

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
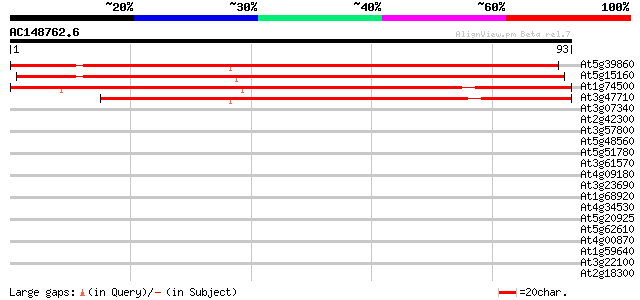
Query= AC148762.6 + phase: 0
(93 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g39860 unknown protein 120 1e-28
At5g15160 unknown protein 118 4e-28
At1g74500 putative DNA-binding protein 117 1e-27
At3g47710 hypothetical protein 93 2e-20
At3g07340 putative bHLH transcription factor (bHLH062) 34 0.014
At2g42300 putative bHLH transcription factor (bHLH048) 33 0.032
At3g57800 bHLH like transcription factor (bHLH060) 32 0.042
At5g48560 putative bHLH transcription factor (bHLH078) 32 0.055
At5g51780 putative bHLH transcription factor (bHLH036) 32 0.071
At3g61570 unknown protein 32 0.071
At4g09180 putative bHLH transcription factor (bHLH081) 31 0.093
At3g23690 bHLH transcription factor like protein (bHLH077) 31 0.093
At1g68920 putative bHLH transcription factor (bHLH049) 31 0.093
At4g34530 putative bHLH transcription factor (bHLH063) 30 0.16
At5g20925 protein kinase (TOUSLED) 30 0.21
At5g62610 putative bHLH transcription factor (bHLH079) 30 0.27
At4g00870 putative bHLH transcription factor (bHLH014) 30 0.27
At1g59640 putative bHLH transcription factor (bHLH031) 30 0.27
At3g22100 hypothetical protein 28 0.60
At2g18300 putative bHLH transcription factor (bHLH064) 28 0.79
>At5g39860 unknown protein
Length = 92
Score = 120 bits (301), Expect = 1e-28
Identities = 66/92 (71%), Positives = 78/92 (84%), Gaps = 2/92 (2%)
Query: 1 MSSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEI-RNRRSDKVPASKVLQETCNYIRNL 59
MS+RRSRQ S + RISD+Q+IDLV KLRQ++PEI + RRSDK ASKVLQETCNYIRNL
Sbjct: 1 MSNRRSRQSSS-APRISDNQMIDLVSKLRQILPEIGQRRRSDKASASKVLQETCNYIRNL 59
Query: 60 QREVDDLSLRLSQLLATIDSDSAEASIIRSLL 91
REVD+LS RLSQLL ++D DS EA++IRSLL
Sbjct: 60 NREVDNLSERLSQLLESVDEDSPEAAVIRSLL 91
>At5g15160 unknown protein
Length = 94
Score = 118 bits (296), Expect = 4e-28
Identities = 65/92 (70%), Positives = 77/92 (83%), Gaps = 2/92 (2%)
Query: 2 SSRRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIR-NRRSDKVPASKVLQETCNYIRNLQ 60
SSRRSRQ S S+RISDDQI DL+ KLRQ +PEIR NRRS+ V ASKVLQETCNYIRNL
Sbjct: 3 SSRRSRQASS-SSRISDDQITDLISKLRQSIPEIRQNRRSNTVSASKVLQETCNYIRNLN 61
Query: 61 REVDDLSLRLSQLLATIDSDSAEASIIRSLLN 92
+E DDLS RL+QLL +ID +S +A++IRSL+N
Sbjct: 62 KEADDLSDRLTQLLESIDPNSPQAAVIRSLIN 93
>At1g74500 putative DNA-binding protein
Length = 93
Score = 117 bits (293), Expect = 1e-27
Identities = 65/95 (68%), Positives = 83/95 (86%), Gaps = 4/95 (4%)
Query: 1 MSSRRSR-QQSGGSTRISDDQIIDLVCKLRQLVPEIRN-RRSDKVPASKVLQETCNYIRN 58
MS RRSR +QS G++RIS+DQI DL+ KL+QL+PE+R+ RRSDKV A++VLQ+TCNYIRN
Sbjct: 1 MSGRRSRSRQSSGTSRISEDQINDLIIKLQQLLPELRDSRRSDKVSAARVLQDTCNYIRN 60
Query: 59 LQREVDDLSLRLSQLLATIDSDSAEASIIRSLLNQ 93
L REVDDLS RLS+LLA +SD+A+A++IRSLL Q
Sbjct: 61 LHREVDDLSERLSELLA--NSDTAQAALIRSLLTQ 93
>At3g47710 hypothetical protein
Length = 78
Score = 93.2 bits (230), Expect = 2e-20
Identities = 52/79 (65%), Positives = 66/79 (82%), Gaps = 3/79 (3%)
Query: 16 ISDDQIIDLVCKLRQLVPEI-RNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
I+D+QI DLV +L +L+PE+ NRRS KV AS+VLQETC+YIRNL +EVDDLS RLSQLL
Sbjct: 2 ITDEQINDLVLQLHRLLPELANNRRSGKVSASRVLQETCSYIRNLSKEVDDLSERLSQLL 61
Query: 75 ATIDSDSAEASIIRSLLNQ 93
+ +DSA+A++IRSLL Q
Sbjct: 62 ES--TDSAQAALIRSLLMQ 78
>At3g07340 putative bHLH transcription factor (bHLH062)
Length = 456
Score = 33.9 bits (76), Expect = 0.014
Identities = 21/64 (32%), Positives = 40/64 (61%), Gaps = 4/64 (6%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP N+ + K + +L E NY+++LQR+V+ LS++LS +
Sbjct: 274 RVRREKISERMKLLQDLVPGC-NKVTGK---ALMLDEIINYVQSLQRQVEFLSMKLSSVN 329
Query: 75 ATID 78
+D
Sbjct: 330 TRLD 333
>At2g42300 putative bHLH transcription factor (bHLH048)
Length = 327
Score = 32.7 bits (73), Expect = 0.032
Identities = 26/68 (38%), Positives = 40/68 (58%), Gaps = 8/68 (11%)
Query: 28 LRQLVPEIRNRRSDKVPASK-VLQETCNYIRNLQREVDDLSLRLSQLLATID--SDSAEA 84
L++LVP DK+ + VL E N+++ LQR+V+ LS+RL+ + ID DS A
Sbjct: 214 LQELVPGC-----DKIQGTALVLDEIINHVQTLQRQVEMLSMRLAAVNPRIDFNLDSILA 268
Query: 85 SIIRSLLN 92
S SL++
Sbjct: 269 SENGSLMD 276
>At3g57800 bHLH like transcription factor (bHLH060)
Length = 379
Score = 32.3 bits (72), Expect = 0.042
Identities = 20/52 (38%), Positives = 33/52 (63%), Gaps = 6/52 (11%)
Query: 28 LRQLVPEIRNRRSDKVPASK-VLQETCNYIRNLQREVDDLSLRLSQLLATID 78
L++LVP DK+ + VL E N++++LQR+V+ LS+RL+ + ID
Sbjct: 233 LQELVPGC-----DKIQGTALVLDEIINHVQSLQRQVEMLSMRLAAVNPRID 279
>At5g48560 putative bHLH transcription factor (bHLH078)
Length = 498
Score = 32.0 bits (71), Expect = 0.055
Identities = 22/77 (28%), Positives = 46/77 (59%), Gaps = 4/77 (5%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP N+ + K + +L E NY+++LQR+V+ LS++LS +
Sbjct: 317 RVRREKIGERMKLLQDLVPGC-NKVTGK---ALMLDEIINYVQSLQRQVEFLSMKLSSVN 372
Query: 75 ATIDSDSAEASIIRSLL 91
T + +A + + ++
Sbjct: 373 DTRLDFNVDALVSKDVM 389
>At5g51780 putative bHLH transcription factor (bHLH036)
Length = 158
Score = 31.6 bits (70), Expect = 0.071
Identities = 20/53 (37%), Positives = 30/53 (55%), Gaps = 6/53 (11%)
Query: 24 LVCKLRQLVPE--IRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
L LR L+P I+ +RS S + E NYI+ LQR++ +LS+R L+
Sbjct: 4 LYASLRSLLPLHFIKGKRS----TSDQVNEAVNYIKYLQRKIKELSVRRDDLM 52
>At3g61570 unknown protein
Length = 712
Score = 31.6 bits (70), Expect = 0.071
Identities = 16/40 (40%), Positives = 25/40 (62%)
Query: 54 NYIRNLQREVDDLSLRLSQLLATIDSDSAEASIIRSLLNQ 93
N IR L+ VDDL+ +L+ L TI+S + E +++ L Q
Sbjct: 431 NQIRKLKDTVDDLNQKLTNCLRTIESKNVELLNLQTALGQ 470
>At4g09180 putative bHLH transcription factor (bHLH081)
Length = 262
Score = 31.2 bits (69), Expect = 0.093
Identities = 15/53 (28%), Positives = 32/53 (60%), Gaps = 4/53 (7%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLS 67
R+ +I D + KL++LVP + + + + +L+E Y++ LQR++ +L+
Sbjct: 200 RVRRTRISDRIRKLQELVPNMDKQTN----TADMLEEAVEYVKVLQRQIQELT 248
>At3g23690 bHLH transcription factor like protein (bHLH077)
Length = 371
Score = 31.2 bits (69), Expect = 0.093
Identities = 20/57 (35%), Positives = 36/57 (63%), Gaps = 4/57 (7%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLS 71
R ++I + + L+ LVP NR + K + +L E NY+++LQR+V+ LS++L+
Sbjct: 207 RARREKISERMTLLQDLVPGC-NRITGK---AVMLDEIINYVQSLQRQVEFLSMKLA 259
>At1g68920 putative bHLH transcription factor (bHLH049)
Length = 486
Score = 31.2 bits (69), Expect = 0.093
Identities = 19/64 (29%), Positives = 36/64 (55%), Gaps = 4/64 (6%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLL 74
R+ ++I + + L+ LVP V +L E NY+++LQR+V+ LS++L+ +
Sbjct: 319 RVRREKISERMKFLQDLVPGCNKVTGKAV----MLDEIINYVQSLQRQVEFLSMKLATVN 374
Query: 75 ATID 78
+D
Sbjct: 375 PQMD 378
>At4g34530 putative bHLH transcription factor (bHLH063)
Length = 335
Score = 30.4 bits (67), Expect = 0.16
Identities = 18/58 (31%), Positives = 35/58 (60%), Gaps = 6/58 (10%)
Query: 15 RISDDQIIDLVCKLRQLVPEIRNRRSDKVPASK-VLQETCNYIRNLQREVDDLSLRLS 71
R+ ++I + + L+ LVP DK+ +L E NY+++LQR+++ LS++L+
Sbjct: 188 RVRREKISERMKFLQDLVPGC-----DKITGKAGMLDEIINYVQSLQRQIEFLSMKLA 240
>At5g20925 protein kinase (TOUSLED)
Length = 688
Score = 30.0 bits (66), Expect = 0.21
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 7/63 (11%)
Query: 4 RRSRQQSGGSTRISDDQIIDLVCKLRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREV 63
R Q G + D+ + L K+ L E+R R D E + +RNL+ EV
Sbjct: 185 RPDGQLRNGECSLQDEDLKSLRAKIAMLEEELRKSRQDS-------SEYHHLVRNLENEV 237
Query: 64 DDL 66
DL
Sbjct: 238 KDL 240
>At5g62610 putative bHLH transcription factor (bHLH079)
Length = 281
Score = 29.6 bits (65), Expect = 0.27
Identities = 13/23 (56%), Positives = 19/23 (82%)
Query: 48 VLQETCNYIRNLQREVDDLSLRL 70
VL E NYI++LQR+V+ LS++L
Sbjct: 198 VLDEIINYIQSLQRQVEFLSMKL 220
>At4g00870 putative bHLH transcription factor (bHLH014)
Length = 423
Score = 29.6 bits (65), Expect = 0.27
Identities = 18/62 (29%), Positives = 35/62 (56%), Gaps = 9/62 (14%)
Query: 28 LRQLVPEIRNRRSDKVPASKVLQETCNYIRNLQREVDDLSLRLSQLLAT----IDSDSAE 83
LR +VP++ R DK + +L + +YI +L+ ++DDL + ++ T +D+ S+
Sbjct: 268 LRAIVPKVS--RMDK---ASLLSDAVSYIESLKSKIDDLETEIKKMKMTETDKLDNSSSN 322
Query: 84 AS 85
S
Sbjct: 323 TS 324
>At1g59640 putative bHLH transcription factor (bHLH031)
Length = 264
Score = 29.6 bits (65), Expect = 0.27
Identities = 13/23 (56%), Positives = 19/23 (82%)
Query: 48 VLQETCNYIRNLQREVDDLSLRL 70
VL E NYI++LQR+V+ LS++L
Sbjct: 181 VLDEIINYIQSLQRQVEFLSMKL 203
>At3g22100 hypothetical protein
Length = 252
Score = 28.5 bits (62), Expect = 0.60
Identities = 25/100 (25%), Positives = 44/100 (44%), Gaps = 12/100 (12%)
Query: 1 MSSRRSRQQSGGSTRISDDQII-------DLVCKLRQLVPEIRNRRSDKVPASKVLQETC 53
++S S SG T +D II D + L +L+P R K+ + L+E+
Sbjct: 119 LNSTSSSTTSGSPTASNDGGIITKRRKISDKIRSLEKLMPWER-----KMNLAMTLEESH 173
Query: 54 NYIRNLQREVDDLSLRLSQLLATIDSDSAEASIIRSLLNQ 93
YI+ LQ ++ L + + + E +++SL Q
Sbjct: 174 KYIKFLQSQIASLRWMPLESVYNTAGEVGETDLLKSLTRQ 213
>At2g18300 putative bHLH transcription factor (bHLH064)
Length = 320
Score = 28.1 bits (61), Expect = 0.79
Identities = 13/39 (33%), Positives = 25/39 (63%)
Query: 48 VLQETCNYIRNLQREVDDLSLRLSQLLATIDSDSAEASI 86
+L E NY++ LQR+V+ LS++L+ L ++ + S+
Sbjct: 213 MLDEIINYVQCLQRQVEFLSMKLAVLNPELELAVEDVSV 251
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.130 0.339
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,667,286
Number of Sequences: 26719
Number of extensions: 53094
Number of successful extensions: 293
Number of sequences better than 10.0: 58
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 22
Number of HSP's that attempted gapping in prelim test: 250
Number of HSP's gapped (non-prelim): 61
length of query: 93
length of database: 11,318,596
effective HSP length: 69
effective length of query: 24
effective length of database: 9,474,985
effective search space: 227399640
effective search space used: 227399640
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC148762.6


