

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148762.7 + phase: 2 /partial
(212 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
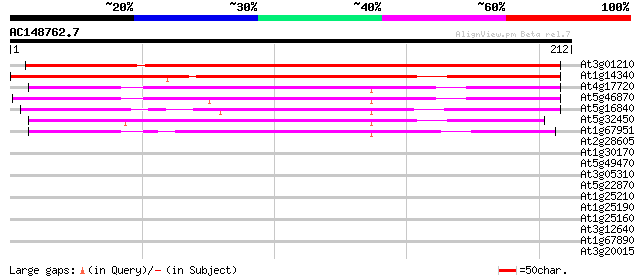
Score E
Sequences producing significant alignments: (bits) Value
At3g01210 unknown protein 218 2e-57
At1g14340 unknown protein 177 4e-45
At4g17720 unknown protein 118 2e-27
At5g46870 putative protein 114 5e-26
At5g16840 unknown protein 104 3e-23
At5g32450 unknown protein 103 5e-23
At1g67951 unknown protein 99 2e-21
At2g28605 unknown protein 30 1.2
At1g30170 hypothetical protein 29 1.6
At5g49470 protein kinase like protein 29 2.1
At3g05310 unknown protein 28 2.7
At5g22870 putative protein 28 3.6
At1g25210 28 4.6
At1g25190 28 4.6
At1g25160 28 4.6
At3g12640 hypothetical protein 27 6.1
At1g67890 putative protein kinase (At1g67890) 27 6.1
At3g20015 putative nucleoid chloroplast DNA-binding protein 27 7.9
>At3g01210 unknown protein
Length = 249
Score = 218 bits (554), Expect = 2e-57
Identities = 112/202 (55%), Positives = 147/202 (72%), Gaps = 3/202 (1%)
Query: 7 TAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMD 66
TAYVTFRDAYAL+ A+LL+G+ I+D+ + IS + Y +SNN L + S+T ++ D
Sbjct: 46 TAYVTFRDAYALDMAVLLSGATIVDQTVWISVYGVYLHESNN---LRQEEDYSVTVTRSD 102
Query: 67 KFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
F SSPGEA+T+AQQVV+TM+AKG+VLSKDA AKA DES+ SS A K+AE+S+ +G
Sbjct: 103 AFASSPGEAITVAQQVVQTMLAKGYVLSKDAIGKAKALDESQRFSSLVATKLAEISHYLG 162
Query: 127 LTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANSSYF 186
LT+ I S +E +S DEKYH +D TKSA VTGT A+ AT+TG+ A AA +++ NS YF
Sbjct: 163 LTQNIQSSMELVRSADEKYHFSDFTKSAVLVTGTAAVAAATITGKVAAAAATSVVNSRYF 222
Query: 187 AKGALWVSDMLSRAAKSTADLG 208
A GALW SD L RAAK+ A +G
Sbjct: 223 ANGALWFSDALGRAAKAAAHIG 244
>At1g14340 unknown protein
Length = 244
Score = 177 bits (449), Expect = 4e-45
Identities = 97/211 (45%), Positives = 136/211 (63%), Gaps = 16/211 (7%)
Query: 1 SSDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHED-- 58
S + TAYV F+D+Y+ ETA+LL G+ ILD+ + I+RW + ++ + WN + ED
Sbjct: 41 SGEQACTAYVMFKDSYSQETAVLLTGATILDQRVCITRWGQHHEEFDFWNATSRGFEDES 100
Query: 59 -SITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
S Y+Q +F + GEA+T AQ+VVK M+A GFVL KDA AKAFDES VS+ A +
Sbjct: 101 DSQHYAQRSEF--NAGEAVTKAQEVVKIMLATGFVLGKDALSKAKAFDESHGVSAAAVAR 158
Query: 118 VAELSNKIGLTETINSGIETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAG 177
V++L +IGLT+ I +G+E + D++YHV+D KSA TGR A AA
Sbjct: 159 VSQLEQRIGLTDKIFTGLEAVRMTDQRYHVSDTAKSA-----------VFATGRTAAAAA 207
Query: 178 SAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
+++ NSSYF+ GALW+S L RAAK+ +DLG
Sbjct: 208 TSVVNSSYFSSGALWLSGALERAAKAASDLG 238
>At4g17720 unknown protein
Length = 313
Score = 118 bits (296), Expect = 2e-27
Identities = 74/205 (36%), Positives = 106/205 (51%), Gaps = 23/205 (11%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDK 67
AYVTF+D ETA+LL+G+ I+D + +S Y L+P S+ +K
Sbjct: 45 AYVTFKDLQGAETAVLLSGATIVDSSVIVSMAPDY--------QLSPEALASLEPKDSNK 96
Query: 68 FVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGL 127
+ L A+ VV +M+AKGF+L KDA AK+ DE ++STA+ KVA KIG
Sbjct: 97 SPKAGDSVLRKAEDVVSSMLAKGFILGKDAIAKAKSVDEKHQLTSTASAKVASFDKKIGF 156
Query: 128 TETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANS 183
T+ IN+G E + VD+KY V++ TKSA T AGSAI +
Sbjct: 157 TDKINTGTVVVGEKVREVDQKYQVSEKTKSAIAAAEQT-----------VSNAGSAIMKN 205
Query: 184 SYFAKGALWVSDMLSRAAKSTADLG 208
Y GA WV+ ++ AK+ ++G
Sbjct: 206 RYVLTGATWVTGAFNKVAKAAEEVG 230
>At5g46870 putative protein
Length = 293
Score = 114 bits (284), Expect = 5e-26
Identities = 76/215 (35%), Positives = 106/215 (48%), Gaps = 27/215 (12%)
Query: 2 SDYESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSIT 61
+D AYVTF+D ETA+LL GS I+D ++++ Y L P+ SI
Sbjct: 37 NDGSKLAYVTFKDLQGAETAVLLTGSTIVDSSVTVTMSPDY--------QLPPDALASIE 88
Query: 62 YSQMDKFVSSPGE----ALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANK 117
+ SSP A+ VV M++KGFVL KDA AK+ DE ++STA+ +
Sbjct: 89 SLKESNKSSSPTREDVSVFRKAEDVVSGMISKGFVLGKDAIAKAKSLDEKHQLTSTASAR 148
Query: 118 VAELSNKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAA 173
V +IG TE IN+G E K VD+K+ VT+ TKSA T
Sbjct: 149 VTSFDKRIGFTEKINTGTTVVSEKVKEVDQKFQVTEKTKSAIAAAEQT-----------V 197
Query: 174 MAAGSAIANSSYFAKGALWVSDMLSRAAKSTADLG 208
AGSAI + Y GA WV+ +R +K+ ++G
Sbjct: 198 SNAGSAIMKNRYVLTGATWVTGAFNRVSKAAEEVG 232
>At5g16840 unknown protein
Length = 259
Score = 104 bits (260), Expect = 3e-23
Identities = 70/209 (33%), Positives = 106/209 (50%), Gaps = 32/209 (15%)
Query: 5 ESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQ 64
E +AYVTF++ ETA+LL+G+ I D+ + I Y+ + P+ E
Sbjct: 41 EHSAYVTFKETQGAETAVLLSGASIADQSVIIELAPNYSPPA------APHAETQ----- 89
Query: 65 MDKFVSSPGEALTM-AQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSN 123
SS E++ A+ VV +M+AKGF+L KDA AKAFDE +STA VA L
Sbjct: 90 -----SSGAESVVQKAEDVVSSMLAKGFILGKDAVGKAKAFDEKHGFTSTATAGVASLDQ 144
Query: 124 KIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSA 179
KIGL++ + +G E K+VD+ + VT+ TKS V + +AGSA
Sbjct: 145 KIGLSQKLTAGTSLVNEKIKAVDQNFQVTERTKS-----------VYAAAEQTVSSAGSA 193
Query: 180 IANSSYFAKGALWVSDMLSRAAKSTADLG 208
+ + Y G W + +R A++ ++G
Sbjct: 194 VMKNRYVLTGVSWAAGAFNRVAQAAGEVG 222
>At5g32450 unknown protein
Length = 267
Score = 103 bits (258), Expect = 5e-23
Identities = 71/204 (34%), Positives = 102/204 (49%), Gaps = 20/204 (9%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETY-----TDDSNNWNNLTPNHEDSITY 62
A+VTF D ALE ALLL+G+ I+D+ ++I+R E Y T + +N P T
Sbjct: 46 AFVTFTDPKALEIALLLSGATIVDQIVTITRAENYVQRRETQEVRMLDNAMPLGLQESTT 105
Query: 63 SQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELS 122
+ ++ AQ VV T++AKG L +DA AKAFDE + + A+ KV+
Sbjct: 106 QTKTNMDGNSRAYVSKAQDVVATVLAKGSALGQDAVNKAKAFDEKHQLRANASAKVSSFD 165
Query: 123 NKIGLTETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGS 178
++GLTE ++ GI E KSVD+K V+D T +A R GS
Sbjct: 166 KRVGLTEKLSVGISAVNEKVKSVDQKLQVSDKTMAA-----------IFAAERKLNDTGS 214
Query: 179 AIANSSYFAKGALWVSDMLSRAAK 202
A+ +S Y GA W S S+ A+
Sbjct: 215 AVKSSRYVTAGAAWFSGAFSKVAR 238
>At1g67951 unknown protein
Length = 278
Score = 99.0 bits (245), Expect = 2e-21
Identities = 67/203 (33%), Positives = 102/203 (50%), Gaps = 29/203 (14%)
Query: 8 AYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDK 67
AYVTF+D+ ETA+LL G++I D +SI+ Y L P +D
Sbjct: 70 AYVTFKDSQGAETAMLLTGAVIADLRVSITPAVNY--------QLPPEA------LALDS 115
Query: 68 FVSSPGEALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIGL 127
S G ++ A+ VV MV +G+ L KDA AKAFD+ ++ S A+ +A L +K+GL
Sbjct: 116 EHSFNGFSVKKAEDVVNIMVGRGYALGKDAMEKAKAFDDRHNLISNASATIASLDDKMGL 175
Query: 128 TETINSGI----ETFKSVDEKYHVTDITKSAATVTGTTAIVVATVTGRAAMAAGSAIANS 183
+E ++ G E + +DE+Y V +ITKSA TAI +A +A+ +
Sbjct: 176 SEKLSIGTTVVNEKLRDIDERYQVREITKSALAAAEETAI-----------SARTALMAN 224
Query: 184 SYFAKGALWVSDMLSRAAKSTAD 206
Y + GA W S+ K+ +
Sbjct: 225 PYVSSGASWFSNAFGAVTKAVKE 247
>At2g28605 unknown protein
Length = 232
Score = 29.6 bits (65), Expect = 1.2
Identities = 25/116 (21%), Positives = 52/116 (44%), Gaps = 11/116 (9%)
Query: 22 LLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDKF-VSSPGEALTMAQ 80
L NG ++ ++ S+ + YTD +N + L P+ +Y++++K ++ E L
Sbjct: 58 LFFNGFLLDNKAKSMEELQRYTDSNNGFTLLIPS-----SYTKVEKAGANALFEELNNGS 112
Query: 81 QVVKTMVAKGFVLSKD-----AFVMAKAFDESRSVSSTAANKVAELSNKIGLTETI 131
+ +V+ + S D FV K + + ST +V + + GL + +
Sbjct: 113 NNIGVVVSPVRIKSLDQFGSPQFVADKLINAEKRKESTKEAEVVSVGERAGLGQQV 168
>At1g30170 hypothetical protein
Length = 366
Score = 29.3 bits (64), Expect = 1.6
Identities = 14/52 (26%), Positives = 26/52 (49%)
Query: 4 YESTAYVTFRDAYALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPN 55
YE+ ++ FR+ E + I D CI +S+ E + ++++ L PN
Sbjct: 272 YETQRFMVFREEETTEERFMCYTDDIGDLCIFVSKSEAFCVPASSYPGLKPN 323
>At5g49470 protein kinase like protein
Length = 770
Score = 28.9 bits (63), Expect = 2.1
Identities = 14/53 (26%), Positives = 29/53 (54%)
Query: 74 EALTMAQQVVKTMVAKGFVLSKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
E T +V ++ +K + SK+A V D + + A+K+++L++K+G
Sbjct: 229 EGETSFSRVTSSVASKLGLDSKEAVVSKLGLDSQQPIQVAIASKISDLASKVG 281
>At3g05310 unknown protein
Length = 648
Score = 28.5 bits (62), Expect = 2.7
Identities = 25/87 (28%), Positives = 42/87 (47%), Gaps = 9/87 (10%)
Query: 41 TYTDDSNNWNNLTPNHEDSITYSQMDKFVSSPGEALTMAQQVVKTMVAKGFVLSKDAF-- 98
+Y DDSNN N T H ++ + +S + L + + +K GF+LSK+A
Sbjct: 451 SYDDDSNNNNGSTDEHY-AVNMVKEPGVISDTDKTLVLKEVRIKD---DGFMLSKEALAA 506
Query: 99 --VMAKAFDESRSVS-STAANKVAELS 122
V +D S S + A + +AE++
Sbjct: 507 CDVAIFIYDSSDEYSWNRAVDMLAEVA 533
>At5g22870 putative protein
Length = 207
Score = 28.1 bits (61), Expect = 3.6
Identities = 23/92 (25%), Positives = 42/92 (45%), Gaps = 19/92 (20%)
Query: 16 YALETALLLNGSMILDRCISISRWETYTDDSNNWNNLTPNHEDSITYSQMDKFVSSPGEA 75
Y +E A + N ++ D +S + +T S+N PNH S+ YS ++ FV +
Sbjct: 58 YTVENASVQNFNLTNDNHMSATF--QFTIQSHN-----PNHRISVYYSSVEIFVKFKDQT 110
Query: 76 LTM------------AQQVVKTMVAKGFVLSK 95
L +Q+ +T++A+ +SK
Sbjct: 111 LAFDTVEPFHQPRMNVKQIDETLIAENVAVSK 142
>At1g25210
Length = 1012
Score = 27.7 bits (60), Expect = 4.6
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Query: 14 DAYALETALLLNGS---MILDRCISISRWETYTDDSNNWN--NLTPNHE 57
D+YAL + N M+ R +S+ R+E Y SN+W +TPN E
Sbjct: 737 DSYALGYDINRNHKILRMVQTRNVSVYRYEIYDLRSNSWRVLEVTPNGE 785
>At1g25190
Length = 1012
Score = 27.7 bits (60), Expect = 4.6
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Query: 14 DAYALETALLLNGS---MILDRCISISRWETYTDDSNNWN--NLTPNHE 57
D+YAL + N M+ R +S+ R+E Y SN+W +TPN E
Sbjct: 737 DSYALGYDINRNHKILRMVQTRNVSVYRYEIYDLRSNSWRVLEVTPNGE 785
>At1g25160
Length = 1020
Score = 27.7 bits (60), Expect = 4.6
Identities = 18/49 (36%), Positives = 26/49 (52%), Gaps = 5/49 (10%)
Query: 14 DAYALETALLLNGS---MILDRCISISRWETYTDDSNNWN--NLTPNHE 57
D+YAL + N M+ R +S+ R+E Y SN+W +TPN E
Sbjct: 745 DSYALGYDINRNHKILRMVQTRNVSVYRYEIYDLRSNSWRVLEVTPNGE 793
>At3g12640 hypothetical protein
Length = 674
Score = 27.3 bits (59), Expect = 6.1
Identities = 10/25 (40%), Positives = 17/25 (68%)
Query: 38 RWETYTDDSNNWNNLTPNHEDSITY 62
+W+ + D+ N NN+ PN+ S+TY
Sbjct: 637 QWKRDSADTGNNNNVAPNNARSLTY 661
>At1g67890 putative protein kinase (At1g67890)
Length = 765
Score = 27.3 bits (59), Expect = 6.1
Identities = 10/33 (30%), Positives = 21/33 (63%)
Query: 94 SKDAFVMAKAFDESRSVSSTAANKVAELSNKIG 126
SK+A V D + + + A+K+++L++K+G
Sbjct: 249 SKEAVVSKLGLDSQQPIQAAIASKISDLASKVG 281
>At3g20015 putative nucleoid chloroplast DNA-binding protein
Length = 386
Score = 26.9 bits (58), Expect = 7.9
Identities = 14/34 (41%), Positives = 19/34 (55%), Gaps = 2/34 (5%)
Query: 6 STAYVTFRDAYALETALL--LNGSMILDRCISIS 37
+ AYV FRD + +TA L +G I D C +S
Sbjct: 276 TAAYVAFRDGFKSQTANLPRASGVSIFDTCYDLS 309
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.122 0.333
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,881,975
Number of Sequences: 26719
Number of extensions: 130921
Number of successful extensions: 431
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 401
Number of HSP's gapped (non-prelim): 19
length of query: 212
length of database: 11,318,596
effective HSP length: 95
effective length of query: 117
effective length of database: 8,780,291
effective search space: 1027294047
effective search space used: 1027294047
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC148762.7


