

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.4 - phase: 0
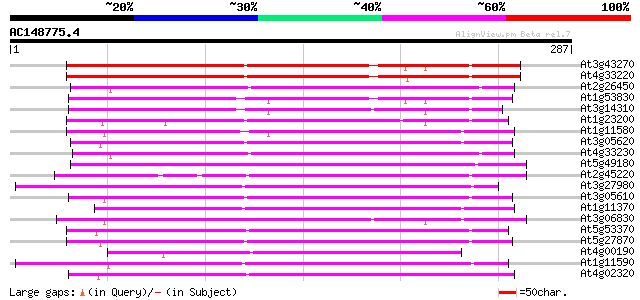
(287 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g43270 pectinesterase like protein 209 1e-54
At4g33220 pectinesterase like protein 194 6e-50
At2g26450 putative pectinesterase 165 3e-41
At1g53830 unknown protein 162 2e-40
At3g14310 pectin methylesterase like protein 159 2e-39
At1g23200 putative pectinesterase 156 1e-38
At1g11580 unknown protein 156 1e-38
At3g05620 putative pectinesterase 155 2e-38
At4g33230 pectinesterase - like protein 154 5e-38
At5g49180 pectin methylesterase 152 2e-37
At2g45220 pectinesterase like protein 152 3e-37
At3g27980 pectin methylesterase, putative 150 1e-36
At3g05610 putative pectinesterase 146 1e-35
At1g11370 similar to flower-specific pectin methylesterase precu... 146 1e-35
At3g06830 pectin methylesterase like protein 144 5e-35
At5g53370 pectinesterase 144 7e-35
At5g27870 pectin methyl-esterase - like protein 143 1e-34
At4g00190 putative pectinesterase 143 1e-34
At1g11590 putative pectin methylesterase 143 1e-34
At4g02320 pectinesterase - like protein 142 2e-34
>At3g43270 pectinesterase like protein
Length = 527
Score = 209 bits (533), Expect = 1e-54
Identities = 121/238 (50%), Positives = 158/238 (65%), Gaps = 12/238 (5%)
Query: 30 KFPSWVKPGDRKLLQASAVP-ADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
KFPSWVKPGDRKLLQ + ADAVVA+DG+GN+ I DAV+AAP+ S KRYVIH+K+GV
Sbjct: 192 KFPSWVKPGDRKLLQTDNITVADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGV 251
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E+V I K N+MM+GDG+ ATVITG+ S+ D T + TF V G GF A+DI+F+
Sbjct: 252 YVENVEIKKKKWNIMMVGDGIDATVITGNRSF-IDGWTTFRSATFAVSGRGFIARDITFQ 310
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLT---SYLV 205
NTA PE HQAVA+ SD+D VFYRC + G+QD+L A HS+R E +T ++
Sbjct: 311 NTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYA----HSMRQFFRECIITGTVDFIF 366
Query: 206 RQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
++ I ++G Q+N+ITAQG + PN P GF QF N+ AD + L +N
Sbjct: 367 GDATAVFQSCQIKAKQGLPNQKNSITAQGRKD-PNEPTGFTIQFSNIAADTDLLLNLN 423
>At4g33220 pectinesterase like protein
Length = 404
Score = 194 bits (492), Expect = 6e-50
Identities = 109/237 (45%), Positives = 152/237 (63%), Gaps = 11/237 (4%)
Query: 30 KFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
+FP WV+P DRKLL+++ D VA DG+GN+ KIMDA+ AP+ S R+VI+IKKG+Y
Sbjct: 70 QFPDWVRPDDRKLLESNGRTYDVSVALDGTGNFTKIMDAIKKAPDYSSTRFVIYIKKGLY 129
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRN 149
E+V I K N++M+GDG+ TVI+G+ S+ D T + TF V G GF A+DI+F+N
Sbjct: 130 LENVEIKKKKWNIVMLGDGIDVTVISGNRSF-IDGWTTFRSATFAVSGRGFLARDITFQN 188
Query: 150 TAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS-----YL 204
TA PE HQAVAL SDSD SVF+RC + G+QD+L H++R E +T +
Sbjct: 189 TAGPEKHQAVALRSDSDLSVFFRCAMRGYQDTLYT----HTMRQFYRECTITGTVDFIFG 244
Query: 205 VRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVN 261
+ IL ++G Q+NTITAQG + N P GF+ QF N+ AD + +P++N
Sbjct: 245 DGTVVFQNCQILAKRGLPNQKNTITAQGRKD-VNQPSGFSIQFSNISADADLVPYLN 300
>At2g26450 putative pectinesterase
Length = 496
Score = 165 bits (417), Expect = 3e-41
Identities = 93/231 (40%), Positives = 134/231 (57%), Gaps = 6/231 (2%)
Query: 32 PSWVKPGDRKLLQASAVPA---DAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
PSWV DR++L+A V A +A VA DGSG++ I DA+ A P + RY+I++K+G+
Sbjct: 161 PSWVSNDDRRMLRAVDVKALKPNATVAKDGSGDFTTINDALRAMPEKYEGRYIIYVKQGI 220
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y+E+V ++ K+NL M+GDG T++TG+ S + K+ T T TF +G GF AQ + FR
Sbjct: 221 YDEYVTVDKKKANLTMVGDGSQKTIVTGNKSHAK-KIRTFLTATFVAQGEGFMAQSMGFR 279
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
NTA PE HQAVA+ SD S+F C G+QD+L A R +
Sbjct: 280 NTAGPEGHQAVAIRVQSDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTIDFIFGDAA 339
Query: 209 SSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
+ F+ +I +RKG GQ+NT+TAQG +K GF C + A+ + P
Sbjct: 340 AIFQNCNIFIRKGLPGQKNTVTAQGRVDKFQTT-GFVVHNCKIAANEDLKP 389
>At1g53830 unknown protein
Length = 587
Score = 162 bits (410), Expect = 2e-40
Identities = 97/235 (41%), Positives = 135/235 (57%), Gaps = 17/235 (7%)
Query: 31 FPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYN 90
+P W+ GDR+LLQ S + ADA VA DGSG++ + AV AAP S KR+VIHIK GVY
Sbjct: 256 WPKWLSVGDRRLLQGSTIKADATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHIKAGVYR 315
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDISF 147
E+V + K+N+M +GDG G T+ITG R+ +D S T+ T G F A+DI+F
Sbjct: 316 ENVEVTKKKTNIMFLGDGRGKTIITG----SRNVVDGSTTFHSATVAAVGERFLARDITF 371
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLT---SYL 204
+NTA P HQAVAL SD S FY+C++ +QD+L HS R + +T ++
Sbjct: 372 QNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYV----HSNRQFFVKCHITGTVDFI 427
Query: 205 VRQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
++ DI R+ +GQ+N +TAQG + PN G Q C + + L
Sbjct: 428 FGNAAAVLQDCDINARRPNSGQKNMVTAQGRSD-PNQNTGIVIQNCRIGGTSDLL 481
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 159 bits (402), Expect = 2e-39
Identities = 95/227 (41%), Positives = 129/227 (55%), Gaps = 11/227 (4%)
Query: 31 FPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYN 90
+P+W+ GDR+LLQ S V ADA VA+DGSG + + AV AAP S KRYVIHIK GVY
Sbjct: 261 WPTWLSAGDRRLLQGSGVKADATVAADGSGTFKTVAAAVAAAPENSNKRYVIHIKAGVYR 320
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSAQDISF 147
E+V + K N+M +GDG T+ITG R+ +D S T+ T G F A+DI+F
Sbjct: 321 ENVEVAKKKKNIMFMGDGRTRTIITG----SRNVVDGSTTFHSATVAAVGERFLARDITF 376
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQ 207
+NTA P HQAVAL SD S FY C++ +QD+L + + + A ++
Sbjct: 377 QNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVH-SNRQFFVKCLIAGTVDFIFGN 435
Query: 208 LSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCA 252
+ DI R+ +GQ+N +TAQG + PN G Q C + A
Sbjct: 436 AAVVLQDCDIHARRPNSGQKNMVTAQGRTD-PNQNTGIVIQKCRIGA 481
>At1g23200 putative pectinesterase
Length = 554
Score = 156 bits (395), Expect = 1e-38
Identities = 100/232 (43%), Positives = 132/232 (56%), Gaps = 9/232 (3%)
Query: 30 KFPSWVKPGDRKLLQAS--AVPADAVVASDGSGNYMKIMDAVMAAPNGSKK--RYVIHIK 85
KFPSW DRKLL+ S AD VVA DGSG+Y I AV AA ++ R VI++K
Sbjct: 227 KFPSWFPLSDRKLLEDSKTTAKADLVVAKDGSGHYTSIQQAVNAAAKLPRRNQRLVIYVK 286
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDI 145
GVY E+V+I S N+M+IGDG+ +T++TG+ + +D T + TF V G GF AQ I
Sbjct: 287 AGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNV-QDGTTTFRSATFAVSGNGFIAQGI 345
Query: 146 SFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLV 205
+F NTA PE HQAVAL S SD SVFY C G+QD+L + +R + ++
Sbjct: 346 TFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLHSSRQFLRNCNIYGTV-DFIF 404
Query: 206 RQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
++ +I RK +GQ+NTITAQ E P+ GF Q V E
Sbjct: 405 GDATAILQNCNIYARKPMSGQKNTITAQSRKE-PDETTGFVIQSSTVATASE 455
>At1g11580 unknown protein
Length = 557
Score = 156 bits (394), Expect = 1e-38
Identities = 96/237 (40%), Positives = 129/237 (53%), Gaps = 13/237 (5%)
Query: 30 KFPSWVKPGDRKLLQASA----VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIK 85
+FPSW+ DRKLL++S V A+ VVA DG+G + + +AV AAP S RYVI++K
Sbjct: 221 RFPSWLTALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVK 280
Query: 86 KGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTY---TFGVEGLGFSA 142
KGVY E + I K NLM++GDG AT+ITG L + +D S T+ T G GF A
Sbjct: 281 KGVYKETIDIGKKKKNLMLVGDGKDATIITGSL----NVIDGSTTFRSATVAANGDGFMA 336
Query: 143 QDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTS 202
QDI F+NTA P HQAVAL +D +V RC I +QD+L + R + +
Sbjct: 337 QDIWFQNTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDF 396
Query: 203 YLVRQLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F+ DI+ R GQ+N +TAQ G E N + Q C + A + P
Sbjct: 397 IFGNSAVVFQNCDIVARNPGAGQKNMLTAQ-GREDQNQNTAISIQKCKITASSDLAP 452
>At3g05620 putative pectinesterase
Length = 543
Score = 155 bits (392), Expect = 2e-38
Identities = 95/231 (41%), Positives = 130/231 (56%), Gaps = 7/231 (3%)
Query: 32 PSWVKPGDRKLLQA---SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
P W+ D L+ S + + VVA DG G Y I +A+ APN S KRYVI++KKGV
Sbjct: 218 PEWLTETDESLMMRHDPSVMHPNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIYVKKGV 277
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E++ + K+N+M++GDG+G T+ITGD ++ L T T T V G GF A+DI+FR
Sbjct: 278 YKENIDLKKKKTNIMLVGDGIGQTIITGDRNF-MQGLTTFRTATVAVSGRGFIAKDITFR 336
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
NTA P+N QAVAL DSD S FYRC + G+QD+L A+ R + +
Sbjct: 337 NTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIFGNGA 396
Query: 209 SSFKTDILVRKGPTG-QQNTITAQGGPEKPNLPFGFAFQFCNVCA-DPEFL 257
+ + + + P Q+ TITAQ G + PN GF Q V A P +L
Sbjct: 397 AVLQNCKIYTRVPLPLQKVTITAQ-GRKSPNQNTGFVIQNSYVLATQPTYL 446
>At4g33230 pectinesterase - like protein
Length = 609
Score = 154 bits (389), Expect = 5e-38
Identities = 93/230 (40%), Positives = 128/230 (55%), Gaps = 6/230 (2%)
Query: 33 SWVKPGDRKLLQASAVPA---DAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVY 89
SW+ +R++L+A V A +A VA DGSGN+ I A+ A P + RY I+IK G+Y
Sbjct: 275 SWLSNKERRMLKAVDVKALKPNATVAKDGSGNFTTINAALKAMPAKYQGRYTIYIKHGIY 334
Query: 90 NEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRN 149
+E V+I+ K N+ M+GDG T++TG+ S + K+ T T TF +G GF AQ + FRN
Sbjct: 335 DESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAK-KIRTFLTATFVAQGEGFMAQSMGFRN 393
Query: 150 TAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLS 209
TA PE HQAVA+ SD SVF C G+QD+L A R + +
Sbjct: 394 TAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLYAYTHRQYYRSCVIIGTVDFIFGDAAA 453
Query: 210 SFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F+ DI +RKG GQ+NT+TAQG +K GF C V + + P
Sbjct: 454 IFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTT-GFVIHNCTVAPNEDLKP 502
>At5g49180 pectin methylesterase
Length = 571
Score = 152 bits (384), Expect = 2e-37
Identities = 89/235 (37%), Positives = 129/235 (54%), Gaps = 3/235 (1%)
Query: 32 PSWVKPGDRKLLQASA-VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYN 90
PSWV P R+L+ V A+ VVA DGSG Y I +A+ A P ++K +VI+IK+GVYN
Sbjct: 239 PSWVGPNTRRLMATKGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYIKQGVYN 298
Query: 91 EHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNT 150
E V + +++ IGDG T ITG L++ K+ T T T + G F+A++I F NT
Sbjct: 299 EKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAKNIGFENT 358
Query: 151 AWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSS 210
A PE HQAVAL +D +VFY C+I G+QD+L + R + +
Sbjct: 359 AGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIFGDGIVV 418
Query: 211 FKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVNLPK 264
+ +I+VRK Q ITAQG +K G Q C++ +P ++P ++ K
Sbjct: 419 LQNCNIVVRKPMKSQSCMITAQGRSDKRE-STGLVLQNCHITGEPAYIPVKSINK 472
>At2g45220 pectinesterase like protein
Length = 511
Score = 152 bits (383), Expect = 3e-37
Identities = 97/241 (40%), Positives = 130/241 (53%), Gaps = 6/241 (2%)
Query: 24 TITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIH 83
T +K FPSWVKPGDRKLLQ+S +AVVA DGSGN+ I +A+ AA R+VI+
Sbjct: 179 TPPEKDGFPSWVKPGDRKLLQSSTPKDNAVVAKDGSGNFKTIKEAIDAASGSG--RFVIY 236
Query: 84 IKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQ 143
+K+GVY+E++ I K N+M+ GDG+G T+ITG S G T + T G GF A+
Sbjct: 237 VKQGVYSENLEIR--KKNVMLRGDGIGKTIITGSKSVG-GGTTTFNSATVAAVGDGFIAR 293
Query: 144 DISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSY 203
I+FRNTA N QAVAL S SD SVFY+C +QD+L + R +
Sbjct: 294 GITFRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVYGTVDFI 353
Query: 204 LVRQLSSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLPFVNLP 263
+ + + + P + NTITAQG + PN G V A + P +
Sbjct: 354 FGNAAAVLQNCNIFARRPRSKTNTITAQGRSD-PNQNTGIIIHNSRVTAASDLRPVLGST 412
Query: 264 K 264
K
Sbjct: 413 K 413
>At3g27980 pectin methylesterase, putative
Length = 497
Score = 150 bits (378), Expect = 1e-36
Identities = 96/248 (38%), Positives = 131/248 (52%), Gaps = 3/248 (1%)
Query: 4 SRIFFLFCFFFRTSSSDILDTITQKGKFPSWVKPGDRKLLQASAVPADAVVASDGSGNYM 63
SR + F S D + I+ PSW+ D+K L +A AD VVA DG+G Y
Sbjct: 141 SRARVVLALFISISLRDNTELISVIPNGPSWLFHVDKKDLYLNAEIADVVVAKDGTGKYS 200
Query: 64 KIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRD 123
+ A+ AAP S+KR+VI+IK G+Y+E V+I N+K NL +IGDG T+ITG+LS +
Sbjct: 201 TVNAAIAAAPQHSQKRFVIYIKTGIYDEIVVIENTKPNLTLIGDGQDLTIITGNLS-ASN 259
Query: 124 KLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLC 183
T T T G GF D+ FRNTA P AVAL D SV YRC + G+QD+L
Sbjct: 260 VRRTYNTATVASNGNGFIGVDMCFRNTAGPAKGPAVALRVSGDMSVIYRCRVEGYQDALY 319
Query: 184 ANIKHHSIRIAKSEARLTSYLVRQLSSFK-TDILVRKGPTGQQNTITAQGGPEKPNLPFG 242
+ R + ++ F+ I+ R+ GQ N ITAQ K ++ G
Sbjct: 320 PHSDRQFYRECFITGTVDFICGNAVAVFQFCQIVARQPKMGQSNVITAQSRATK-DVKSG 378
Query: 243 FAFQFCNV 250
F+ Q CN+
Sbjct: 379 FSIQNCNI 386
>At3g05610 putative pectinesterase
Length = 669
Score = 146 bits (369), Expect = 1e-35
Identities = 88/232 (37%), Positives = 124/232 (52%), Gaps = 7/232 (3%)
Query: 31 FPSWVKPGDRKLLQASA----VPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKK 86
FPSWV RKLLQA+A V D VVA DGSG Y I +A+ P +V+HIK
Sbjct: 231 FPSWVDQRGRKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKA 290
Query: 87 GVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
G+Y E+V +N + S+L+ IGDG T+I+G+ ++ +D + T T T + G F A++I
Sbjct: 291 GLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNY-KDGITTYRTATVAIVGNYFIAKNIG 349
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F NTA HQAVA+ SD S+F+ C G+QD+L + R +
Sbjct: 350 FENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGD 409
Query: 207 QLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
+ F+ +LVRK Q ITA G + P GF FQ C + +P++L
Sbjct: 410 AAAVFQNCTLLVRKPLPNQACPITAHGRKD-PRESTGFVFQGCTIAGEPDYL 460
>At1g11370 similar to flower-specific pectin methylesterase
precursor gi|3342904; similar to ESTs gb|H36111,
gb|T76576, gb|AI099976, gb|T42118
Length = 288
Score = 146 bits (368), Expect = 1e-35
Identities = 87/216 (40%), Positives = 119/216 (54%), Gaps = 3/216 (1%)
Query: 44 QASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLM 103
QA AD +VA DGSGN+ + +AV AAP K +VI+IK+G+Y E + I K+NL
Sbjct: 38 QALKDKADLIVAKDGSGNFTTVNEAVAAAPENGVKPFVIYIKEGLYKEVIRIGKKKTNLT 97
Query: 104 MIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLS 163
++GDG TV++GDL+ G D + T + T V+ GF AQD+ RNTA PE QAVAL
Sbjct: 98 LVGDGRDLTVLSGDLN-GVDGIKTFDSATLAVDESGFMAQDLCIRNTAGPEKRQAVALRI 156
Query: 164 DSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFK-TDILVRKGPT 222
+D ++ YRC I +QD+L A R + R + F+ I RK
Sbjct: 157 STDMTIIYRCRIDAYQDTLYAYSGRQFYRDCYITGTVDFIFGRAAAVFQYCQIEARKPGI 216
Query: 223 GQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
GQ N +TAQ E+ GF+FQ CN+ A + P
Sbjct: 217 GQTNILTAQ-SREEDTATSGFSFQKCNISASSDLTP 251
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 144 bits (363), Expect = 5e-35
Identities = 86/247 (34%), Positives = 130/247 (51%), Gaps = 9/247 (3%)
Query: 25 ITQKGKFPSWVKPGDRKLLQASA-----VPADAVVASDGSGNYMKIMDAVMAAPNGSKKR 79
++ + P+WV P R+L+ A V A+AVVA DG+G + I DA+ A P G+K
Sbjct: 226 LSTEDSIPTWVGPEARRLMAAQGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGNKVP 285
Query: 80 YVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLG 139
++IHIK+G+Y E V + ++ IGDG T+ITG L++G K+ T T T +EG
Sbjct: 286 FIIHIKEGIYKEKVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGDH 345
Query: 140 FSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEAR 199
F+A++I NTA PE QAVAL +D +VF+ C+I G QD+L + H + +
Sbjct: 346 FTAKNIGIENTAGPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVH-SHRQFYRDCTVSG 404
Query: 200 LTSYLVRQLSSF--KTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
++ I+VRK GQ +TAQ G G C++ DP ++
Sbjct: 405 TVDFIFGDAKCILQNCKIVVRKPNKGQTCMVTAQ-GRSNVRESTGLVLHGCHITGDPAYI 463
Query: 258 PFVNLPK 264
P ++ K
Sbjct: 464 PMKSVNK 470
>At5g53370 pectinesterase
Length = 587
Score = 144 bits (362), Expect = 7e-35
Identities = 89/230 (38%), Positives = 124/230 (53%), Gaps = 6/230 (2%)
Query: 30 KFPSWVKPGDRKLL--QASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKG 87
+ P+W+K DR+LL SA+ AD V+ DGSG + I +A+ AP S +R+VI++K G
Sbjct: 250 ELPNWLKREDRELLGTPTSAIQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAG 309
Query: 88 VYNE-HVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDIS 146
Y E ++ + K+NLM IGDG G TVITG S D L T +T TF G GF +D++
Sbjct: 310 RYEEENLKVGRKKTNLMFIGDGKGKTVITGGKSIA-DDLTTFHTATFAATGAGFIVRDMT 368
Query: 147 FRNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVR 206
F N A P HQAVAL D +V YRC I G+QD+L + R + +
Sbjct: 369 FENYAGPAKHQAVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGN 428
Query: 207 QLSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPE 255
++ +I RK Q+ TITAQ + PN G + C + A P+
Sbjct: 429 AAVILQSCNIYARKPMAQQKITITAQNRKD-PNQNTGISIHACKLLATPD 477
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 143 bits (360), Expect = 1e-34
Identities = 87/231 (37%), Positives = 125/231 (53%), Gaps = 5/231 (2%)
Query: 30 KFPSWVKPGDRKLLQA--SAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKG 87
+FPSW+ R+LL A S V D VVA DGSG Y I +A+ P +V+HIK+G
Sbjct: 229 EFPSWMDARARRLLNAPMSEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEG 288
Query: 88 VYNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISF 147
+Y E+V +N S ++L+ IGDG TVI+G S+ +D + T T T + G F A++I+F
Sbjct: 289 IYKEYVQVNRSMTHLVFIGDGPDKTVISGSKSY-KDGITTYKTATVAIVGDHFIAKNIAF 347
Query: 148 RNTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQ 207
NTA HQAVA+ +D S+FY C+ G+QD+L A+ R +
Sbjct: 348 ENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDA 407
Query: 208 LSSFKT-DILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFL 257
+ F+ +LVRK Q ITA G + P GF Q C + +P++L
Sbjct: 408 AAVFQNCTLLVRKPLLNQACPITAHGRKD-PRESTGFVLQGCTIVGEPDYL 457
>At4g00190 putative pectinesterase
Length = 474
Score = 143 bits (360), Expect = 1e-34
Identities = 83/183 (45%), Positives = 106/183 (57%), Gaps = 3/183 (1%)
Query: 51 DAVVASDGSGNYMKIMDAVMAAPNGSK--KRYVIHIKKGVYNEHVMINNSKSNLMMIGDG 108
D VVA DGSG+Y I +AV A K RYVIH+K+GVY E+V + +N+M+ GDG
Sbjct: 164 DVVVAQDGSGDYKTIQEAVNGAGERLKGSPRYVIHVKQGVYEEYVNVGIKSNNIMITGDG 223
Query: 109 MGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTS 168
+G T+ITGD S GR T + TF EG GF +DI+ RNTA PENHQAVAL S+SD S
Sbjct: 224 IGKTIITGDKSKGRG-FSTYKSATFVAEGDGFVGRDITIRNTAGPENHQAVALRSNSDMS 282
Query: 169 VFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQLSSFKTDILVRKGPTGQQNTI 228
VFYRC I G+QD+L + R + + + + + P NTI
Sbjct: 283 VFYRCSIEGYQDTLYVHSGRQFFRECDIYGTVDFIFGNAAAVLQNCRIFARNPPNGVNTI 342
Query: 229 TAQ 231
TAQ
Sbjct: 343 TAQ 345
>At1g11590 putative pectin methylesterase
Length = 524
Score = 143 bits (360), Expect = 1e-34
Identities = 96/257 (37%), Positives = 130/257 (50%), Gaps = 7/257 (2%)
Query: 4 SRIFFLFCFFFRTSSSDILDTITQKGKFPSWVKPGDRKLLQASAVP----ADAVVASDGS 59
SR F S D + I+ PSW+ D+K L +A AD VVA DG+
Sbjct: 164 SRARIALALFISISPRDNTELISVIPNSPSWLFHVDKKDLYLNAEALKKIADVVVAKDGT 223
Query: 60 GNYMKIMDAVMAAPNGSKKRYVIHIKKGVYNEHVMINNSKSNLMMIGDGMGATVITGDLS 119
G Y + A+ AAP S+KR+VI+IK G+Y+E V+I N+K NL +IGDG T+IT +LS
Sbjct: 224 GKYSTVNAAIAAAPQHSQKRFVIYIKTGIYDEIVVIENTKPNLTLIGDGQDLTIITSNLS 283
Query: 120 WGRDKLDTSYTYTFGVEGLGFSAQDISFRNTAWPENHQAVALLSDSDTSVFYRCEISGFQ 179
+ T T T G GF D+ FRNTA P AVAL D SV YRC + G+Q
Sbjct: 284 -ASNVRRTFNTATVASNGNGFIGVDMCFRNTAGPAKGPAVALRVSGDMSVIYRCRVEGYQ 342
Query: 180 DSLCANIKHHSIRIAKSEARLTSYLVRQLSSFK-TDILVRKGPTGQQNTITAQGGPEKPN 238
D+L + R + ++ F+ I+ R+ GQ N ITAQ K +
Sbjct: 343 DALYPHSDRQFYRECFITGTVDFICGNAVAVFQFCQIVARQPKMGQSNVITAQSRAFK-D 401
Query: 239 LPFGFAFQFCNVCADPE 255
+ GF Q CN+ A +
Sbjct: 402 IYSGFTIQKCNITASSD 418
>At4g02320 pectinesterase - like protein
Length = 518
Score = 142 bits (359), Expect = 2e-34
Identities = 86/230 (37%), Positives = 117/230 (50%), Gaps = 3/230 (1%)
Query: 31 FPSWVKPGDRKLLQ--ASAVPADAVVASDGSGNYMKIMDAVMAAPNGSKKRYVIHIKKGV 88
FP WV DR LLQ + VVA +G+GNY I +A+ AAPN S+ R+VI+IK G
Sbjct: 185 FPMWVSGSDRNLLQDPVDETKVNLVVAQNGTGNYTTIGEAISAAPNSSETRFVIYIKCGE 244
Query: 89 YNEHVMINNSKSNLMMIGDGMGATVITGDLSWGRDKLDTSYTYTFGVEGLGFSAQDISFR 148
Y E++ I K+ +M IGDG+G TVI + S+ D ++ T GV G GF A+D+SF
Sbjct: 245 YFENIEIPREKTMIMFIGDGIGRTVIKANRSYA-DGWTAFHSATVGVRGSGFIAKDLSFV 303
Query: 149 NTAWPENHQAVALLSDSDTSVFYRCEISGFQDSLCANIKHHSIRIAKSEARLTSYLVRQL 208
N A PE HQAVAL S SD S +YRC +QD++ + R +
Sbjct: 304 NYAGPEKHQAVALRSSSDLSAYYRCSFESYQDTIYVHSHKQFYRECDIYGTVDFIFGDAS 363
Query: 209 SSFKTDILVRKGPTGQQNTITAQGGPEKPNLPFGFAFQFCNVCADPEFLP 258
F+ L + P Q I G E P G + + A P+ +P
Sbjct: 364 VVFQNCSLYARRPNPNQKIIYTAQGRENSREPTGISIISSRILAAPDLIP 413
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,374,536
Number of Sequences: 26719
Number of extensions: 264658
Number of successful extensions: 857
Number of sequences better than 10.0: 72
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 651
Number of HSP's gapped (non-prelim): 79
length of query: 287
length of database: 11,318,596
effective HSP length: 98
effective length of query: 189
effective length of database: 8,700,134
effective search space: 1644325326
effective search space used: 1644325326
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148775.4


