99), Expect = 8e-05
Identities = 36/126 (28%), Positives = 57/126 (44%), Gaps = 5/126 (3%)
Query: 30 VKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSSEIRDLKDCLMA 89
V+E L+ K E I QL ++AA +IL + Q + S + E++DLK ++
Sbjct: 744 VREKLYSK---EKEIEQLQAELAAAVRGNEILRCEV-QSSLDNLSVTTHELKDLKHQMLK 799
Query: 90 EQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNW-QVETLKQKIMKLRKENEVLKRK 148
++E L +Q KE+ A + E W + + +K M L ENE LK
Sbjct: 800 KEESIRRLESNLQEAAKEMARLNALLSKVSNERGQIWSEYKQYGEKNMLLNSENETLKGM 859
Query: 149 FSHIEE 154
+EE
Sbjct: 860 VEKLEE 865
>At2g21380 putative kinesin heavy chain
Length = 1058
Score = 42.0 bits (97), Expect = 1e-04
Identities = 37/143 (25%), Positives = 70/143 (48%), Gaps = 11/143 (7%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQI------LANRLDQVHF 70
E +I LE ++E + S E I++ A+ + +++ L + ++ F
Sbjct: 622 ENSKTQIQNLENDIQEKQRQMKSLEQRITESGEASIANASSIEMQEKVMRLMTQCNEKSF 681
Query: 71 EVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVET 130
E+E S++ R L++ L + E N+L+ K+ LLE+ L KA + ++ + V+
Sbjct: 682 ELEI-ISADNRILQEQLQTKCTENNELHEKVHLLEQRLSSQKATL--SCCDVVTEEYVDE 738
Query: 131 LKQKIMKLRKENEVLKRKFSHIE 153
LK+K+ ENE K K H++
Sbjct: 739 LKKKVQSQEIENE--KLKLEHVQ 759
>At5g27220 putative protein
Length = 1181
Score = 41.6 bits (96), Expect = 2e-04
Identities = 42/159 (26%), Positives = 78/159 (48%), Gaps = 18/159 (11%)
Query: 7 GHRRALKLLEEEYNEKIARLEAQ-------VKESLHEKASYEATISQLHGDIAAHKNHMQ 59
GH ++KLL EE++E++A E + V++ E S E TI QL + + +
Sbjct: 434 GHNESIKLLLEEHSEELAIKEERHNEIAEAVRKLSLEIVSKEKTIQQLSEKQHSKQTKLD 493
Query: 60 ILANRLDQVHFEVESKYSSEIRDLKD----CLMAEQEEKNDLN------RKMQLLEKELL 109
L++ E+ SK +E+ +KD CL + ++ +L +K+Q K+
Sbjct: 494 STEKCLEETTAELVSK-ENELCSVKDTYRECLQNWEIKEKELKSFQEEVKKIQDSLKDFQ 552
Query: 110 LFKAKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRK 148
+A++V ++ +T + + LK+K + +R E LK K
Sbjct: 553 SKEAELVKLKESLTEHEKELGLKKKQIHVRSEKIELKDK 591
Score = 35.4 bits (80), Expect = 0.013
Identities = 27/137 (19%), Positives = 66/137 (47%), Gaps = 7/137 (5%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKY 76
EE E++ L ++E E+ S +S++ + + + + L Q+ +E +Y
Sbjct: 105 EEKREELGCLRKSLEECSVEERSKRGQLSEIVELLRKSQVDLDLKGEELRQMVTHLE-RY 163
Query: 77 SSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIM 136
E+++ K+ L + +L +++ K+L L K+VD + ++ET +++
Sbjct: 164 RVEVKEEKEHLRRTDNGRRELEEEIERKTKDLTLVMNKIVDCDK------RIETRSLELI 217
Query: 137 KLRKENEVLKRKFSHIE 153
K + E E+ +++ ++
Sbjct: 218 KTQGEVELKEKQLDQMK 234
Score = 33.1 bits (74), Expect = 0.062
Identities = 34/148 (22%), Positives = 63/148 (41%), Gaps = 18/148 (12%)
Query: 6 QGHRRALKLLEEEYNEKIARLEAQVK-----ESLHEKASYEATISQLHGDIAAHKNHMQI 60
Q HRR L EEE K L + E L E+ S E +Q +++
Sbjct: 255 QTHRRKL---EEEIERKTKDLTLVMDKIAECEKLFERRSLELIKTQ---------GEVEL 302
Query: 61 LANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ 120
+L+Q+ ++E ++ E+ + + L Q +L +++ KEL K + +
Sbjct: 303 KGKQLEQMDIDLE-RHRGEVNVVMEHLEKSQTRSRELAEEIERKRKELTAVLDKTAEYGK 361
Query: 121 EMTSNWQVETLKQKIMKLRKENEVLKRK 148
+ + L+QK++ +R V K+K
Sbjct: 362 TIELVEEELALQQKLLDIRSSELVSKKK 389
>At5g66030 Golgi-localized protein GRIP
Length = 788
Score = 39.7 bits (91), Expect = 7e-04
Identities = 34/152 (22%), Positives = 72/152 (47%), Gaps = 12/152 (7%)
Query: 17 EEYNEKIARLEAQVKESLHEK----ASYEATISQLHGDIAAHKNHMQILANRLDQVHFEV 72
+EY+ K +++E ++ + + E+ A +A ++LH +Q + LD EV
Sbjct: 127 QEYSSKFSQVEQKLDQEIKERDEKYADLDAKFTRLHKRAKQRIQEIQKEKDDLDARFREV 186
Query: 73 ES---KYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQ-- 127
+ SS+ ++ L +++ N+ + M ++L K+ D +E+ + Q
Sbjct: 187 NETAERASSQHSSMQQELERTRQQANEALKAMDAERQQLRSANNKLRDTIEELRGSLQPK 246
Query: 128 ---VETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+ETL+Q ++ + E LK++ +EE K
Sbjct: 247 ENKIETLQQSLLDKDQILEDLKKQLQAVEERK 278
Score = 29.3 bits (64), Expect = 0.90
Identities = 36/172 (20%), Positives = 70/172 (39%), Gaps = 31/172 (18%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANR 64
E+ + A+ L ++ + + LEAQV ++L E+ TIS L Q+L
Sbjct: 275 EERKQIAVTELSAKHQKNLEGLEAQVVDALSERDKAAETISSL-----------QVLLAE 323
Query: 65 LDQVHFEVESKYSSEIRDLKDC----------LMAEQEEKND--------LNRKMQLLEK 106
+ E+E+ + E L+ L +E E++ + L K+++ E
Sbjct: 324 KESKIAEMEAAATGEAARLRAAAETLKGELAHLKSENEKEKETWEASCDALKSKLEIAES 383
Query: 107 ELLLFKAKMVDQQQEMTSNWQVET--LKQKIMKLRKENEVLKRKFSHIEEGK 156
L + ++ + ++ S ++T L K +L+ E + R S K
Sbjct: 384 NYLQAEIEVAKMRSQLGSEMSMQTQILSTKDAELKGAREEINRLQSEFSSYK 435
>At1g05320 unknown protein
Length = 841
Score = 38.9 bits (89), Expect = 0.001
Identities = 35/142 (24%), Positives = 66/142 (45%), Gaps = 16/142 (11%)
Query: 17 EEYNEKIARLEAQVKESLHEKAS----YEATISQLHGDIAAHKNHMQILANRLDQVHFEV 72
E+ N +A + ++ + L + S ++A +S L + +QI L +
Sbjct: 494 EKENGDLAEVNIKLNQKLANQGSETDDFQAKLSVLEAEKYQQAKELQITIEDLTK----- 548
Query: 73 ESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM-VDQQQEMTSNWQVETL 131
+ +SE L+ + + +EEKN +N Q + EL+ +A++ VD+ + Q+E
Sbjct: 549 --QLTSERERLRSQISSLEEEKNQVNEIYQSTKNELVKLQAQLQVDKSKSDDMVSQIE-- 604
Query: 132 KQKIMKLRKENEVLKRKFSHIE 153
K+ L E VL+ KF +E
Sbjct: 605 --KLSALVAEKSVLESKFEQVE 624
Score = 29.6 bits (65), Expect = 0.69
Identities = 32/160 (20%), Positives = 70/160 (43%), Gaps = 23/160 (14%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATI------SQLHGDIAAHKNHM 58
E+G + ++++ +EY EK+++LE+ + +S + E + H DI
Sbjct: 194 EEGKKSSIQM--QEYQEKVSKLESSLNQSSARNSELEEDLRIALQKGAEHEDIGNVSTKR 251
Query: 59 QILANRLDQVHFEVESKYSSEIRD----------LKDCLMAEQEEKNDLNRKMQLLEKEL 108
+ L Q K +++D L+ L E++ DL+ + + ++L
Sbjct: 252 SVELQGLFQTSQLKLEKAEEKLKDLEAIQVKNSSLEATLSVAMEKERDLSENLNAVMEKL 311
Query: 109 LLFKAKMVDQQQEM----TSNWQVETL-KQKIMKLRKENE 143
+ ++ Q +E+ T + ++E L K +K++K E
Sbjct: 312 KSSEERLEKQAREIDEATTRSIELEALHKHSELKVQKTME 351
>At4g14760 centromere protein homolog
Length = 1676
Score = 38.5 bits (88), Expect = 0.001
Identities = 37/154 (24%), Positives = 70/154 (45%), Gaps = 11/154 (7%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
RRA K++ E + +++ + S + EA L D K + N+LD
Sbjct: 59 RRAHKVMVEAFPNQMSFDMIEDSASSSSEPRTEADTEALQKDGTKSKRSFSQM-NKLDGT 117
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEM------ 122
E+ SE+ LK L+ Q EK LN + QL+ ++ F+ ++ D Q+++
Sbjct: 118 SDSHEA--DSEVETLKRTLLELQTEKEALNLQYQLILSKVSRFEKELNDAQKDVKGFDER 175
Query: 123 --TSNWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
++ +++ LK+ + KL E + ++S E
Sbjct: 176 ACKADIEIKILKESLAKLEVERDTGLLQYSQAIE 209
Score = 34.3 bits (77), Expect = 0.028
Identities = 30/140 (21%), Positives = 67/140 (47%), Gaps = 12/140 (8%)
Query: 21 EKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVES--KYSS 78
E +A+LE + L + + I+ L I+ + + + L NR+ + E S K S
Sbjct: 188 ESLAKLEVERDTGLLQYSQAIERIADLEASISHGQEYAKGLTNRVSEAEREAMSLKKELS 247
Query: 79 EIRDLKDCLMAEQEEK----NDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQK 134
++ K+ + + + L + ++ E+ + +F+ DQ ++ + +++ LKQ+
Sbjct: 248 RLQSEKEAGLLRYNKSLELISSLEKTIRDAEESVRVFR----DQSEQAET--EIKALKQE 301
Query: 135 IMKLRKENEVLKRKFSHIEE 154
++KL + NE L ++ E
Sbjct: 302 LLKLNEVNEDLNVRYQQCLE 321
Score = 30.4 bits (67), Expect = 0.40
Identities = 32/130 (24%), Positives = 63/130 (47%), Gaps = 14/130 (10%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVK--ESLHEKASYEATI--SQLHGDIAAHKNHMQI 60
+Q L+ + +E + + L A ++ ESLH ++ E + S+LH I + +++
Sbjct: 389 KQNEIEKLQAVMQEEQLRFSELGASLRNLESLHSQSQEEQKVLTSELHSRIQMLRE-LEM 447
Query: 61 LANRLD-QVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQ 119
++L+ + + E++ SEI D L ++ E + L + + LE+E V +Q
Sbjct: 448 RNSKLEGDISSKEENRNLSEINDTSISLEIQKNEISCLKKMKEKLEEE--------VAKQ 499
Query: 120 QEMTSNWQVE 129
+S QVE
Sbjct: 500 MNQSSALQVE 509
Score = 27.7 bits (60), Expect = 2.6
Identities = 25/134 (18%), Positives = 62/134 (45%), Gaps = 9/134 (6%)
Query: 25 RLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVES------KYSS 78
+LE +V + +++ ++ + I + G+I + Q L +++ F+ ES K
Sbjct: 491 KLEEEVAKQMNQSSALQVEIHCVKGNIDSMNRRYQKLIDQVSLTGFDPESLSYSVKKLQD 550
Query: 79 EIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIMKL 138
E L + +++E N + K+ ++ + K ++ + SN +++ ++K L
Sbjct: 551 ENSKLVELCTNQRDENNAVTGKLCEMDS---ILKRNADLEKLLLESNTKLDGSREKAKDL 607
Query: 139 RKENEVLKRKFSHI 152
+ E L+ + S +
Sbjct: 608 IERCESLRGEKSEL 621
Score = 27.3 bits (59), Expect = 3.4
Identities = 12/37 (32%), Positives = 22/37 (59%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAA 53
++ E + LE+QV E + ++Y+ I+ L GD+ A
Sbjct: 1279 KQMKETVGFLESQVTELKSQLSAYDPVIASLAGDVKA 1315
Score = 27.3 bits (59), Expect = 3.4
Identities = 35/167 (20%), Positives = 74/167 (43%), Gaps = 31/167 (18%)
Query: 13 KLLEEEYNEKIARLEAQVK----ESLHE-----KASYEATISQ---LHGDIAAHKNHMQI 60
KL++ E E+ R E + + ESLH+ + Y T++ L + K+ M +
Sbjct: 962 KLIKREQQEQKLRAELKFENLKFESLHDSYMVLQQDYSYTLNDNKTLLLKFSEFKDGMHV 1021
Query: 61 LANRLDQVHFEVE-------------SKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKE 107
+ D + E S+ + E+ D + + + +E L RK++ LEK+
Sbjct: 1022 VEEENDAILQEAVALSNTCVVYRSFGSEMAEEVEDFVETVSSLREISTGLKRKVETLEKK 1081
Query: 108 LLLFKAKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
L + K +++ N +E L++ + + +L+ + S+++E
Sbjct: 1082 L---EGK---EKESQGLNKMLENLQEGLEEDNFLTGLLEHQVSNVDE 1122
>At4g32190 unknown protein
Length = 783
Score = 38.1 bits (87), Expect = 0.002
Identities = 35/124 (28%), Positives = 61/124 (48%), Gaps = 13/124 (10%)
Query: 29 QVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSSEIRDLKDCLM 88
+ KE L ++ E TIS+ A H + L L + + E+ S+ + EI +LK L
Sbjct: 144 RAKEELEKR---EKTISE------ASLKH-ESLQEELKRANVELASQ-AREIEELKHKLR 192
Query: 89 AEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRK 148
EE+ L + L E+EL + ++ ++ +E+ + + + K L K NEV+KR+
Sbjct: 193 ERDEERAALQSSLTLKEEELEKMRQEIANRSKEV--SMAISEFESKSQLLSKANEVVKRQ 250
Query: 149 FSHI 152
I
Sbjct: 251 EGEI 254
>At3g58840 unknown protein
Length = 318
Score = 37.7 bits (86), Expect = 0.003
Identities = 35/156 (22%), Positives = 67/156 (42%), Gaps = 21/156 (13%)
Query: 7 GHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEAT----------ISQLHGDIAAHKN 56
G +K +E E N++ +E +++E EK + EA +S LH D+ N
Sbjct: 55 GEIEEMKDVEAEMNQRFGEMEKEIEEYEEEKKALEAISTRAVELETEVSNLHDDLITSLN 114
Query: 57 HMQILANRLDQVH---FEVESKYSS---EIRDLKDCLMAEQEEKNDLNRKMQLLEKELLL 110
+ A + ++ E+ K E L+ ++ DL RK+ +LE +
Sbjct: 115 GVDKTAEEVAELKKALAEIVEKLEGCEKEAEGLRKDRAEVEKRVRDLERKIGVLEVREME 174
Query: 111 FKAKMVDQQQEM-----TSNWQVETLKQKIMKLRKE 141
K+K + ++EM ++E L++ ++ L E
Sbjct: 175 EKSKKLRSEEEMREIDDEKKREIEELQKTVIVLNLE 210
Score = 26.9 bits (58), Expect = 4.5
Identities = 29/139 (20%), Positives = 59/139 (41%), Gaps = 8/139 (5%)
Query: 18 EYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYS 77
E KI +E + +E E + + +L G+I K+ + R ++ E+E
Sbjct: 24 ELERKIEDMENKNQELTRENRELKERLERLTGEIEEMKDVEAEMNQRFGEMEKEIE---- 79
Query: 78 SEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQ--VETLKQKI 135
E + K L A +L ++ L +L+ VD+ E + + + + +K+
Sbjct: 80 -EYEEEKKALEAISTRAVELETEVSNLHDDLIT-SLNGVDKTAEEVAELKKALAEIVEKL 137
Query: 136 MKLRKENEVLKRKFSHIEE 154
KE E L++ + +E+
Sbjct: 138 EGCEKEAEGLRKDRAEVEK 156
>At1g63300 hypothetical protein
Length = 1029
Score = 37.7 bits (86), Expect = 0.003
Identities = 41/174 (23%), Positives = 80/174 (45%), Gaps = 32/174 (18%)
Query: 9 RRALKLLEE--EYNEKIARLEAQVKESLHE----KASYEATISQLHGDIAAHKNHMQILA 62
+ A+K + E E + +LE +K++ E +A YEA + +L ++ + M+ +
Sbjct: 643 KMAMKAMTEANELRMQKRQLEEMIKDANDELRANQAEYEAKLHELSEKLSFKTSQMERML 702
Query: 63 NRLDQVHFEVESK-----------------YSSEIRDLK---DCLMAEQEEKNDLNRKMQ 102
LD+ E++++ EI +LK D LM + E+ +L ++
Sbjct: 703 ENLDEKSNEIDNQKRHEEDVTANLNQEIKILKEEIENLKKNQDSLMLQAEQAENLRVDLE 762
Query: 103 LLEKELLLFKAKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIEEGK 156
+K ++ +A + Q+E N + L+ KI +RKE+E L + I+ K
Sbjct: 763 KTKKSVMEAEASL---QRE---NMKKIELESKISLMRKESESLAAELQVIKLAK 810
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 37.4 bits (85), Expect = 0.003
Identities = 30/146 (20%), Positives = 74/146 (50%), Gaps = 19/146 (13%)
Query: 9 RRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQV 68
+++L + E +++ + +++V+E + E A + T++Q ++++ +V
Sbjct: 515 KKSLSSMILEITDELKQAQSKVQELVTELAESKDTLTQKENELSSFV-----------EV 563
Query: 69 HFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQV 128
H + SS++++L+ + + +E+ +LN+ + E+E K + QQ + ++
Sbjct: 564 HEAHKRDSSSQVKELEARVESAEEQVKELNQNLNSSEEE------KKILSQQISEMSIKI 617
Query: 129 ETLKQKIMKLRKENEVLKRKFSHIEE 154
+ + I +L E+E LK SH E+
Sbjct: 618 KRAESTIQELSSESERLKG--SHAEK 641
Score = 29.3 bits (64), Expect = 0.90
Identities = 24/135 (17%), Positives = 64/135 (46%), Gaps = 8/135 (5%)
Query: 17 EEYNEKIARLEAQVKESLHE-KASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESK 75
EE N+ ++ +++ + + + + + + IS+L G++ + + L ++H E +
Sbjct: 5 EEENKSLSLKVSEISDVIQQGQTTIQELISEL-GEMKEKYKEKESEHSSLVELHKTHERE 63
Query: 76 YSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQKI 135
SS++++L+ + + ++ D + + E+E L K+ + E ++ + +
Sbjct: 64 SSSQVKELEAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNE------IQEAQNTM 117
Query: 136 MKLRKENEVLKRKFS 150
+L E+ LK S
Sbjct: 118 QELMSESGQLKESHS 132
Score = 28.5 bits (62), Expect = 1.5
Identities = 33/149 (22%), Positives = 65/149 (43%), Gaps = 13/149 (8%)
Query: 17 EEYNEKIARLEAQVKESLH----EKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEV 72
EE E +L A++ ++L+ EK I++L +I +N +Q L + Q+ E
Sbjct: 248 EEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLK-ES 306
Query: 73 ESKYSSEIRDLKDCLMAEQEEK----NDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQV 128
S ++ L+D Q E ++L +++ E+ + + D ++E N +
Sbjct: 307 HSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEE---NKAI 363
Query: 129 ETLKQKIM-KLRKENEVLKRKFSHIEEGK 156
+ +IM KL + +K + E K
Sbjct: 364 SSKNLEIMDKLEQAQNTIKELMDELGELK 392
Score = 28.5 bits (62), Expect = 1.5
Identities = 34/161 (21%), Positives = 69/161 (42%), Gaps = 24/161 (14%)
Query: 17 EEYNEKIARLEAQVKESLH----EKASYEATISQLHGDIAAHKNHMQILANR-------- 64
E + E +L A +SL+ EK I++L +I +N MQ L +
Sbjct: 72 EAHIESSEKLVADFTQSLNNAEEEKKLLSQKIAELSNEIQEAQNTMQELMSESGQLKESH 131
Query: 65 ---------LDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKM 115
L +H + S+ +L+ L + +++ +DL+ ++ E+E +K
Sbjct: 132 SVKERELFSLRDIHEIHQRDSSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKN 191
Query: 116 VD--QQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
V+ + E T N ++ L ++ KL+ + + + S + E
Sbjct: 192 VETMNKLEQTQN-TIQELMAELGKLKDSHREKESELSSLVE 231
Score = 28.1 bits (61), Expect = 2.0
Identities = 29/144 (20%), Positives = 63/144 (43%), Gaps = 20/144 (13%)
Query: 13 KLLEEEYNE---KIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVH 69
K+L ++ +E KI R E+ ++E E +L G A N + L + +H
Sbjct: 604 KILSQQISEMSIKIKRAESTIQELSSES-------ERLKGSHAEKDNELFSLRD----IH 652
Query: 70 FEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVE 129
+ + S+++R L+ L + + +L+ ++ E+E K+ + E+ E
Sbjct: 653 ETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISETSDEL------E 706
Query: 130 TLKQKIMKLRKENEVLKRKFSHIE 153
+ + +L ++ LK + + E
Sbjct: 707 RTQIMVQELTADSSKLKEQLAEKE 730
Score = 27.3 bits (59), Expect = 3.4
Identities = 29/143 (20%), Positives = 64/143 (44%), Gaps = 13/143 (9%)
Query: 11 ALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLD---Q 67
+LK EEE N+ I+ + L + + TI +L ++ K+ + + L +
Sbjct: 176 SLKAAEEE-NKAISSKNVETMNKLEQTQN---TIQELMAELGKLKDSHREKESELSSLVE 231
Query: 68 VHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQ 127
VH + S +++L++ + + ++ +LN+ + E+E + K+ + E
Sbjct: 232 VHETHQRDSSIHVKELEEQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNE------ 285
Query: 128 VETLKQKIMKLRKENEVLKRKFS 150
++ + I +L E+ LK S
Sbjct: 286 IKEAQNTIQELVSESGQLKESHS 308
>At2g36200 putative kinesin-related cytokinesis protein
Length = 1056
Score = 37.4 bits (85), Expect = 0.003
Identities = 28/130 (21%), Positives = 63/130 (47%), Gaps = 7/130 (5%)
Query: 14 LLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVE 73
L+++ Y E I RL+A+V S + Y ++ +++A +++Q+ ++E
Sbjct: 374 LIKDLYGE-IERLKAEVYASREKNGVYMPKERYYQ-----EESERKVMAEQIEQMGGQIE 427
Query: 74 SKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQ 133
+ Y ++ +L+D + + E +DL K+ + EK L + +E+ + K
Sbjct: 428 N-YQKQLEELQDKYVGQVRECSDLTTKLDITEKNLSQTCKVLASTNEELKKSQYAMKEKD 486
Query: 134 KIMKLRKENE 143
I+ +K++E
Sbjct: 487 FIISEQKKSE 496
>At5g07660 SMC-like protein
Length = 1058
Score = 37.0 bits (84), Expect = 0.004
Identities = 38/150 (25%), Positives = 73/150 (48%), Gaps = 15/150 (10%)
Query: 16 EEEYNEKIARLEAQVKESLHE--------KASYEATISQLHGDIAAHKNHMQILANRLDQ 67
++E EK + LE ++++SL E KASYE G+I A + L + D+
Sbjct: 727 QKEIEEKESLLE-KLQDSLKEAELKANELKASYENLYESAKGEIEALEKAEDELKEKEDE 785
Query: 68 VH-FEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS-- 124
+H E E + +I +KD ++ E ++ + +++++ +E KA ++ + E+ +
Sbjct: 786 LHSAETEKNHYEDI--MKDKVLPEIKQAETIYKELEMKRQESNK-KASIICPESEIKALG 842
Query: 125 NWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
W T Q ++ K N LKR+ + E
Sbjct: 843 PWDGPTPLQLSAQINKINHRLKRENENYSE 872
>At4g33200 myosin - like protein
Length = 1374
Score = 37.0 bits (84), Expect = 0.004
Identities = 38/145 (26%), Positives = 69/145 (47%), Gaps = 21/145 (14%)
Query: 16 EEEYNEKIARLEAQVKE-SLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVES 74
EE + +I++L+ ++ SL A+ ATI++ N +L +LD + + +S
Sbjct: 909 EEAKSSEISKLQKTLESFSLKLDAARLATINEC--------NKNAVLEKQLD-ISMKEKS 959
Query: 75 KYSSEIR---DLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTS------- 124
E+ +LK + N L +K ++LEKELL K + Q++
Sbjct: 960 AVERELNGMVELKKDNALLKNSMNSLEKKNRVLEKELLNAKTNCNNTLQKLKEAEKRCSE 1019
Query: 125 -NWQVETLKQKIMKLRKENEVLKRK 148
V++L++K+ L EN+VL +K
Sbjct: 1020 LQTSVQSLEEKLSHLENENQVLMQK 1044
>At3g20350 unknown protein
Length = 673
Score = 37.0 bits (84), Expect = 0.004
Identities = 37/150 (24%), Positives = 70/150 (46%), Gaps = 10/150 (6%)
Query: 6 QGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRL 65
Q R +K LE E + +LE +K+ E+A++ S+ H + A + M+ N+
Sbjct: 224 QEARACIKDLESEKRSQKKKLEQFLKKVSEERAAWR---SREHEKVRAIIDDMKADMNQE 280
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDL--NRKMQLLEKELLLFKAKMVDQQQEMT 123
+ +E S + +L D +A + +D RK + L +E+ AK +++ +
Sbjct: 281 KKTRQRLEIVNSKLVNELADSKLAVKRYMHDYQQERKARELIEEVCDELAKEIEEDKA-- 338
Query: 124 SNWQVETLKQKIMKLRKENEVLKRKFSHIE 153
++E LK + M LR+E + +R E
Sbjct: 339 ---EIEALKSESMNLREEVDDERRMLQMAE 365
>At2g39300 hypothetical protein
Length = 768
Score = 36.2 bits (82), Expect = 0.007
Identities = 37/152 (24%), Positives = 67/152 (43%), Gaps = 20/152 (13%)
Query: 15 LEEEYNEKIARLEAQVKESLHEK---ASYEATISQLHGDIAAHKNHMQILANRLDQVHFE 71
LE+ Y E R++ + E + EK + + IS L GDI + LA FE
Sbjct: 257 LEKRYKEAEKRVKL-LSEEMEEKKFLSDCDFDISSLVGDIRQMEEERVGLA-------FE 308
Query: 72 VESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNW----- 126
V S S++ + + KND + ++ LEKE + ++ + +S W
Sbjct: 309 VLSLLRSQMDERASTREDIRRVKNDWDLLLKRLEKEKTELQVQLETELDRRSSEWTSKVE 368
Query: 127 ----QVETLKQKIMKLRKENEVLKRKFSHIEE 154
+ + L++++ +L + N L+R+ S E
Sbjct: 369 SFKVEEKRLRERVRELAEHNVSLQREISTFHE 400
Score = 34.3 bits (77), Expect = 0.028
Identities = 26/97 (26%), Positives = 50/97 (50%), Gaps = 5/97 (5%)
Query: 60 ILANRLDQVHFEVESKYSSEIRDLKDC--LMAEQEEKNDLNRKMQLLEKELLLFKAKMVD 117
+L + +++ ES SS R ++ + E N + +Q KELL K+++
Sbjct: 637 LLLEKSNEMASNSESSCSSAARPSSRSVEMVKKDENINRMEINLQEAAKELLTLP-KVLE 695
Query: 118 QQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHIEE 154
+++EM +V+ +++ M L E E+LK+K +EE
Sbjct: 696 EREEMWK--EVKECRKRNMDLESEKEMLKKKVEKLEE 730
Score = 30.0 bits (66), Expect = 0.53
Identities = 25/132 (18%), Positives = 54/132 (39%), Gaps = 6/132 (4%)
Query: 20 NEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSSE 79
+E + L A +E E +S+L + + + ++ E + + S
Sbjct: 413 DETVTELSATAEEMREENLFLMQNLSKLQESYTGSTDDLDYVRRNFEEKDMECKELHKSV 472
Query: 80 IRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQ------EMTSNWQVETLKQ 133
R L+ C E+ + + + ++K+ K + +Q E++ +VE++K
Sbjct: 473 TRLLRTCKEQEKTIQGLRDGFSEEIKKQPSEHVDKKLQMEQLRLVGVELSLRKEVESMKL 532
Query: 134 KIMKLRKENEVL 145
+ LR+EN L
Sbjct: 533 EAESLRRENNCL 544
>At5g46070 putative protein
Length = 1060
Score = 35.8 bits (81), Expect = 0.010
Identities = 38/160 (23%), Positives = 71/160 (43%), Gaps = 12/160 (7%)
Query: 5 EQGHRRALKLLEEEYNEKIARLEAQVKESLHEK-----ASYEATISQLHGDI-AAHKNHM 58
E+ +A KL + E + + R + +V ES E AS T+S++ + +A+K +
Sbjct: 676 EEITEKATKLEKAEQSLTVLRSDLKVAESKLESFEVELASLRLTLSEMTDKLDSANKKAL 735
Query: 59 --QILANRLDQVHFEVESKYSSEIRDLKD----CLMAEQEEKNDLNRKMQLLEKELLLFK 112
+ AN+L+Q +E KY SE + + C AE E K + + K
Sbjct: 736 AYEKEANKLEQEKIRMEQKYRSEFQRFDEVKERCKAAEIEAKRATELADKARTDAVTSQK 795
Query: 113 AKMVDQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHI 152
K Q+ M Q+E ++++ L ++ L+ + +
Sbjct: 796 EKSESQRLAMERLAQIERAERQVENLERQKTDLEDELDRL 835
Score = 30.8 bits (68), Expect = 0.31
Identities = 31/156 (19%), Positives = 64/156 (40%), Gaps = 7/156 (4%)
Query: 4 SEQGHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILAN 63
+ Q + + L E +I R E QV+ +K E + +L + + IL
Sbjct: 792 TSQKEKSESQRLAMERLAQIERAERQVENLERQKTDLEDELDRLRVSEMEAVSKVTILEA 851
Query: 64 RLDQVHFEVES-------KYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMV 116
R+++ E+ S + + ++ L+ L E++ NR+ + L EL +A +
Sbjct: 852 RVEEREKEIGSLIKETNAQRAHNVKSLEKLLDEERKAHIAANRRAEALSLELQAAQAHVD 911
Query: 117 DQQQEMTSNWQVETLKQKIMKLRKENEVLKRKFSHI 152
+ QQE+ ET ++ + + +F +
Sbjct: 912 NLQQELAQARLKETALDNKIRAASSSHGKRSRFEDV 947
>At3g22790 unknown protein
Length = 1694
Score = 35.8 bits (81), Expect = 0.010
Identities = 32/141 (22%), Positives = 67/141 (46%), Gaps = 5/141 (3%)
Query: 7 GHRRALKLLEEEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHK-NHMQILANRL 65
G ++L L E +I LE + L E + +S ++ + + K + A L
Sbjct: 1041 GKNKSLHLKFSELKGEICILEEENGAILEEAIALN-NVSVVYQSLGSEKAEQAEAFAKNL 1099
Query: 66 DQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSN 125
+ + + S ++ L++ L ++ + +LN K++ L++ L +A ++ E
Sbjct: 1100 NSLQ-NINSGLKQKVETLEEILKGKEVDSQELNSKLEKLQESLE--EANELNDLLEHQIL 1156
Query: 126 WQVETLKQKIMKLRKENEVLK 146
+ ETL+QK ++L + E+LK
Sbjct: 1157 VKEETLRQKAIELLEAEEMLK 1177
Score = 28.1 bits (61), Expect = 2.0
Identities = 28/140 (20%), Positives = 64/140 (45%), Gaps = 12/140 (8%)
Query: 21 EKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVES--KYSS 78
E +A+LEA+ +L I++L + + ++ L NR + EVE+ + S
Sbjct: 223 EALAKLEAERDAALLRYNESMQKITELEESFSHAQEDVKGLTNRATKAETEVENLKQAHS 282
Query: 79 EIRDLKDCLMAEQ----EEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQVETLKQK 134
+ K+ +AE E ++L +K++ E+ F + + E ++ L+ +
Sbjct: 283 RLHSEKEAGLAEYNRCLEMISNLEKKVRDAEENAQNFSNQSAKAEDE------IKALRHE 336
Query: 135 IMKLRKENEVLKRKFSHIEE 154
++K+ + + L+ ++ E
Sbjct: 337 LVKVNEVKDGLRLRYQQCLE 356
Score = 27.7 bits (60), Expect = 2.6
Identities = 26/119 (21%), Positives = 55/119 (45%), Gaps = 17/119 (14%)
Query: 25 RLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVES---------K 75
+LE +V +++ ++++ I +L +I + Q + +++ + +S
Sbjct: 528 KLEEEVARHINQSSAFQEEIRRLKDEIDSLNKRYQAIMEQVNLAGLDPKSLACSVRKLQD 587
Query: 76 YSSEIRDL-------KDCLMAEQEEKNDLNRKMQLLEKELLLFKAKMVDQQQEMTSNWQ 127
+S++ +L KD L + E +++ RK LEK LL K+ D +E T + Q
Sbjct: 588 ENSKLTELCNHQSDDKDALTEKLRELDNILRKNVCLEKLLLESNTKL-DGSREKTKDLQ 645
Score = 26.9 bits (58), Expect = 4.5
Identities = 27/145 (18%), Positives = 64/145 (43%), Gaps = 14/145 (9%)
Query: 17 EEYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVE--- 73
++ E+ L + E + E+A+ + + + ++ +L L + E++
Sbjct: 642 KDLQERCESLRGEKYEFIAERANLLSQLQIMTENMQKLLEKNSLLETSLSGANIELQCVK 701
Query: 74 ------SKYSSEIRDLKDCLMAEQE----EKNDLNRKMQLLEKELLLFKAKMVD-QQQEM 122
++ +++ K L+ E+E + N + K+ +LEK+ + K D Q+++
Sbjct: 702 EKSKCFEEFFQLLKNDKAELIKERESLISQLNAVKEKLGVLEKKFTELEGKYADLQREKQ 761
Query: 123 TSNWQVETLKQKIMKLRKENEVLKR 147
N QVE L+ + ++E +R
Sbjct: 762 FKNLQVEELRVSLATEKQERASYER 786
Score = 26.2 bits (56), Expect = 7.6
Identities = 22/81 (27%), Positives = 37/81 (45%), Gaps = 2/81 (2%)
Query: 18 EYNEKIARLEAQVKESLHEKASYEATISQLHGDIAAHKNHMQILANRLDQVHFEVE--SK 75
E E + L KES K + E S+L ++IL+N + + EV+ K
Sbjct: 1185 ELCEAVEELRKDCKESRKLKGNLEKRNSELCDLAGRQDEEIKILSNLKENLESEVKLLHK 1244
Query: 76 YSSEIRDLKDCLMAEQEEKND 96
E R ++ L +E +EK++
Sbjct: 1245 EIQEHRVREEFLSSELQEKSN 1265
>At3g02930 unknown protein
Length = 806
Score = 35.8 bits (81), Expect = 0.010
Identities = 28/118 (23%), Positives = 58/118 (48%), Gaps = 8/118 (6%)
Query: 44 ISQLHGDIAAHKNHMQILANRLDQVHFEVESKYSSEIRDLKDCLMAEQEEKNDLNRKMQL 103
+ + ++++ M L N + + E ++ + E + ++DCL ++E L ++
Sbjct: 545 VKEFDEEVSSMGKEMNRLGNLVKRTKEEADASWEKESQ-MRDCLKEVEDEVIYLQETLRE 603
Query: 104 LEKELLLFKAKMVDQQQEMTS----NWQVETLKQKIMKLRKE-NEVLKRKFS--HIEE 154
+ E L K KM+D++ E S N ++ + +K KE +E+L+ + HIEE
Sbjct: 604 AKAETLKLKGKMLDKETEFQSIVHENDELRVKQDDSLKKIKELSELLEEALAKKHIEE 661
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.127 0.334
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,123,417
Number of Sequences: 26719
Number of extensions: 117363
Number of successful extensions: 811
Number of sequences better than 10.0: 196
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 146
Number of HSP's that attempted gapping in prelim test: 645
Number of HSP's gapped (non-prelim): 301
length of query: 156
length of database: 11,318,596
effective HSP length: 91
effective length of query: 65
effective length of database: 8,887,167
effective search space: 577665855
effective search space used: 577665855
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC148819.6


