

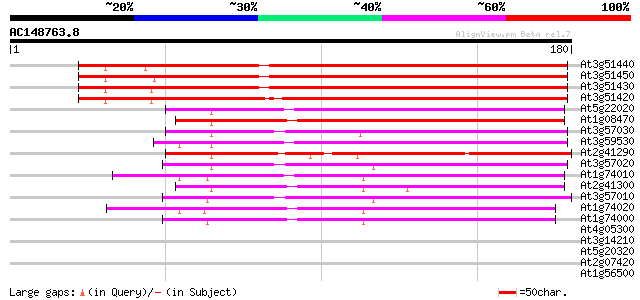
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148763.8 - phase: 1 /pseudo
(180 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g51440 mucin -like protein 172 9e-44
At3g51450 mucin -like protein 168 1e-42
At3g51430 mucin -like protein 167 2e-42
At3g51420 mucin-like protein 166 6e-42
At5g22020 putative strictosidine synthase - like 101 2e-22
At1g08470 unknown protein 99 9e-22
At3g57030 unknown protein 88 2e-18
At3g59530 unknown protein 83 7e-17
At2g41290 strictosidine synthase like protein 80 6e-16
At3g57020 unknown protein 79 2e-15
At1g74010 strictosidine synthase like protein 74 6e-14
At2g41300 putative strictosidine synthase 72 2e-13
At3g57010 unknown protein 69 1e-12
At1g74020 putative strictosidine synthase 68 3e-12
At1g74000 strictosidine synthase AtSS-3 (SS) 67 5e-12
At4g05300 putative protein 29 1.6
At3g14210 myrosinase-associated protein like 28 3.5
At5g20320 CAF-like protein 27 4.5
At2g07420 putative retroelement pol polyprotein 27 4.5
At1g56500 unknown protein 27 5.9
>At3g51440 mucin -like protein
Length = 371
Score = 172 bits (436), Expect = 9e-44
Identities = 85/162 (52%), Positives = 118/162 (72%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVA----DSVVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
E++ +IY GC DGW+KR+ VA DS+VE+ V++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 EDSGFIYTGCVDGWVKRVKVAESVNDSLVEDLVNTGGRPLGIAFGIHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L E DG++FKL D V V ++G +YFT+ +YKYNL+ F DILEG+
Sbjct: 137 NIS---GDGKKTELLTEEADGVRFKLPDAVTVADNGVLYFTDGSYKYNLHQFSFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P TK +L R+LYFAN V+++PDQ +V+CET
Sbjct: 194 PHGRLMSFDPTTKVTRVLLRDLYFANGVSLSPDQTHLVFCET 235
>At3g51450 mucin -like protein
Length = 371
Score = 168 bits (426), Expect = 1e-42
Identities = 82/162 (50%), Positives = 116/162 (70%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADSV----VENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
+E++ IY GC DGW+KR+ VADSV VE+WV++ GRPLG+A GE+IVAD H GLL
Sbjct: 77 KESNLIYTGCVDGWVKRVKVADSVNDSVVEDWVNTGGRPLGIAFGIHGEVIVADVHKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L +E DG+KFKLTD V V ++G +YFT+A+YKY L D+LEG+
Sbjct: 137 NIS---GDGKKTELLTDEADGVKFKLTDAVTVADNGVLYFTDASYKYTLNQLSLDMLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
P GR +S++P T+ +L ++LYFAN + I+PDQ +++CET
Sbjct: 194 PFGRLLSFDPTTRVTKVLLKDLYFANGITISPDQTHLIFCET 235
>At3g51430 mucin -like protein
Length = 371
Score = 167 bits (424), Expect = 2e-42
Identities = 81/162 (50%), Positives = 120/162 (74%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS VVE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 QDSNLIYTGCIDGWVKRVSVHDSANDSVVEDWVNTGGRPLGIAFGVHGEVIVADAYKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ G K E+L ++ +G+KFKLTD V V ++G +YFT+A+YKY L+ DILEG+
Sbjct: 137 NIS---GDGKKTELLTDQAEGVKFKLTDVVAVADNGVLYFTDASYKYTLHQVKFDILEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN V+++PDQ +++CET
Sbjct: 194 PHGRLMSFDPTTRVTRVLLKDLYFANGVSMSPDQTHLIFCET 235
>At3g51420 mucin-like protein
Length = 370
Score = 166 bits (420), Expect = 6e-42
Identities = 81/162 (50%), Positives = 119/162 (73%), Gaps = 8/162 (4%)
Query: 23 EEASYIY-GCEDGWIKRITVADS----VVENWVHSSGRPLGLALEKTGELIVADAHLGLL 77
++++ IY GC DGW+KR++V DS +VE+WV++ GRPLG+A GE+IVADA+ GLL
Sbjct: 77 KDSNLIYTGCVDGWVKRVSVHDSANDSIVEDWVNTGGRPLGIAFGLHGEVIVADANKGLL 136
Query: 78 RVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGE 137
++ GK K E+L +E DG++FKLTD V V ++G +YFT+A+ KY+ Y F D LEG+
Sbjct: 137 SISDG-GK--KTELLTDEADGVRFKLTDAVTVADNGVLYFTDASSKYDFYQFIFDFLEGK 193
Query: 138 PHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCET 179
PHGR MS++P T+ +L ++LYFAN ++++PDQ V+CET
Sbjct: 194 PHGRVMSFDPTTRATRVLLKDLYFANGISMSPDQTHFVFCET 235
>At5g22020 putative strictosidine synthase - like
Length = 395
Score = 101 bits (251), Expect = 2e-22
Identities = 51/129 (39%), Positives = 77/129 (59%), Gaps = 4/129 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL +K TG+L +ADA++GLL+V + G L E +G+ T+ +D+
Sbjct: 133 HICGRPLGLRFDKRTGDLYIADAYMGLLKVGPEGGLATP---LVTEAEGVPLGFTNDLDI 189
Query: 110 GEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAP 169
+DGT+YFT+++ Y +F + G+ GR + Y+P KK +L NL F N V+I+
Sbjct: 190 ADDGTVYFTDSSISYQRRNFLQLVFSGDNTGRVLKYDPVAKKAVVLVSNLQFPNGVSISR 249
Query: 170 DQKFVVYCE 178
D F V+CE
Sbjct: 250 DGSFFVFCE 258
>At1g08470 unknown protein
Length = 390
Score = 99.4 bits (246), Expect = 9e-22
Identities = 49/126 (38%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL +K G+L +ADA+LG+++V + G V NE DG+ + T+ +D+ ++
Sbjct: 130 GRPLGLRFDKKNGDLYIADAYLGIMKVGPEGGLATSV---TNEADGVPLRFTNDLDIDDE 186
Query: 113 GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPDQK 172
G +YFT+++ + F I+ GE GR + YNP TK+ T L RNL F N +++ D
Sbjct: 187 GNVYFTDSSSFFQRRKFMLLIVSGEDSGRVLKYNPKTKETTTLVRNLQFPNGLSLGKDGS 246
Query: 173 FVVYCE 178
F ++CE
Sbjct: 247 FFIFCE 252
>At3g57030 unknown protein
Length = 374
Score = 88.2 bits (217), Expect = 2e-18
Identities = 51/131 (38%), Positives = 78/131 (58%), Gaps = 5/131 (3%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDV 109
H GRPLGL +K TG+L +ADA+ GLL V G + L E +G F+ T+ +D+
Sbjct: 111 HVCGRPLGLRFDKKTGDLYIADAYFGLLVVGPAGGL---AKPLVTEAEGQPFRFTNDLDI 167
Query: 110 GE-DGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIA 168
E + IYFT+ + ++ F +L + GRF+ Y+ ++KK T+L + L FAN VA++
Sbjct: 168 DEQEDVIYFTDTSARFQRRQFLAAVLNVDKTGRFIKYDRSSKKATVLLQGLAFANGVALS 227
Query: 169 PDQKFVVYCET 179
D+ FV+ ET
Sbjct: 228 KDRSFVLVVET 238
>At3g59530 unknown protein
Length = 403
Score = 83.2 bits (204), Expect = 7e-17
Identities = 47/136 (34%), Positives = 75/136 (54%), Gaps = 6/136 (4%)
Query: 47 ENWVHSS--GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKL 103
+ W H GRPLGL K TG L +ADA+ GLL V + G LA +G
Sbjct: 141 KQWKHEKLCGRPLGLRFHKETGNLYIADAYYGLLVVGPEGGIATP---LATHVEGKPILF 197
Query: 104 TDGVDVGEDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFAN 163
+ +D+ +G+I+FT+ + +Y+ + + +LEGE GR + Y+P TK ++ L F N
Sbjct: 198 ANDLDIHRNGSIFFTDTSKRYDRANHFFILLEGESTGRLLRYDPPTKTTHIVLEGLAFPN 257
Query: 164 RVAIAPDQKFVVYCET 179
+ ++ DQ F+++ ET
Sbjct: 258 GIQLSKDQSFLLFTET 273
>At2g41290 strictosidine synthase like protein
Length = 376
Score = 80.1 bits (196), Expect = 6e-16
Identities = 50/133 (37%), Positives = 81/133 (60%), Gaps = 8/133 (6%)
Query: 51 HSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANE-HDGLKFKLTDGVD 108
H GRPLGLA +K TG+L +ADA++GLL+V G ++L E ++ L+F T+ +D
Sbjct: 107 HVCGRPLGLAFDKSTGDLYIADAYMGLLKVGPTGGVA--TQVLPRELNEALRF--TNSLD 162
Query: 109 VG-EDGTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAI 167
+ G +YFT+++ Y ++ ++ G+ GR M Y+ TK+VT L NL F N VA+
Sbjct: 163 INPRTGVVYFTDSSSVYQRRNYIGAMMSGDKTGRLMKYD-NTKQVTTLLSNLAFVNGVAL 221
Query: 168 APDQKFVVYCETA 180
+ + +++ ETA
Sbjct: 222 SQNGDYLLVVETA 234
>At3g57020 unknown protein
Length = 370
Score = 78.6 bits (192), Expect = 2e-15
Identities = 44/132 (33%), Positives = 76/132 (57%), Gaps = 5/132 (3%)
Query: 50 VHSSGRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVD 108
V + GRPLGL EK TG+L + D +LGL++V + G E++ +E +G K + D
Sbjct: 104 VPTCGRPLGLTFEKKTGDLYICDGYLGLMKVGPEGGL---AELIVDEAEGRKVMFANQGD 160
Query: 109 VGEDGTI-YFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAI 167
+ E+ + YF +++ KY+ D + + GE GR + Y+ TK+ ++ NL N +A+
Sbjct: 161 IDEEEDVFYFNDSSDKYHFRDVFFVAVSGERSGRVIRYDKKTKEAKVIMDNLVCNNGLAL 220
Query: 168 APDQKFVVYCET 179
D+ F++ CE+
Sbjct: 221 NKDRSFLITCES 232
>At1g74010 strictosidine synthase like protein
Length = 325
Score = 73.6 bits (179), Expect = 6e-14
Identities = 54/156 (34%), Positives = 76/156 (48%), Gaps = 14/156 (8%)
Query: 34 GWIKRITVADSVVENWVHSS---------GRPLGLALE-KTGELIVADAHLGLLRVTQKE 83
G++ + DS W + + GRP G+AL KTG+L VADA LGL ++
Sbjct: 66 GYVDFAQITDSSNSAWCNGALGTAFAGKCGRPAGIALNSKTGDLYVADAPLGLHVISPAG 125
Query: 84 GKEPKVEILANEHDGLKFKLTDGVDVGED-GTIYFTEATYKYNLYDFYNDILEGEPHGRF 142
G K LA+ DG FK DG+DV G +YFT + K+ + + + G+
Sbjct: 126 GLATK---LADSVDGKPFKFLDGLDVDPTTGVVYFTSFSSKFGPREVLIAVGLKDASGKL 182
Query: 143 MSYNPATKKVTLLARNLYFANRVAIAPDQKFVVYCE 178
Y+PATK VT L L A A++ D FV+ E
Sbjct: 183 FKYDPATKAVTELMEGLSGAAGCAVSSDGSFVLVSE 218
>At2g41300 putative strictosidine synthase
Length = 395
Score = 72.0 bits (175), Expect = 2e-13
Identities = 45/128 (35%), Positives = 73/128 (56%), Gaps = 6/128 (4%)
Query: 54 GRPLGLALEK-TGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVDVGED 112
GRPLGL+ EK +G+L D +LG+++V K G KV +E +G K + +D+ E+
Sbjct: 133 GRPLGLSFEKKSGDLYFCDGYLGVMKVGPKGGLAEKV---VDEVEGQKVMFANQMDIDEE 189
Query: 113 -GTIYFTEATYKYNL-YDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAIAPD 170
IYF +++ Y+ D + L GE GR + Y+ TK+ ++ L+F N +A++ D
Sbjct: 190 EDAIYFNDSSDTYHFGRDVFYAFLCGEKTGRAIRYDKKTKEAKVIMDRLHFPNGLALSID 249
Query: 171 QKFVVYCE 178
FV+ CE
Sbjct: 250 GSFVLSCE 257
>At3g57010 unknown protein
Length = 376
Score = 69.3 bits (168), Expect = 1e-12
Identities = 42/133 (31%), Positives = 73/133 (54%), Gaps = 5/133 (3%)
Query: 50 VHSSGRPLGLALE-KTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVD 108
V S GRPLGL+ E KTG+L + D + G+++V + G E++ +E +G K + D
Sbjct: 105 VPSCGRPLGLSFERKTGDLYICDGYFGVMKVGPEGGL---AELVVDEAEGRKVMFANQGD 161
Query: 109 VGEDGTI-YFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAI 167
+ E+ I YF +++ Y+ D + L G GR + Y+ K+ ++ L N +A+
Sbjct: 162 IDEEEDIFYFNDSSDTYHFRDVFYVSLSGTKVGRVIRYDMKKKEAKVIMDKLRLPNGLAL 221
Query: 168 APDQKFVVYCETA 180
+ + FVV CE++
Sbjct: 222 SKNGSFVVTCESS 234
>At1g74020 putative strictosidine synthase
Length = 335
Score = 67.8 bits (164), Expect = 3e-12
Identities = 48/155 (30%), Positives = 76/155 (48%), Gaps = 14/155 (9%)
Query: 32 EDGWIKRITVADSVVENWVHSS---------GRPLGLAL-EKTGELIVADAHLGLLRVTQ 81
E G++ + +S +W + GRP G+A EKTG+L VADA LGL ++
Sbjct: 66 ETGYVDFAQITESSNSSWCDGTIGTALAGRCGRPAGIAFNEKTGDLYVADAPLGLHVISP 125
Query: 82 KEGKEPKVEILANEHDGLKFKLTDGVDVGED-GTIYFTEATYKYNLYDFYNDILEGEPHG 140
G K+ + DG FK DG+DV G +YFT + +++ + + G
Sbjct: 126 AGGLATKI---TDSVDGKPFKFLDGLDVDPTTGVVYFTSFSSRFSPIQVLIALGLKDATG 182
Query: 141 RFMSYNPATKKVTLLARNLYFANRVAIAPDQKFVV 175
+ Y+P+TK VT+L L + A++ D FV+
Sbjct: 183 KLYKYDPSTKVVTVLMEGLSGSAGCAVSSDGSFVL 217
>At1g74000 strictosidine synthase AtSS-3 (SS)
Length = 329
Score = 67.0 bits (162), Expect = 5e-12
Identities = 44/128 (34%), Positives = 65/128 (50%), Gaps = 5/128 (3%)
Query: 50 VHSSGRPLGLALE-KTGELIVADAHLGLLRVTQKEGKEPKVEILANEHDGLKFKLTDGVD 108
V GRP G+A KTG+L VADA LGL + ++ G K+ A+ G F DG+D
Sbjct: 95 VEKCGRPAGIAFNTKTGDLYVADAALGLHVIPRRGGLAKKI---ADSVGGKPFLFLDGLD 151
Query: 109 VGED-GTIYFTEATYKYNLYDFYNDILEGEPHGRFMSYNPATKKVTLLARNLYFANRVAI 167
V G +YFT + + D + + G+F Y+P+ K VT+L L + A+
Sbjct: 152 VDPTTGVVYFTSFSSTFGPRDVLKAVATKDSTGKFFKYDPSKKVVTVLMEGLSGSAGCAV 211
Query: 168 APDQKFVV 175
+ D FV+
Sbjct: 212 SSDGSFVL 219
>At4g05300 putative protein
Length = 387
Score = 28.9 bits (63), Expect = 1.6
Identities = 19/77 (24%), Positives = 33/77 (42%), Gaps = 5/77 (6%)
Query: 34 GWIKRITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVTQKEGKEPKVEILA 93
GWI++ + + H+ G P+ L GE + H T + PK +
Sbjct: 298 GWIEKSVRGSTRISASEHTQGSPMDAILPVKGE-STKNGH----GETDQRSVRPKGDSTD 352
Query: 94 NEHDGLKFKLTDGVDVG 110
N+ + L+++ D DVG
Sbjct: 353 NQRESLRWEPMDKTDVG 369
>At3g14210 myrosinase-associated protein like
Length = 392
Score = 27.7 bits (60), Expect = 3.5
Identities = 21/79 (26%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query: 72 AHLGLLRVTQKEGKEPK-----VEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNL 126
A LG L + ++E K + LA +H+G G + E I + +++ +
Sbjct: 211 APLGCLPIVRQEYKTGNECYELLNDLAKQHNG-----KIGPMLNEFAKISTSPYGFQFTV 265
Query: 127 YDFYNDILEGEPHGRFMSY 145
+DFYN +L GR ++Y
Sbjct: 266 FDFYNAVLRRIATGRSLNY 284
>At5g20320 CAF-like protein
Length = 1589
Score = 27.3 bits (59), Expect = 4.5
Identities = 16/43 (37%), Positives = 21/43 (48%), Gaps = 1/43 (2%)
Query: 38 RITVADSVVENWVHSSGRPLGLALEKTGELIVADAHLGLLRVT 80
R+T D V VHS R LG+ TGE+ + H L + T
Sbjct: 1122 RVT-CDEVASKEVHSLNRDLGILESNTGEIRCSKGHHWLYKKT 1163
>At2g07420 putative retroelement pol polyprotein
Length = 1664
Score = 27.3 bits (59), Expect = 4.5
Identities = 18/62 (29%), Positives = 31/62 (49%), Gaps = 4/62 (6%)
Query: 88 KVEILANEHDGLKFKLTDGVDVGEDGTIYFTEATYKYNLYDF-YNDILEGEPHGRFMSYN 146
K EIL G+K+ +++ + ++ Y+LY F +N G HGRF+ Y+
Sbjct: 1431 KKEILKLVDAGVKYPISESTWIS---LVHCVPKKEVYDLYIFGFNRGDSGGLHGRFLGYS 1487
Query: 147 PA 148
P+
Sbjct: 1488 PS 1489
>At1g56500 unknown protein
Length = 1055
Score = 26.9 bits (58), Expect = 5.9
Identities = 17/46 (36%), Positives = 27/46 (57%), Gaps = 3/46 (6%)
Query: 56 PLGLALEKTGELIVADAHLGLLR-VTQKEGKEPKVEILANEHDGLK 100
P GLA+ + G L VAD + L+R + +G++ EIL E G++
Sbjct: 860 PAGLAITENGRLFVADTNNSLIRYIDLNKGEDS--EILTLELKGVQ 903
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.330 0.148 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,966,771
Number of Sequences: 26719
Number of extensions: 161574
Number of successful extensions: 562
Number of sequences better than 10.0: 23
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 516
Number of HSP's gapped (non-prelim): 24
length of query: 180
length of database: 11,318,596
effective HSP length: 93
effective length of query: 87
effective length of database: 8,833,729
effective search space: 768534423
effective search space used: 768534423
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148763.8


