

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148819.1 + phase: 0
(244 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
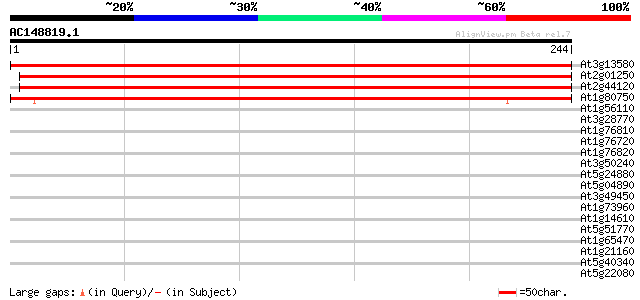
Score E
Sequences producing significant alignments: (bits) Value
At3g13580 putative ribosomal protein 416 e-117
At2g01250 60S ribosomal protein L7 413 e-116
At2g44120 At2g44120/F6E13.25 408 e-114
At1g80750 ribosomal protein L7, putative 197 4e-51
At1g56110 SAR like DNA binding protein 37 0.007
At3g28770 hypothetical protein 33 0.11
At1g76810 translation initiation factor IF-2 like protein 33 0.18
At1g76720 putative translation initiation factor IF-2 33 0.18
At1g76820 putative translation initiation factor IF-2 32 0.24
At3g50240 kinesin-related protein (kicp-02) 32 0.31
At5g24880 glutamic acid-rich protein 32 0.40
At5g04890 unknown protein 32 0.40
At3g49450 putative protein 31 0.52
At1g73960 unknown protein 31 0.52
At1g14610 valyl tRNA synthetase (valRS) 31 0.52
At5g51770 putative protein 31 0.69
At1g65470 hypothetical protein, 3' partial 31 0.69
At1g21160 transcription factor, putative 31 0.69
At5g40340 unknown protein 30 0.90
At5g22080 unknown protein 30 0.90
>At3g13580 putative ribosomal protein
Length = 244
Score = 416 bits (1070), Expect = e-117
Identities = 200/244 (81%), Positives = 228/244 (92%)
Query: 1 MAKEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDS 60
M + E KTVVPESVLKK+KR EEWAL KKQE E+AKK+ +E RKLI++RAKQY+KEY +
Sbjct: 1 MTEAESKTVVPESVLKKRKREEEWALAKKQELEAAKKQNAEKRKLIFNRAKQYSKEYQEK 60
Query: 61 QKELIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLK 120
++ELI LKREAKLKGGFYVDPEAKLLFIIRIRGINA+DPK++KILQLLRLRQI NGVFLK
Sbjct: 61 ERELIQLKREAKLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLK 120
Query: 121 VNKATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGI 180
VNKAT+NML RVEPYVTYGYPNLKSV+ELIYKRG+GK++ QR ALTDNSI++Q LGKHGI
Sbjct: 121 VNKATINMLRRVEPYVTYGYPNLKSVKELIYKRGFGKLNHQRTALTDNSIVDQGLGKHGI 180
Query: 181 ICIEDLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELI 240
IC+EDLIHEI+TVGPHF+EANNFLWPF+LKAPLGGMKKKRNHYVEGGDAGNRE++INEL+
Sbjct: 181 ICVEDLIHEIMTVGPHFKEANNFLWPFQLKAPLGGMKKKRNHYVEGGDAGNRENFINELV 240
Query: 241 RRMN 244
RRMN
Sbjct: 241 RRMN 244
>At2g01250 60S ribosomal protein L7
Length = 242
Score = 413 bits (1061), Expect = e-116
Identities = 201/240 (83%), Positives = 224/240 (92%)
Query: 5 EVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKEL 64
E K VVPESVLKK+KR EEWAL KKQ E+AKKK +E RKLI+ RA+QY+KEY + +KEL
Sbjct: 3 ESKVVVPESVLKKRKREEEWALEKKQNVEAAKKKNAENRKLIFKRAEQYSKEYAEKEKEL 62
Query: 65 IALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKA 124
I+LKREAKLKGGFYVDPEAKLLFIIRIRGINA+DPK++KILQLLRLRQI NGVFLKVNKA
Sbjct: 63 ISLKREAKLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKA 122
Query: 125 TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIE 184
T+NML RVEPYVTYG+PNLKSV+ELIYKRGYGK++ QRIALTDNSI+EQ LGKHGIIC E
Sbjct: 123 TMNMLRRVEPYVTYGFPNLKSVKELIYKRGYGKLNHQRIALTDNSIVEQALGKHGIICTE 182
Query: 185 DLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRMN 244
DLIHEILTVGPHF+EANNFLWPF+LKAPLGG+KKKRNHYVEGGDAGNRE++INELIRRMN
Sbjct: 183 DLIHEILTVGPHFKEANNFLWPFQLKAPLGGLKKKRNHYVEGGDAGNRENFINELIRRMN 242
>At2g44120 At2g44120/F6E13.25
Length = 242
Score = 408 bits (1048), Expect = e-114
Identities = 199/240 (82%), Positives = 219/240 (90%)
Query: 5 EVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKEL 64
E K VVPESVLKK KR EEWAL KK E +AKKK E RKLI+ RA+QYAKEY + EL
Sbjct: 3 ESKVVVPESVLKKIKRQEEWALAKKDEAVAAKKKSVEARKLIFKRAEQYAKEYAEKDNEL 62
Query: 65 IALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVFLKVNKA 124
I LKREAKLKGGFYVDPEAKLLFIIRIRGINA+DPK++KILQLLRLRQI NGVFLKVNKA
Sbjct: 63 IRLKREAKLKGGFYVDPEAKLLFIIRIRGINAIDPKTKKILQLLRLRQIFNGVFLKVNKA 122
Query: 125 TVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKHGIICIE 184
TVNML RVEPYVTYGYPNLKSV+ELIYKRGYGK++ QRIALTDNSI++Q LGKHGIIC+E
Sbjct: 123 TVNMLRRVEPYVTYGYPNLKSVKELIYKRGYGKLNHQRIALTDNSIVDQALGKHGIICVE 182
Query: 185 DLIHEILTVGPHFREANNFLWPFKLKAPLGGMKKKRNHYVEGGDAGNREDYINELIRRMN 244
DLIHEI+TVGPHF+EANNFLWPF+LKAPLGG+KKKRNHYVEGGDAGNRE++INEL+RRMN
Sbjct: 183 DLIHEIMTVGPHFKEANNFLWPFQLKAPLGGLKKKRNHYVEGGDAGNRENFINELVRRMN 242
>At1g80750 ribosomal protein L7, putative
Length = 247
Score = 197 bits (501), Expect = 4e-51
Identities = 105/247 (42%), Positives = 156/247 (62%), Gaps = 3/247 (1%)
Query: 1 MAKEEVKTV--VPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYD 58
MA+EE K + +PE VLKK+K +E A ++K++ E + + + R + + E+
Sbjct: 1 MAEEEAKGLDYIPEIVLKKRKNRDELAFIRKKQLELGNSGKKKKKVSDIKRPEDFVHEFR 60
Query: 59 DSQKELIALKREAKLKGGFYVDPEAKLLFIIRIRGINAMDPKSRKILQLLRLRQICNGVF 118
+ ++I +K+ K ++ L+FIIRI+G N M PK+++IL L+L+ + GVF
Sbjct: 61 AKEVDMIRMKQRVKRPKSSPPPVKSDLVFIIRIQGKNDMHPKTKRILNNLQLKSVFTGVF 120
Query: 119 LKVNKATVNMLHRVEPYVTYGYPNLKSVRELIYKRGYGKVDRQRIALTDNSIIEQVLGKH 178
K + L +V+PYVTYGYPN KSV++LIYK+G ++ + LTDN+IIEQ LG+H
Sbjct: 121 AKATDSLFQKLLKVQPYVTYGYPNDKSVKDLIYKKGCTIIEGNPVPLTDNNIIEQALGEH 180
Query: 179 GIICIEDLIHEILTVGPHFREANNFLWPFKLKAPLGG-MKKKRNHYVEGGDAGNREDYIN 237
I+ IEDL++EI VG HFRE FL P KL P+ + +K+ + EGGD GNRED IN
Sbjct: 181 KILGIEDLVNEIARVGDHFREVMRFLGPLKLNKPVADVLHRKKQVFSEGGDTGNREDKIN 240
Query: 238 ELIRRMN 244
+LI +MN
Sbjct: 241 DLISKMN 247
>At1g56110 SAR like DNA binding protein
Length = 522
Score = 37.4 bits (85), Expect = 0.007
Identities = 27/74 (36%), Positives = 38/74 (50%), Gaps = 11/74 (14%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
KE V V +S KK K EE +V +E +S KKK+ E RK+ + A+E + S+K
Sbjct: 438 KEPVDASVKKSKKKKAKGEEEEEVVAMEEDKSEKKKKKEKRKM------ETAEENEKSEK 491
Query: 63 ELIALKREAKLKGG 76
K+ K K G
Sbjct: 492 -----KKTKKSKAG 500
>At3g28770 hypothetical protein
Length = 2081
Score = 33.5 bits (75), Expect = 0.11
Identities = 20/59 (33%), Positives = 34/59 (56%), Gaps = 7/59 (11%)
Query: 16 KKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLK 74
KK+K KK+E+E+ +KK SE K S+ K+ KE++D++ ++K+E K
Sbjct: 1049 KKEKEESRDLKAKKKEEETKEKKESENHK---SKKKEDKKEHEDNK----SMKKEEDKK 1100
Score = 31.2 bits (69), Expect = 0.52
Identities = 18/70 (25%), Positives = 37/70 (52%), Gaps = 1/70 (1%)
Query: 4 EEVKTVVPESVLKKQKRNEEWALVKKQEQE-SAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
EE K+ E K++K++++ +K +E +KK++ E+R L + ++ KE +S+
Sbjct: 1016 EEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESEN 1075
Query: 63 ELIALKREAK 72
K + K
Sbjct: 1076 HKSKKKEDKK 1085
Score = 28.5 bits (62), Expect = 3.4
Identities = 23/79 (29%), Positives = 35/79 (44%), Gaps = 10/79 (12%)
Query: 4 EEVKTVVPESVLKKQKRNEEWALVKKQEQE----------SAKKKRSETRKLIWSRAKQY 53
E+ K++ E K++K++EE KK+E + S KKK + K K
Sbjct: 1088 EDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKLEDQNSNKKKEDKNEKKKSQHVKLV 1147
Query: 54 AKEYDDSQKELIALKREAK 72
KE D +K+ K E K
Sbjct: 1148 KKESDKKEKKENEEKSETK 1166
Score = 27.3 bits (59), Expect = 7.6
Identities = 20/78 (25%), Positives = 41/78 (51%), Gaps = 6/78 (7%)
Query: 3 KEEVKTVVPESVLKKQKRNE------EWALVKKQEQESAKKKRSETRKLIWSRAKQYAKE 56
KEE K + LKKQ+ N+ E + +K++ +++ +KK SE K+Y ++
Sbjct: 959 KEEDKKEYVNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEK 1018
Query: 57 YDDSQKELIALKREAKLK 74
+++E K++++ K
Sbjct: 1019 KSKTKEEAKKEKKKSQDK 1036
Score = 27.3 bits (59), Expect = 7.6
Identities = 17/72 (23%), Positives = 37/72 (50%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
KE+ ++ S +++K EE K+E + KKK + ++ ++ +K+ + +
Sbjct: 997 KEKKESEDSASKNREKKEYEEKKSKTKEEAKKEKKKSQDKKREEKDSEERKSKKEKEESR 1056
Query: 63 ELIALKREAKLK 74
+L A K+E + K
Sbjct: 1057 DLKAKKKEEETK 1068
Score = 27.3 bits (59), Expect = 7.6
Identities = 17/61 (27%), Positives = 29/61 (46%), Gaps = 3/61 (4%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
+ E K+ E K ++NE KK ++ KKK E ++ S K+ K +D +K
Sbjct: 1158 ENEEKSETKEIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKE---SEEKKLKKNEEDRKK 1214
Query: 63 E 63
+
Sbjct: 1215 Q 1215
Score = 27.3 bits (59), Expect = 7.6
Identities = 24/77 (31%), Positives = 37/77 (47%), Gaps = 7/77 (9%)
Query: 6 VKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQ---YAKEYD-DSQ 61
V T V K +++ VK ++ES KKKR E ++ S K+ +A D D Q
Sbjct: 848 VDTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQ 907
Query: 62 K---ELIALKREAKLKG 75
K E + K++ K +G
Sbjct: 908 KGSGESVKYKKDEKKEG 924
>At1g76810 translation initiation factor IF-2 like protein
Length = 1280
Score = 32.7 bits (73), Expect = 0.18
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 1/72 (1%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
K++ K E + K R + AL ++QE E KKK E KL ++ +E ++Q
Sbjct: 397 KKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEE-EKLRKEEEERRRQEELEAQA 455
Query: 63 ELIALKREAKLK 74
E KR+ K K
Sbjct: 456 EEAKRKRKEKEK 467
Score = 27.3 bits (59), Expect = 7.6
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query: 10 VPESVLKKQ----KRNEEWALVKKQEQESAKKKRSETRKL--IWSRAKQYAKEYDDSQKE 63
+P+ V + Q +R E KK+E+E +K+ E R+ + ++A++ ++ + +KE
Sbjct: 409 IPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKE 468
Query: 64 -LIALKREAKL 73
L+ K E KL
Sbjct: 469 KLLRKKLEGKL 479
>At1g76720 putative translation initiation factor IF-2
Length = 1224
Score = 32.7 bits (73), Expect = 0.18
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 1/72 (1%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
K++ K E + K R + AL ++QE E KKK E KL ++ +E ++Q
Sbjct: 373 KKDAKGKAAEKKIPKHVREMQEALARRQEAEERKKKEEE-EKLRKEEEERRRQEELEAQA 431
Query: 63 ELIALKREAKLK 74
E KR+ K K
Sbjct: 432 EEAKRKRKEKEK 443
Score = 27.3 bits (59), Expect = 7.6
Identities = 20/71 (28%), Positives = 39/71 (54%), Gaps = 7/71 (9%)
Query: 10 VPESVLKKQ----KRNEEWALVKKQEQESAKKKRSETRKL--IWSRAKQYAKEYDDSQKE 63
+P+ V + Q +R E KK+E+E +K+ E R+ + ++A++ ++ + +KE
Sbjct: 385 IPKHVREMQEALARRQEAEERKKKEEEEKLRKEEEERRRQEELEAQAEEAKRKRKEKEKE 444
Query: 64 -LIALKREAKL 73
L+ K E KL
Sbjct: 445 KLLRKKLEGKL 455
>At1g76820 putative translation initiation factor IF-2
Length = 1146
Score = 32.3 bits (72), Expect = 0.24
Identities = 23/72 (31%), Positives = 35/72 (47%), Gaps = 1/72 (1%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
K++ K E + K R + AL ++QE + KKK E KL ++ +E D+Q
Sbjct: 283 KKDAKGKAAEKKIPKHVREIQEALARRQEAKERKKKEEE-EKLRKEEEERRRQEELDAQA 341
Query: 63 ELIALKREAKLK 74
E KR+ K K
Sbjct: 342 EEAKRKRKEKEK 353
Score = 27.3 bits (59), Expect = 7.6
Identities = 18/67 (26%), Positives = 39/67 (57%), Gaps = 6/67 (8%)
Query: 10 VPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKL--IWSRAKQYAKEYDDSQKE-LIA 66
+ E++ ++Q+ E KK+E+E +K+ E R+ + ++A++ ++ + +KE L+
Sbjct: 302 IQEALARRQEAKER---KKKEEEEKLRKEEEERRRQEELDAQAEEAKRKRKEKEKEKLLR 358
Query: 67 LKREAKL 73
K E KL
Sbjct: 359 KKLEGKL 365
>At3g50240 kinesin-related protein (kicp-02)
Length = 1051
Score = 32.0 bits (71), Expect = 0.31
Identities = 16/59 (27%), Positives = 36/59 (60%)
Query: 14 VLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAK 72
VLK+++++E+ A K E + K ++ + ++ + A+Q+ + +KEL+ LK+E +
Sbjct: 610 VLKQKQKSEDAAKRLKTEIQCIKAQKVQLQQKMKQEAEQFRQWKASQEKELLQLKKEGR 668
>At5g24880 glutamic acid-rich protein
Length = 443
Score = 31.6 bits (70), Expect = 0.40
Identities = 19/75 (25%), Positives = 39/75 (51%), Gaps = 2/75 (2%)
Query: 3 KEEVKTVVPESVL--KKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDS 60
+E+VK + E+ K ++E V++ QE ++ + E ++ + K+ K +D
Sbjct: 302 EEDVKKKIDENETPEKVDTESKEVESVEETTQEKEEEVKEEGKERVEEEEKEKEKVKEDD 361
Query: 61 QKELIALKREAKLKG 75
QKE + + + K+KG
Sbjct: 362 QKEKVEEEEKEKVKG 376
Score = 27.7 bits (60), Expect = 5.8
Identities = 19/69 (27%), Positives = 35/69 (50%), Gaps = 5/69 (7%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
KEE K V E +K+K E+ K +E+E K K E ++ + + + + +K
Sbjct: 340 KEEGKERVEEEEKEKEKVKEDDQKEKVEEEEKEKVKGDEEKEKV-----KEEESAEGKKK 394
Query: 63 ELIALKREA 71
E++ K+E+
Sbjct: 395 EVVKGKKES 403
>At5g04890 unknown protein
Length = 366
Score = 31.6 bits (70), Expect = 0.40
Identities = 19/60 (31%), Positives = 36/60 (59%), Gaps = 5/60 (8%)
Query: 15 LKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLK 74
+KKQ E+ AL++K ++E+ K+ +E RKL ++ AK +++ + + + EAK K
Sbjct: 151 MKKQLLEEKEALIRKLQEEAKAKEEAEMRKL-----QEEAKAKEEAAAKKLQEEIEAKEK 205
>At3g49450 putative protein
Length = 397
Score = 31.2 bits (69), Expect = 0.52
Identities = 20/70 (28%), Positives = 30/70 (42%), Gaps = 17/70 (24%)
Query: 178 HGIICIEDLIH---------EILTVGPHFREANNFLWP--------FKLKAPLGGMKKKR 220
HG ICI ++ E +TV H ++ N P K LGG++ K
Sbjct: 231 HGSICINGVLFYLAMKSESKEYMTVSFHMKDENFMFIPNQDLLSTLINYKGRLGGIRHKS 290
Query: 221 NHYVEGGDAG 230
+++GGD G
Sbjct: 291 FGFMDGGDVG 300
>At1g73960 unknown protein
Length = 1273
Score = 31.2 bits (69), Expect = 0.52
Identities = 23/84 (27%), Positives = 37/84 (43%), Gaps = 4/84 (4%)
Query: 1 MAKEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDS 60
+ KE+ K E KK+KR + L KK+ ++ K+K E KL+ S K+ +
Sbjct: 1168 IGKEKEKKKDKEKKEKKRKREDPVYLEKKRLKKEKKRKEKEMAKLVSSTTDPAKKKIE-- 1225
Query: 61 QKELIALKREAKLKGGFYVDPEAK 84
+ +K E G + E K
Sbjct: 1226 --SVAEVKEEEPSDGAMLIKVEPK 1247
>At1g14610 valyl tRNA synthetase (valRS)
Length = 1108
Score = 31.2 bits (69), Expect = 0.52
Identities = 28/88 (31%), Positives = 47/88 (52%), Gaps = 9/88 (10%)
Query: 1 MAKEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDS 60
M++ E K + E + +K+K+ E K +E+E K+K E +L +AKQ AK+ +
Sbjct: 45 MSESEKKILTEEELERKKKKEE-----KAKEKELKKQKALEKERLAELKAKQ-AKDGTNV 98
Query: 61 QKELI--ALKREAKLKG-GFYVDPEAKL 85
K+ + KR+A + +VDPE L
Sbjct: 99 PKKSAKKSSKRDASEENPEDFVDPETPL 126
>At5g51770 putative protein
Length = 654
Score = 30.8 bits (68), Expect = 0.69
Identities = 20/65 (30%), Positives = 30/65 (45%), Gaps = 5/65 (7%)
Query: 16 KKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQKELIALKREAKLKG 75
KK + EW L +E+E KKK+ + W +Y KE K + K++ L+
Sbjct: 383 KKSSKRLEWWLSLDEEKEKGKKKKRRMVREWWK--DEYRKEL---AKRMKKKKKKKTLES 437
Query: 76 GFYVD 80
FY D
Sbjct: 438 EFYSD 442
>At1g65470 hypothetical protein, 3' partial
Length = 618
Score = 30.8 bits (68), Expect = 0.69
Identities = 18/75 (24%), Positives = 44/75 (58%), Gaps = 2/75 (2%)
Query: 2 AKEEVKTVVPESVLKKQKRNEEWALVKKQ--EQESAKKKRSETRKLIWSRAKQYAKEYDD 59
A++E K + + + +K ++ +E L++K ++ + +K+ +E+RK I + + KE
Sbjct: 259 AEKEKKRMERQVLKEKLQQEKEQKLLQKAIVDENNKEKEETESRKRIKKQQDESEKEQKR 318
Query: 60 SQKELIALKREAKLK 74
+KE LK++ +++
Sbjct: 319 REKEQAELKKQLQVQ 333
>At1g21160 transcription factor, putative
Length = 1088
Score = 30.8 bits (68), Expect = 0.69
Identities = 21/70 (30%), Positives = 35/70 (50%), Gaps = 1/70 (1%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
K++VK V E + K R ++ L + +E E KKK E R L ++ +E + +
Sbjct: 206 KKDVKIKVAEKKVPKHVREKQETLARWKEAEDGKKKEEEER-LRKEEEERRIEEEREREA 264
Query: 63 ELIALKREAK 72
E I KR+ +
Sbjct: 265 EEIRQKRKIR 274
>At5g40340 unknown protein
Length = 1008
Score = 30.4 bits (67), Expect = 0.90
Identities = 22/82 (26%), Positives = 40/82 (47%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
KEE +T + K++KR + + + E E +K+ +E+ K R K +K+ D ++
Sbjct: 717 KEENETDKHGKMKKERKRKKSESKKEGGEGEETQKEANESTKKERKRKKSESKKQSDGEE 776
Query: 63 ELIALKREAKLKGGFYVDPEAK 84
E E+ K +PE+K
Sbjct: 777 ETQKEPSESTKKERKRKNPESK 798
Score = 27.3 bits (59), Expect = 7.6
Identities = 17/59 (28%), Positives = 30/59 (50%), Gaps = 4/59 (6%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQ 61
KE V++ E KK K +EE + ++ E KKK+ E + S+ K+ E+ ++
Sbjct: 811 KESVESTKKERKRKKPKHDEEEVPNETEKPEKKKKKKREGK----SKKKETETEFSGAE 865
>At5g22080 unknown protein
Length = 246
Score = 30.4 bits (67), Expect = 0.90
Identities = 15/73 (20%), Positives = 43/73 (58%), Gaps = 1/73 (1%)
Query: 3 KEEVKTVVPESVLKKQKRNEEWALVKKQEQESAKKKRSETRKLIWSRAKQYAKEYDDSQK 62
++E+K V E + ++ R + A+ +E+ KK +E +K IW + +++ ++++ +++
Sbjct: 145 QKELKLKVREILTDQEWRRRKMAMRISEEEGRLKKDEAE-QKEIWKKKREHEEQWEGTRE 203
Query: 63 ELIALKREAKLKG 75
+ ++ R+ + G
Sbjct: 204 KRVSSWRDFQKAG 216
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.138 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,580,603
Number of Sequences: 26719
Number of extensions: 235367
Number of successful extensions: 1257
Number of sequences better than 10.0: 78
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 1054
Number of HSP's gapped (non-prelim): 188
length of query: 244
length of database: 11,318,596
effective HSP length: 97
effective length of query: 147
effective length of database: 8,726,853
effective search space: 1282847391
effective search space used: 1282847391
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148819.1


