

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148775.1 - phase: 0
(207 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
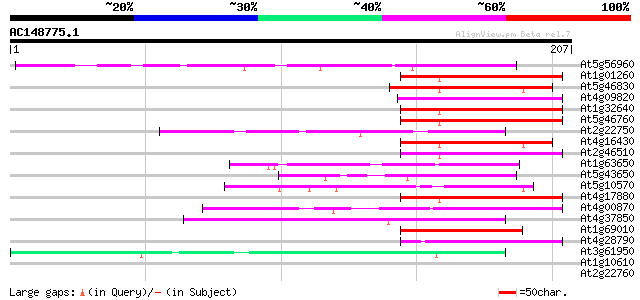
At5g56960 putative protein 91 3e-19
At1g01260 transcription factor MYC7E, putative bHLH transcriptio... 45 3e-05
At5g46830 putative transcription factor bHLH28 45 4e-05
At4g09820 putative protein 44 5e-05
At1g32640 transcription factor like protein, BHLH6 44 5e-05
At5g46760 putative transcription factor BHLH5 43 1e-04
At2g22750 putative bHLH transcription factor (bHLH018) 43 1e-04
At4g16430 putative transcription factor BHLH3 42 3e-04
At2g46510 putative bHLH transcription factor 42 3e-04
At1g63650 putative transcription factor (BHLH2) 42 3e-04
At5g43650 putative protein 41 4e-04
At5g10570 putative bHLH transcription factor (bHLH061) 41 4e-04
At4g17880 putative transcription factor BHLH4 41 4e-04
At4g00870 putative bHLH transcription factor (bHLH014) 41 5e-04
At4g37850 putative bHLH transcription factor (bHLH025) 40 7e-04
At1g69010 putative bHLH transcription factor (bHLH102) 40 7e-04
At4g28790 bHLH transcription factor like protein (bHLH023) 40 9e-04
At3g61950 bHLH transcription factor (bHLH067) like protein 40 9e-04
At1g10610 similar to PDR5-like ABC transporter emb|CAA94437 39 0.003
At2g22760 putative bHLH transcription factor (bHLH019) 38 0.004
>At5g56960 putative protein
Length = 466
Score = 91.3 bits (225), Expect = 3e-19
Identities = 76/211 (36%), Positives = 103/211 (48%), Gaps = 45/211 (21%)
Query: 3 SLSTAGSPEYSSLIFNIPPGTSPPQFSPDLTQLGGVNIPPMRPVSNTLLPLQLQQLPQIT 62
SLS EYSSL+F + P S T VN+P + P L P+ +
Sbjct: 126 SLSPNNISEYSSLLFPLIPKPS--------TTTEAVNVPVLPP----LAPINMIHPQHQE 173
Query: 63 PTQLYPTTQIENDVIMRAIQNVL---STPPSHQHQPQQNYVAHPEASAFGRYRH---DKS 116
P L+ Q E + + +AI VL S+PPS PQ+ A+AF RY D+
Sbjct: 174 P--LFRNRQREEEAMTQAILAVLTGPSSPPSTSSSPQRK----GRATAFKRYYSMISDRG 227
Query: 117 PNIGSNFRRQSLMKRSFAFFRSLNLMRMRDR--------------------NQAARPSSN 156
+ R+QS+M R+ +F+ LN+ R+R + PS+
Sbjct: 228 RAPLPSVRKQSMMTRAMSFYNRLNI-NQRERFTRENATTHGEGSGGSGGGGRYTSGPSAT 286
Query: 157 QLHHMISERRRREKLNENFQALRALLPQGTK 187
QL HMISER+RREKLNE+FQALR+LLP GTK
Sbjct: 287 QLQHMISERKRREKLNESFQALRSLLPPGTK 317
>At1g01260 transcription factor MYC7E, putative bHLH transcription
factor (AtbHLH013)
Length = 590
Score = 45.1 bits (105), Expect = 3e-05
Identities = 24/63 (38%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + RP++ + L+H+ +ER+RREKLN+ F ALR+++P +K+ L+ I
Sbjct: 416 RPRKRGRRPANGRAEALNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAVSYI 475
Query: 202 NAL 204
N L
Sbjct: 476 NEL 478
>At5g46830 putative transcription factor bHLH28
Length = 511
Score = 44.7 bits (104), Expect = 4e-05
Identities = 24/64 (37%), Positives = 41/64 (63%), Gaps = 4/64 (6%)
Query: 141 LMRMRDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKV-RTKLLISS 196
L + + + + +P+ + L+H+ +ER RREKLN F ALRA++P +K+ +T LL +
Sbjct: 322 LEKKKGKKRGRKPAHGRDKPLNHVEAERMRREKLNHRFYALRAVVPNVSKMDKTSLLEDA 381
Query: 197 CCFI 200
C+I
Sbjct: 382 VCYI 385
>At4g09820 putative protein
Length = 379
Score = 44.3 bits (103), Expect = 5e-05
Identities = 22/61 (36%), Positives = 34/61 (55%)
Query: 144 MRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFIINA 203
+ D + R L H+++ERRRREKLNE F LR+++P TK+ ++ +N
Sbjct: 209 LHDNTKDKRLPREDLSHVVAERRRREKLNEKFITLRSMVPFVTKMDKVSILGDTIAYVNH 268
Query: 204 L 204
L
Sbjct: 269 L 269
>At1g32640 transcription factor like protein, BHLH6
Length = 623
Score = 44.3 bits (103), Expect = 5e-05
Identities = 23/63 (36%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R + + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ L+ I
Sbjct: 435 RPKKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRAVVPNVSKMDKASLLGDAIAYI 494
Query: 202 NAL 204
N L
Sbjct: 495 NEL 497
>At5g46760 putative transcription factor BHLH5
Length = 592
Score = 43.1 bits (100), Expect = 1e-04
Identities = 22/63 (34%), Positives = 39/63 (60%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
+ R + +P++ + L+H+ +ER+RREKLN+ F +LRA++P +K+ L+ I
Sbjct: 398 KPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYSLRAVVPNVSKMDKASLLGDAISYI 457
Query: 202 NAL 204
N L
Sbjct: 458 NEL 460
>At2g22750 putative bHLH transcription factor (bHLH018)
Length = 304
Score = 43.1 bits (100), Expect = 1e-04
Identities = 34/132 (25%), Positives = 62/132 (46%), Gaps = 15/132 (11%)
Query: 56 QQLPQITPTQLYPTTQIENDVIMRAIQNVLSTPPSHQHQPQQNYVAHPEASAFGRYRHDK 115
+ L + P+++ TT I ++ + N PP +HQP ++ + +
Sbjct: 30 KSLVEERPSKILKTTHISPNLHPFSSSN----PPPPKHQPSSRILSFEKTGL--HVMNHN 83
Query: 116 SPNIGSNFRRQSL----MKRSFAFFRSLNLMRMRDRNQAARPSSNQLHHMISERRRREKL 171
SPN+ + + + + K++ R + R+Q SN H+++ER+RREKL
Sbjct: 84 SPNLIFSPKDEEIGLPEHKKAELIIRGTKRAQSLTRSQ-----SNAQDHILAERKRREKL 138
Query: 172 NENFQALRALLP 183
+ F AL AL+P
Sbjct: 139 TQRFVALSALIP 150
>At4g16430 putative transcription factor BHLH3
Length = 467
Score = 41.6 bits (96), Expect = 3e-04
Identities = 22/60 (36%), Positives = 40/60 (66%), Gaps = 4/60 (6%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKV-RTKLLISSCCFI 200
+ R + +P++ + L+H+ +ER+RREKLN+ F ALRA++P +K+ + LL + +I
Sbjct: 303 KPRKRGRKPANGREEALNHVEAERQRREKLNQRFYALRAVVPNISKMDKASLLADAITYI 362
>At2g46510 putative bHLH transcription factor
Length = 662
Score = 41.6 bits (96), Expect = 3e-04
Identities = 22/63 (34%), Positives = 38/63 (59%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
R R + +P++ + L+H+ +ER+RREKLN+ F ALR+++P +K+ L+ I
Sbjct: 378 RPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYALRSVVPNISKMDKASLLGDAISYI 437
Query: 202 NAL 204
L
Sbjct: 438 KEL 440
>At1g63650 putative transcription factor (BHLH2)
Length = 596
Score = 41.6 bits (96), Expect = 3e-04
Identities = 34/112 (30%), Positives = 55/112 (48%), Gaps = 13/112 (11%)
Query: 82 QNVLSTPPSHQHQ----PQ-QNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFF 136
Q V+ST HQ PQ QN+ + S+F R++ S ++ + K F
Sbjct: 331 QGVISTIFKTTHQLILGPQFQNF---DKRSSFTRWKRSSSVKTLGEKSQKMIKKILF--- 384
Query: 137 RSLNLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKV 188
+ LM ++ P +H +SE++RREKLNE F LR+++P +K+
Sbjct: 385 -EVPLMNKKEELLPDTPEETG-NHALSEKKRREKLNERFMTLRSIIPSISKI 434
>At5g43650 putative protein
Length = 247
Score = 41.2 bits (95), Expect = 4e-04
Identities = 33/96 (34%), Positives = 46/96 (47%), Gaps = 16/96 (16%)
Query: 100 VAHPEASAFGRYRHDK-----SPNIGSNFRRQSLMKRSFAFFRSLNLMRMR---DRNQAA 151
V+ P SAF Y D SP I S+ + ++ KR +NL+R +N A
Sbjct: 30 VSVPIRSAFRSYMKDTELRMMSPKISSS--KVNVKKRM------VNLLRKNWEEKKNTVA 81
Query: 152 RPSSNQLHHMISERRRREKLNENFQALRALLPQGTK 187
HM+ ER RREK +++ AL +LLP TK
Sbjct: 82 PEKERSRRHMLKERTRREKQKQSYLALHSLLPFATK 117
>At5g10570 putative bHLH transcription factor (bHLH061)
Length = 315
Score = 41.2 bits (95), Expect = 4e-04
Identities = 38/134 (28%), Positives = 63/134 (46%), Gaps = 25/134 (18%)
Query: 80 AIQNVLSTPPSHQHQPQQN----YVAHPEASAFG--------RYRHDKSPNI-------G 120
++ N++S PP HQP Q Y + P +SAF + P I
Sbjct: 58 SLMNLISQPPPLLHQPPQPSSPLYDSPPLSSAFDYPFLEDIIHSSYSPPPLILPASQENT 117
Query: 121 SNFRRQSLMKRSFAFFRSLNLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRA 180
+N+ +SF N R + + +PS N +++ERRRR++LN+ LR+
Sbjct: 118 NNYSPLMEESKSFISIGETNKKRSNKKLEG-QPSKN----LMAERRRRKRLNDRLSLLRS 172
Query: 181 LLPQGTKV-RTKLL 193
++P+ TK+ RT +L
Sbjct: 173 IVPKITKMDRTSIL 186
>At4g17880 putative transcription factor BHLH4
Length = 589
Score = 41.2 bits (95), Expect = 4e-04
Identities = 21/63 (33%), Positives = 39/63 (61%), Gaps = 3/63 (4%)
Query: 145 RDRNQAARPSSNQ---LHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFII 201
+ R + +P++ + L+H+ +ER+RREKLN+ F +LRA++P +K+ L+ I
Sbjct: 399 KPRKRGRKPANGREEPLNHVEAERQRREKLNQRFYSLRAVVPNVSKMDKASLLGDAISYI 458
Query: 202 NAL 204
+ L
Sbjct: 459 SEL 461
>At4g00870 putative bHLH transcription factor (bHLH014)
Length = 423
Score = 40.8 bits (94), Expect = 5e-04
Identities = 35/135 (25%), Positives = 61/135 (44%), Gaps = 18/135 (13%)
Query: 72 IENDVIMRAIQNVLSTPPSHQHQPQQNYVAHPEASAFGRYRHDKSPN--IGSNFRRQSLM 129
I+N + ++++ + + +H Q P S H KS N GS +R+
Sbjct: 176 IQNRNFINRVKSIFGSGKTTKHTNQTGSYPKPAVSD-----HSKSGNQQFGSERKRR--- 227
Query: 130 KRSFAFFRSLNLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVR 189
R L R+ + + L H+ +E++RREKLN F ALRA++P+ +++
Sbjct: 228 -------RKLETTRVAAATKEKHHPA-VLSHVEAEKQRREKLNHRFYALRAIVPKVSRMD 279
Query: 190 TKLLISSCCFIINAL 204
L+S I +L
Sbjct: 280 KASLLSDAVSYIESL 294
>At4g37850 putative bHLH transcription factor (bHLH025)
Length = 304
Score = 40.4 bits (93), Expect = 7e-04
Identities = 31/121 (25%), Positives = 52/121 (42%), Gaps = 2/121 (1%)
Query: 65 QLYPTTQIENDVIMRAIQNVLSTPPSHQHQPQQNYVAHPEASAFGRYRHDKSPNIGSNFR 124
++ P +E I + ++ P H H+ + + E H+ SP ++
Sbjct: 32 EMKPPKILETTYISPSSHLPPNSKPHHIHRHSSSRILSFEDYGSNDMEHEYSPTYLNSIF 91
Query: 125 RQSLMKRSFAFFRS--LNLMRMRDRNQAARPSSNQLHHMISERRRREKLNENFQALRALL 182
L + +S N + +R SN H+I+ER+RREKL + F AL AL+
Sbjct: 92 SPKLEAQVQPHQKSDEFNRKGTKRAQPFSRNQSNAQDHIIAERKRREKLTQRFVALSALV 151
Query: 183 P 183
P
Sbjct: 152 P 152
>At1g69010 putative bHLH transcription factor (bHLH102)
Length = 311
Score = 40.4 bits (93), Expect = 7e-04
Identities = 18/45 (40%), Positives = 28/45 (62%)
Query: 145 RDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVR 189
RD + + S+ + H ++E+RRR K+NE FQ LR L+P + R
Sbjct: 35 RDSKENDKASAIRSKHSVTEQRRRSKINERFQILRELIPNSEQKR 79
>At4g28790 bHLH transcription factor like protein (bHLH023)
Length = 413
Score = 40.0 bits (92), Expect = 9e-04
Identities = 22/60 (36%), Positives = 34/60 (56%), Gaps = 1/60 (1%)
Query: 145 RDRNQAARPSSNQLHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFIINAL 204
RD + R S + H +SERRRR+K+NE +AL+ LLP+ TK ++ + +L
Sbjct: 268 RDSTSSKR-SRAAIMHKLSERRRRQKINEMMKALQELLPRCTKTDRSSMLDDVIEYVKSL 326
>At3g61950 bHLH transcription factor (bHLH067) like protein
Length = 358
Score = 40.0 bits (92), Expect = 9e-04
Identities = 44/192 (22%), Positives = 72/192 (36%), Gaps = 16/192 (8%)
Query: 1 MRSLSTAGSPEYSSLIFNIPPGTSPPQFSPDLTQLGGVNIPPMRPVS--NTLLPLQLQQL 58
+RSL G E S F S P L L + + N L L LQ L
Sbjct: 19 VRSLEVQGFAEAQSFAFKEKEEESLQDTVPFLQMLQSEDPSSFFSIKEPNFLTLLSLQTL 78
Query: 59 PQITPTQLYPTTQIENDVIMRAIQNVLSTPPSHQHQPQQNYVAHPEASAFGRYRHDKSPN 118
+ P +L +E+ +Q+ +++ N + F + +
Sbjct: 79 KE--PWELERYLSLEDSQFHSPVQS-----ETNRFMEGANQAVSSQEIPFSQANMTLPSS 131
Query: 119 IGSNFRRQSLMKRSFAFFRSLNLMRMRDRNQAARPSSN-------QLHHMISERRRREKL 171
S S KR + R + + + +PS N +++H+ ER RR ++
Sbjct: 132 TSSPLSAHSRRKRKINHLLPQEMTREKRKRRKTKPSKNNEEIENQRINHIAVERNRRRQM 191
Query: 172 NENFQALRALLP 183
NE+ +LRALLP
Sbjct: 192 NEHINSLRALLP 203
>At1g10610 similar to PDR5-like ABC transporter emb|CAA94437
Length = 421
Score = 38.5 bits (88), Expect = 0.003
Identities = 41/179 (22%), Positives = 79/179 (43%), Gaps = 31/179 (17%)
Query: 43 MRPVSNTLLPLQLQQLPQITPTQLYPTTQIENDVIMRAIQNV---------------LST 87
MRP +++ L + + T + +P +++ +I RA +++ +S
Sbjct: 147 MRPFDESMVHLIMSRCT--TFFEPFPEQRLQFRIIPRAEESMSSGVNLSVEGGGSSSVSN 204
Query: 88 PPSHQHQPQQNYVAHPEASAFGRYRHDKSPNIGSNFRRQSLMKRSFAFFRSLNLMRMRDR 147
P S NY P AS R +++P + N + +++ +
Sbjct: 205 PSSETQNLFGNY---PNASCVEILREEQTPCLIMNKEKDVVVQNA----------NDSKA 251
Query: 148 NQAARPSSN-QLHHMISERRRREKLNENFQALRALLPQGTKVRTKLLISSCCFIINALL 205
N+ P+ N + ++ SER+RRE++N+ LRA++P+ TK+ + S IN LL
Sbjct: 252 NKKLLPTENFKSKNLHSERKRRERINQAMYGLRAVVPKITKLNKIGIFSDAVDYINELL 310
>At2g22760 putative bHLH transcription factor (bHLH019)
Length = 295
Score = 37.7 bits (86), Expect = 0.004
Identities = 20/35 (57%), Positives = 26/35 (74%), Gaps = 1/35 (2%)
Query: 160 HMISERRRREKLNENFQALRALLPQGTKVRTKLLI 194
H+++ER+RREKL+E F AL ALLP G K K+ I
Sbjct: 120 HVLAERKRREKLSEKFIALSALLP-GLKKADKVTI 153
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.133 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,616,748
Number of Sequences: 26719
Number of extensions: 193973
Number of successful extensions: 757
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 649
Number of HSP's gapped (non-prelim): 126
length of query: 207
length of database: 11,318,596
effective HSP length: 95
effective length of query: 112
effective length of database: 8,780,291
effective search space: 983392592
effective search space used: 983392592
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148775.1


