

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.5 + phase: 0 /pseudo
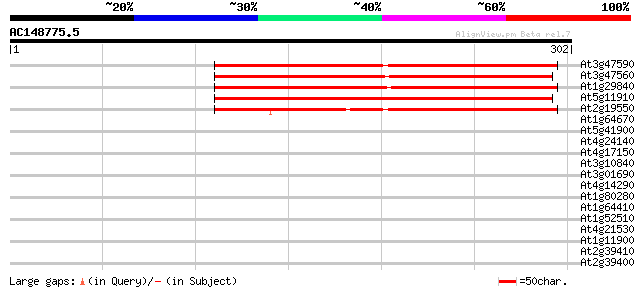
(302 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g47590 unknown protein 266 8e-72
At3g47560 unknown protein 260 6e-70
At1g29840 hypothetical protein 257 5e-69
At5g11910 putative esterase - like protein 246 1e-65
At2g19550 putative esterase 214 6e-56
At1g64670 unknown protein 40 0.002
At5g41900 unknown protein 38 0.006
At4g24140 unknown protein 37 0.013
At4g17150 unknown protein 35 0.065
At3g10840 alpha/beta hydrolase like protein 34 0.085
At3g01690 unknown protein 31 0.72
At4g14290 hypothetical protein 29 3.6
At1g80280 unknown protein 29 3.6
At1g64410 unknown protein 29 3.6
At1g52510 unknown protein (At1g52510) 29 3.6
At4g21530 hypothetical protein 28 4.7
At1g11900 hypothetical protein 28 4.7
At2g39410 putative phospholipase 28 8.0
At2g39400 phospholipase like protein 28 8.0
>At3g47590 unknown protein
Length = 309
Score = 266 bits (681), Expect = 8e-72
Identities = 126/185 (68%), Positives = 150/185 (80%), Gaps = 2/185 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+++ V+N
Sbjct: 123 ESEGSFYYGNYNHEADDLHSVVQYFSNKNRVVPIILGHSKGGDVVLLYASKYHDVRNVIN 182
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ GF DV GK YRVTE+SLMDRL T++HEACL
Sbjct: 183 LSGRYDLKKGIRERLGEDFLERIKQQGFIDV--GDGKSGYRVTEKSLMDRLSTDIHEACL 240
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE+IPV+DA EFAKIIPNHKL I+EGA+H Y HQ +L S M F
Sbjct: 241 KIDKECRVLTVHGSEDEVIPVEDAKEFAKIIPNHKLEIVEGANHGYTEHQSQLVSTVMEF 300
Query: 291 IKETI 295
IK I
Sbjct: 301 IKTVI 305
>At3g47560 unknown protein
Length = 265
Score = 260 bits (665), Expect = 6e-70
Identities = 125/182 (68%), Positives = 147/182 (80%), Gaps = 2/182 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ESEGSF YGNY E DDLH+V Q+F NRV+ I+GHSKGGDVVLLYASKYH+I V+N
Sbjct: 74 ESEGSFYYGNYNYEADDLHSVIQYFSNLNRVVTIILGHSKGGDVVLLYASKYHDIPNVIN 133
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+D+LERI++ G+ DVK G YRVTEESLMDRL T+MHEACL
Sbjct: 134 LSGRYDLKKGIGERLGEDFLERIKQQGYIDVK--DGDSGYRVTEESLMDRLNTDMHEACL 191
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS DE +PV+DA EFAKIIPNH+L I+EGADH Y N+Q +L M F
Sbjct: 192 KIDKECRVLTVHGSGDETVPVEDAKEFAKIIPNHELQIVEGADHCYTNYQSQLVLTVMEF 251
Query: 291 IK 292
IK
Sbjct: 252 IK 253
>At1g29840 hypothetical protein
Length = 263
Score = 257 bits (657), Expect = 5e-69
Identities = 121/185 (65%), Positives = 150/185 (80%), Gaps = 2/185 (1%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF +GNY E DDLH+V ++F NRV+ I+GHSKGGDVVL+YASKY +I+ V+N
Sbjct: 77 ESKGSFYFGNYNYEADDLHSVIRYFTNMNRVVPIIIGHSKGGDVVLVYASKYQDIRNVIN 136
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
LSGRYDLK GI ERLG+DYLERI++ GF D+K G +RVTEESLM+RL T+MHEACL
Sbjct: 137 LSGRYDLKRGIGERLGEDYLERIKQQGFIDIKE--GNAGFRVTEESLMERLNTDMHEACL 194
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+IDK+CR+LT+HGS+DE+IP++DA EFAKIIPNHKL I+EGADH Y HQ +L + M F
Sbjct: 195 KIDKECRVLTVHGSADEVIPLEDAKEFAKIIPNHKLEIVEGADHCYTKHQSQLITNVMEF 254
Query: 291 IKETI 295
IK I
Sbjct: 255 IKTVI 259
>At5g11910 putative esterase - like protein
Length = 297
Score = 246 bits (628), Expect = 1e-65
Identities = 115/182 (63%), Positives = 148/182 (81%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
ES+GSF+YGNY +EV+DL +V QH R NRVI AI+GHSKGG+VVLLYA+KY++++TVVN
Sbjct: 86 ESQGSFQYGNYRREVEDLRSVLQHLRGVNRVISAIIGHSKGGNVVLLYAAKYNDVQTVVN 145
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
+SGR+ L GIE RLGKDY +RI+ +GF DV GK +YRVTEESLMDRL TN HEACL
Sbjct: 146 ISGRFFLDRGIEFRLGKDYFKRIKDNGFIDVSNRKGKFEYRVTEESLMDRLTTNAHEACL 205
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
I ++CR+LT+HGS+D I+ V +A EFAK I NHKL++IEGADH + +HQ +L+S+ +SF
Sbjct: 206 SIRENCRVLTVHGSNDRIVHVTEASEFAKQIKNHKLYVIEGADHEFTSHQHQLASIVLSF 265
Query: 291 IK 292
K
Sbjct: 266 FK 267
>At2g19550 putative esterase
Length = 332
Score = 214 bits (544), Expect = 6e-56
Identities = 108/190 (56%), Positives = 140/190 (72%), Gaps = 9/190 (4%)
Query: 111 ESEGSFEYGNYWKEV-DDLHAVAQHFRESN---RVIRAIVGHSKGGDVVLLYASKYHE-I 165
+SEG+F YGN+ E DDLH V QH SN R++ I+GHSKGGDVVLLYASK+ + I
Sbjct: 68 DSEGTFYYGNFNSEAEDDLHYVIQHLSSSNIMNRLVPVILGHSKGGDVVLLYASKFPDYI 127
Query: 166 KTVVNLSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNM 225
+ VVN+SGR+DLK + RLG Y+E+I++ GF D + GK +RVT+ESLMDRL T+M
Sbjct: 128 RNVVNISGRFDLKNDV--RLGDGYIEKIKEQGFIDA--TEGKSCFRVTQESLMDRLNTDM 183
Query: 226 HEACLQIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSS 285
H+ACL IDK C++LT+HGS D ++P +DA EFAK+IPNHKL I+EGA+H Y HQ EL S
Sbjct: 184 HQACLNIDKQCKVLTVHGSDDTVVPGEDAKEFAKVIPNHKLEIVEGANHGYTKHQKELVS 243
Query: 286 VFMSFIKETI 295
+ + F K I
Sbjct: 244 IAVEFTKTAI 253
>At1g64670 unknown protein
Length = 469
Score = 40.0 bits (92), Expect = 0.002
Identities = 13/40 (32%), Positives = 24/40 (59%)
Query: 235 DCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADH 274
DC + HG DE+IPV+ ++ + +P ++H++ DH
Sbjct: 399 DCEVAVFHGGRDELIPVECSYGVKRKVPRARIHVVPDKDH 438
>At5g41900 unknown protein
Length = 471
Score = 38.1 bits (87), Expect = 0.006
Identities = 14/40 (35%), Positives = 22/40 (55%)
Query: 235 DCRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADH 274
DC + HG DE+IPV+ ++ +P +H+I DH
Sbjct: 407 DCDVAIFHGGKDELIPVECSYSVKSKVPRATVHVIPDKDH 446
>At4g24140 unknown protein
Length = 498
Score = 37.0 bits (84), Expect = 0.013
Identities = 14/39 (35%), Positives = 23/39 (58%)
Query: 236 CRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADH 274
C + HG DE+IPV+ ++ + IP ++ +IE DH
Sbjct: 436 CNVTIFHGGDDELIPVECSYNVKQRIPRARVKVIEHKDH 474
>At4g17150 unknown protein
Length = 502
Score = 34.7 bits (78), Expect = 0.065
Identities = 43/172 (25%), Positives = 68/172 (39%), Gaps = 11/172 (6%)
Query: 122 WKEVDDLHAVAQHFRESNRVIR-AIVGHSKGGDVVLLYASKYHEIKTVVNLSGRYDLKAG 180
W E DDL V + R SN+V R + G S G LLY ++ I +V L +
Sbjct: 125 WHEKDDLKTVVSYLRNSNQVSRIGLWGRSMGAVTSLLYGAEDPSIAGMV-LDSAFSNLFD 183
Query: 181 IEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQIDKDCRILT 240
+ L Y R+ K F VK + + + +++ + + N C+++ I
Sbjct: 184 LMMELVDVYKIRLPK---FTVKVAVQYMRRIIQKKAKFNIMDLN----CVKVSPKTFIPA 236
Query: 241 I--HGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQDELSSVFMSF 290
+ H S D+ I + K K I DH + Q SV + F
Sbjct: 237 LFGHASGDKFIQPHHSDLILKCYAGDKNIIKFDGDHNSSRPQSYYDSVLVFF 288
>At3g10840 alpha/beta hydrolase like protein
Length = 429
Score = 34.3 bits (77), Expect = 0.085
Identities = 16/57 (28%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Query: 236 CRILTIHGSSDEIIPVQDAHEFAKIIPNHKLHIIEGADHAYNNHQ-DELSSVFMSFI 291
C +L + G +D I+P +A A+ IP +I+ H + DE S+ F+
Sbjct: 352 CPVLIVTGDTDRIVPAWNAERLARAIPGSVFEVIKKCGHLPQEEKPDEFISIVAKFL 408
>At3g01690 unknown protein
Length = 361
Score = 31.2 bits (69), Expect = 0.72
Identities = 34/182 (18%), Positives = 72/182 (38%), Gaps = 33/182 (18%)
Query: 111 ESEGSFEYGNYWKEVDDLHAVAQHFRESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVVN 170
+S G N + +++ ++ + S + + G S G L AS+ +++ VV
Sbjct: 109 QSTGKPSEHNTYADIEAVYKCLEETFGSKQEGVILYGQSVGSGPTLDLASRLPQLRAVVL 168
Query: 171 LSGRYDLKAGIEERLGKDYLERIRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACL 230
S + +G+ + ++K +FD+ ++ K+ Y
Sbjct: 169 HS---PILSGLR------VMYSVKKTYWFDIYKNIDKIPY-------------------- 199
Query: 231 QIDKDCRILTIHGSSDEIIPVQDAHEFAKIIPN-HKLHIIEGADHAYNNHQDELSSVFMS 289
DC +L IHG+SDE++ + ++ + ++ ++G +H H E
Sbjct: 200 ---VDCPVLIIHGTSDEVVDCSHGKQLWELCKDKYEPLWVKGGNHCDLEHYPEYIRHLKK 256
Query: 290 FI 291
FI
Sbjct: 257 FI 258
>At4g14290 hypothetical protein
Length = 505
Score = 28.9 bits (63), Expect = 3.6
Identities = 16/49 (32%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 122 WKEVDDLHAVAQHFR-ESNRVIRAIVGHSKGGDVVLLYASKYHEIKTVV 169
W E DDL AV ++ R + N + + G S G L+Y ++ I +V
Sbjct: 163 WNEKDDLKAVVEYLRTDGNVSLIGLWGRSMGAVTSLMYGAEDPSIAAMV 211
>At1g80280 unknown protein
Length = 647
Score = 28.9 bits (63), Expect = 3.6
Identities = 35/148 (23%), Positives = 62/148 (41%), Gaps = 23/148 (15%)
Query: 145 IVGHSKGGDVVLLYASKYHEIKTVVNLSGRYDLKAGIEER-------------LGKDYLE 191
+VGH GG + L A + E K + + G L + LGK +L
Sbjct: 453 LVGHDDGGLLALKAAQRLLETKDPIKVKGVVLLNVSLTREVVPAFARILLHTSLGKKHLV 512
Query: 192 R--IRKDGFFDVKRSSGKLDYRVTEESLMDRLGTNMHEACLQIDKDCRILTIHG--SSDE 247
R +R + V R + ++T + L +++A L ++ L G SS+
Sbjct: 513 RPLLRTEIAQVVNRRAWYDPAKMTTDVL------RLYKAPLHVEGWDEALHEIGRLSSEM 566
Query: 248 IIPVQDAHEFAKIIPNHKLHIIEGADHA 275
++P Q+A K + N + ++ GA+ A
Sbjct: 567 VLPTQNALSLLKAVENLPVLVVAGAEDA 594
>At1g64410 unknown protein
Length = 753
Score = 28.9 bits (63), Expect = 3.6
Identities = 18/70 (25%), Positives = 38/70 (53%), Gaps = 9/70 (12%)
Query: 117 EYGNYWKEV-----DDLHAVAQHFRES-NRVIRAIV---GHSKGGDVVLLYASKYHEIKT 167
++G KE D+ H +++H ES +R +R I+ G G V+++ + ++ +
Sbjct: 615 DFGELVKEANLIIWDEAHMMSKHCFESLDRTLRDIMNNPGDKPFGGKVIVFGGDFRQVLS 674
Query: 168 VVNLSGRYDL 177
V+N +GR ++
Sbjct: 675 VINGAGREEI 684
>At1g52510 unknown protein (At1g52510)
Length = 380
Score = 28.9 bits (63), Expect = 3.6
Identities = 16/33 (48%), Positives = 20/33 (60%), Gaps = 1/33 (3%)
Query: 243 GSSDEIIPVQDAHEFAKIIP-NHKLHIIEGADH 274
G +D+ +P A EF K P N KL +IEGA H
Sbjct: 330 GIADKYLPQSIAEEFEKQNPQNVKLRLIEGAGH 362
>At4g21530 hypothetical protein
Length = 493
Score = 28.5 bits (62), Expect = 4.7
Identities = 16/54 (29%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Query: 121 YWKEVDDLHAVAQ---HFRESNRVIRA--IVGHSKGGDVVLLYASKYHEIKTVV 169
+WK +LH VAQ + + VIRA V + + D + + K+H + T++
Sbjct: 233 FWKRKYELHQVAQQASNIEDLTEVIRASLSVMNKQWADAMKTFHEKFHSLSTLI 286
>At1g11900 hypothetical protein
Length = 364
Score = 28.5 bits (62), Expect = 4.7
Identities = 14/51 (27%), Positives = 26/51 (50%), Gaps = 3/51 (5%)
Query: 137 ESNRVIRAIVGHSKGGDVVLL---YASKYHEIKTVVNLSGRYDLKAGIEER 184
+ +++ IV HS+ G ++ Y + + NLSG YDL ++E+
Sbjct: 46 DEEELLKKIVNHSESGSKIISKIDYTNLVEKFTRDGNLSGAYDLLQSLQEK 96
>At2g39410 putative phospholipase
Length = 311
Score = 27.7 bits (60), Expect = 8.0
Identities = 22/55 (40%), Positives = 31/55 (56%), Gaps = 4/55 (7%)
Query: 111 ESEGSFEY-GNYWKEVDDL--HAVAQHFRESNR-VIRAIVGHSKGGDVVLLYASK 161
+S+G Y N+ + VDD+ H A RE N+ +R ++G S GG VVLL K
Sbjct: 71 KSDGLSAYISNFDRLVDDVSTHYTAICEREENKWKMRFMLGESMGGAVVLLLGRK 125
>At2g39400 phospholipase like protein
Length = 311
Score = 27.7 bits (60), Expect = 8.0
Identities = 24/58 (41%), Positives = 32/58 (54%), Gaps = 10/58 (17%)
Query: 111 ESEGSFEY-GNYWKEVDDLHAVAQHF-----RESNR-VIRAIVGHSKGGDVVLLYASK 161
+SEG Y N+ VDD V+ H+ RE N+ +R ++G S GG VVLL A K
Sbjct: 71 KSEGLNGYISNFDDLVDD---VSNHYSTICEREENKGKMRFLLGESMGGAVVLLLARK 125
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.337 0.147 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,565,683
Number of Sequences: 26719
Number of extensions: 280686
Number of successful extensions: 937
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 919
Number of HSP's gapped (non-prelim): 19
length of query: 302
length of database: 11,318,596
effective HSP length: 99
effective length of query: 203
effective length of database: 8,673,415
effective search space: 1760703245
effective search space used: 1760703245
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148775.5


