

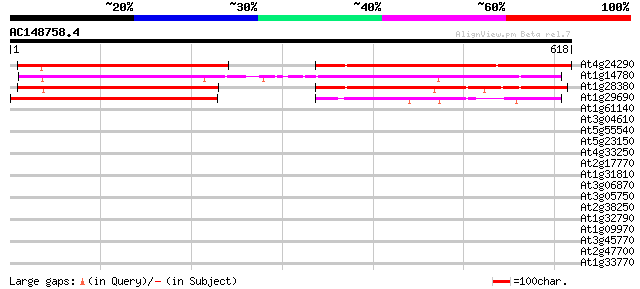
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148758.4 - phase: 0 /pseudo
(618 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g24290 unknown protein 399 e-111
At1g14780 unknown protein 382 e-106
At1g28380 unknown protein 278 5e-75
At1g29690 unknown protein 198 6e-51
At1g61140 unknown protein 31 2.3
At3g04610 putative RNA-binding protein 30 3.0
At5g55540 putative protein 30 4.0
At5g23150 transcription factor-like protein (gb|AAD31171.1) 30 5.2
At4g33250 unknown protein 30 5.2
At2g17770 bZIP transcription factor atbzip27 30 5.2
At1g31810 hypothetical protein 30 5.2
At3g06870 unknown protein 29 6.8
At3g05750 hypothetical protein 29 6.8
At2g38250 putative GT-1-like transcription factor 29 6.8
At1g32790 RNA-binding protein, putative 29 6.8
At1g09970 leucine-rich repeat receptor-like kinase At1g09970 29 6.8
At3g45770 nuclear receptor binding factor-like protein 29 8.9
At2g47700 unknown protein 29 8.9
At1g33770 protein kinase, putative 29 8.9
>At4g24290 unknown protein
Length = 606
Score = 399 bits (1025), Expect = e-111
Identities = 198/282 (70%), Positives = 228/282 (80%), Gaps = 2/282 (0%)
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IEELHQFLEFQLPRQWAPVF EL LGP RK+ QS +SLQFSF GPKLYVNT+PV VG
Sbjct: 322 KPPIEELHQFLEFQLPRQWAPVFSELPLGPQRKQ-QSCASLQFSFFGPKLYVNTTPVDVG 380
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNVSDASSERKYYEKVQWKS 456
+P+TG+RLYLEG++SN LAIHLQHLSSLPK +QL+D+ NR++ S +R+YYEKV WK+
Sbjct: 381 KRPITGMRLYLEGRRSNRLAIHLQHLSSLPKIYQLEDDLNRSIRQESHDRRYYEKVNWKN 440
Query: 457 FSHICTAPVESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAPEWDGSPGL 516
+SH+CT PVES DD +VVTGA V G K VLFLRL F +V AT V+ EWD + G
Sbjct: 441 YSHVCTEPVESDDDLSVVTGAQLHVESHGFKNVLFLRLCFSRVVGATLVKNSEWDEAVGF 500
Query: 517 TQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLKFVDTTEMTR 576
KSG+IST IS F+ QK PPP+P+DVN+NSA+YPGGPPVP QAPKLLKFVDT+EMTR
Sbjct: 501 APKSGLISTLISHHFTAAQK-PPPRPADVNINSAIYPGGPPVPTQAPKLLKFVDTSEMTR 559
Query: 577 GPQDLPGYWVVSGARLYVEKGKISLKVKYSLLTVILQDEETE 618
GPQ+ PGYWVVSGARL VEKGKISLKVKYSL T IL DE E
Sbjct: 560 GPQESPGYWVVSGARLLVEKGKISLKVKYSLFTPILGDEVIE 601
Score = 265 bits (676), Expect = 7e-71
Identities = 136/238 (57%), Positives = 178/238 (74%), Gaps = 5/238 (2%)
Query: 9 SVEDVIKSVGLGYDLTNDLRLKFCK---YDSKLIAI-DHDNLRTVELPGRVSIPNVPKSI 64
+ E I S+G GYDL DLRLK+CK DS+L+ I + D+ + LPG +SIPNV KSI
Sbjct: 10 AAEVAIGSIGCGYDLAIDLRLKYCKGGSKDSRLLDIKEGDDNCEIVLPGGISIPNVSKSI 69
Query: 65 NCDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTKSLA 124
CDKG+RMR SD+L FQQM+EQFNQE+SL+GKIP+G FN+ F+FS WQ+DAA TK+LA
Sbjct: 70 KCDKGERMRFRSDILPFQQMAEQFNQELSLAGKIPSGLFNAMFEFSSCWQKDAAYTKNLA 129
Query: 125 FDGVSITLYNIALDKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGTDII 184
FDGV I+LY++ALDK+ V+L +HVK+AVPS+WDPAALARFI+ YGTH +V VK+GG D+I
Sbjct: 130 FDGVFISLYSVALDKSQVLLREHVKQAVPSTWDPAALARFIDIYGTHIIVSVKMGGKDVI 189
Query: 185 YAKQQYSSPLQPSDVQKKLKDMADELF-RGQAGQNNANDGTFNSKEKFMRDNGLGFLD 241
YAKQQ+SS LQP D+QK+LK++AD+ F N ++ S + ++ L F D
Sbjct: 190 YAKQQHSSKLQPEDLQKRLKEVADKRFVEASVVHNTGSERVQASSKVETKEQRLRFAD 247
>At1g14780 unknown protein
Length = 627
Score = 382 bits (981), Expect = e-106
Identities = 244/635 (38%), Positives = 352/635 (55%), Gaps = 61/635 (9%)
Query: 10 VEDVIKSVGLGYDLTNDLRLKFCKY------DSKLIAIDHDNLRTVELPGRVSIPNVPKS 63
+E +KS+G G+DLT D RLK+CK D +L+ +D R + +PG NV
Sbjct: 9 IETAVKSLGKGFDLTADFRLKYCKDGDGSAGDDRLVVLDQTQNRELHIPGFGVFQNVSAD 68
Query: 64 INCDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQF-SGVWQRDAANTKS 122
INCDKG+R R SD+L F +MSE FNQ S++GKIP+G+FN+ F F SG W DAAN KS
Sbjct: 69 INCDKGERTRFRSDILDFNKMSEYFNQRSSVTGKIPSGNFNATFGFQSGSWATDAANVKS 128
Query: 123 LAFDGVSITLYNIAL-DKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGT 181
L D +TL+N+ + + + L+D V+ AVPSSWDP LARFIE+YGTH + GV +GG
Sbjct: 129 LGLDASVVTLFNLHIHNPNRLRLTDRVRNAVPSSWDPQLLARFIERYGTHVITGVSVGGQ 188
Query: 182 DIIYAKQQYSSPLQPSDVQKKLKDMADELFRG----QAGQNNANDGTFNSKEKFMRDNGL 237
D++ +Q SS L ++ L D+ D+LF G + N +S+ KF +
Sbjct: 189 DVVVVRQDKSSDLDNDLLRHHLYDLGDQLFTGSCLLSTRRLNKAYHHSHSQPKFPEAFNV 248
Query: 238 GFLDIQAQSYRETEKNTTNVLAIYPLLVKHSRCRI*SLCAR----GKVEMGNKISATMSG 293
F D Q ++ N+ N + + +CA+ G+ + ++ T+
Sbjct: 249 -FDDKQTVAFNNFSINSQNGITV--------------ICAKRGGDGRAKSHSEWLITVPD 293
Query: 294 AKLFCLNLM*YQCHSYQLHLYLVE*MGVDI*LMP*ISIYDTADKPAIEELHQFLEFQLPR 353
K +N + + L + G + L +S+Y P + +L FL+F PR
Sbjct: 294 -KPDAINF-----NFIPITSLLKDVPGSGL-LSHAMSLYLRYKPP-LMDLQYFLDFSGPR 345
Query: 354 QWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVGMKPVTGLRLYLEGKKSN 413
WAPV +L G + + +L +FMGPKLYVNT+PV PVTG+R +LEGKK N
Sbjct: 346 AWAPVHNDLPFGAAPNMASAYPALHINFMGPKLYVNTTPVTSEKNPVTGMRFFLEGKKCN 405
Query: 414 CLAIHLQHLSSLPKTF--QLKDETNRNVSDASSER-KYYEKVQWKSFSHICTAPVESYDD 470
LAIHLQHL + T ++ DE SD ++ +Y+E + K FSH+CT PV+ YD
Sbjct: 406 RLAIHLQHLDNTRTTVGEKITDEHIWRGSDQITDNDRYFEPLNGKKFSHVCTVPVK-YDP 464
Query: 471 N---------------AVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAPEWDGSPG 515
N +VTGA EV + G K VL LRL + KV+D V+ G G
Sbjct: 465 NWIKTTSNHKSQNDVAFIVTGAQLEVKKHGSKSVLHLRLRYTKVSDHYVVQNSWVHGPIG 524
Query: 516 LTQKSGMISTFISTRFSGP--QKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLKFVDTTE 573
+QKSG+ S+ SG + ++V ++S ++PGGPPVPA K++KFVD ++
Sbjct: 525 TSQKSGIFSSMSMPLTSGSVHHNMIQKDKNEVVLDSGVFPGGPPVPAN-NKIVKFVDLSQ 583
Query: 574 MTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
+ RGPQ PG+W+V+G RLY++KGK+ L VK++LL
Sbjct: 584 LCRGPQHSPGHWLVTGVRLYLDKGKLCLHVKFALL 618
>At1g28380 unknown protein
Length = 612
Score = 278 bits (712), Expect = 5e-75
Identities = 160/286 (55%), Positives = 191/286 (65%), Gaps = 12/286 (4%)
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IEELHQFLEFQLPRQWAPV+G+L LG R K QSS SLQFS MGPKLYVNTS V G
Sbjct: 328 KPPIEELHQFLEFQLPRQWAPVYGDLPLGLRRSK-QSSPSLQFSLMGPKLYVNTSKVDSG 386
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNVSDASSERKYYEKVQWKS 456
+PVTGLR +LEGKK N LAIHLQHLS+ P + L + + E+ YY V+W
Sbjct: 387 ERPVTGLRFFLEGKKGNHLAIHLQHLSACPPSLHLSHDDTYEPIEEPVEKGYYVPVKWGI 446
Query: 457 FSHICTAPVE-----SYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVADATRVRAPEWD 511
FSH+CT PV+ S D ++VT A EV G++KVLFLRL F A A R WD
Sbjct: 447 FSHVCTYPVQYNGARSDDTASIVTKAWLEVKGMGMRKVLFLRLGFSLDASAV-TRKSCWD 505
Query: 512 GSPGLTQKSG---MISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLKF 568
++KSG MIST +ST S PQ S +++NSA+YP GP P + PKLL
Sbjct: 506 NLSTNSRKSGVFSMISTRLSTGLSPNPATTKPQ-SKIDINSAVYPRGPSPPVK-PKLLSL 563
Query: 569 VDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLLTVILQD 614
VDT E+ RGP++ PGYWVV+GA+L VE GKIS+K KYSLLTVI +D
Sbjct: 564 VDTKEVMRGPEEQPGYWVVTGAKLCVEAGKISIKAKYSLLTVISED 609
Score = 214 bits (544), Expect = 1e-55
Identities = 108/225 (48%), Positives = 147/225 (65%), Gaps = 3/225 (1%)
Query: 9 SVEDVIKSVGLGYDLTNDLRLKFCKYD---SKLIAIDHDNLRTVELPGRVSIPNVPKSIN 65
+ E + +GLGYDL +D+R CK S+L+ ID R + PG + + NV SI
Sbjct: 14 AAEKAVSVIGLGYDLCSDVRFSACKTTPDGSRLVEIDPTRNRDLIFPGGIVVNNVSSSIK 73
Query: 66 CDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTKSLAF 125
CDKG+R RL SD+LSF QMSE+FNQ++ LSGKIP+G FN+ F FS W +DA++ K+LA+
Sbjct: 74 CDKGERTRLRSDILSFNQMSEKFNQDMCLSGKIPSGMFNNMFAFSKCWPKDASSVKTLAY 133
Query: 126 DGVSITLYNIALDKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGTDIIY 185
DG I+LY++ + + + L D VKR VPSSWD AALA FIEKYGTH VVGV +GG D+I+
Sbjct: 134 DGWFISLYSVEIVRKQLTLRDEVKREVPSSWDSAALAGFIEKYGTHVVVGVTMGGKDVIH 193
Query: 186 AKQQYSSPLQPSDVQKKLKDMADELFRGQAGQNNANDGTFNSKEK 230
KQ S +P ++QK LK DE F ++ + ++ K K
Sbjct: 194 VKQMRKSNHEPEEIQKMLKHWGDERFCVDPVESKSPASVYSGKPK 238
>At1g29690 unknown protein
Length = 561
Score = 198 bits (504), Expect = 6e-51
Identities = 121/292 (41%), Positives = 162/292 (55%), Gaps = 56/292 (19%)
Query: 337 KPAIEELHQFLEFQLPRQWAPVFGELALGPDRKKSQSSSSLQFSFMGPKLYVNTSPVVVG 396
KP IE+L FL++Q+ R WAP L ++K SSLQFS MGPKL+++ V VG
Sbjct: 304 KPPIEDLQYFLDYQIARAWAPEQSNL-----QRKEPVCSSLQFSLMGPKLFISADQVTVG 358
Query: 397 MKPVTGLRLYLEGKKSNCLAIHLQHLSSLPKTFQLKDETNRNV------SDASSERKYYE 450
KPVTGLRL LEG K N L+IHLQHL SLPK Q +++ + + +++E
Sbjct: 359 RKPVTGLRLSLEGSKQNRLSIHLQHLVSLPKILQPHWDSHVPIGAPKWQGPEEQDSRWFE 418
Query: 451 KVQWKSFSHICTAPVESYDDNA-------VVTGAHFEVGETGLKKVLFLRLHFCKVADAT 503
++WK+FSH+ T+P+E + + +VTGA V + G K VL L+L F KV T
Sbjct: 419 PIKWKNFSHVSTSPIEHTETHIGDLSGVHIVTGAQLGVWDFGSKNVLHLKLLFSKVPGCT 478
Query: 504 RVRAPEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGP------- 556
+R WD +P V + L PGGP
Sbjct: 479 -IRRSVWDHTP------------------------------VASSGRLEPGGPSTSSSTE 507
Query: 557 PVPAQAPKLLKFVDTTEMTRGPQDLPGYWVVSGARLYVEKGKISLKVKYSLL 608
V Q+ KL K VD++EM +GPQDLPG+W+V+GA+L VEKGKI L+VKYSLL
Sbjct: 508 EVSGQSGKLAKIVDSSEMLKGPQDLPGHWLVTGAKLGVEKGKIVLRVKYSLL 559
Score = 185 bits (470), Expect = 6e-47
Identities = 95/230 (41%), Positives = 140/230 (60%), Gaps = 2/230 (0%)
Query: 2 SSKKKVLSVEDVIKSVGLGYDLTNDLRLKFCKY--DSKLIAIDHDNLRTVELPGRVSIPN 59
SS+ ++ + I+++G G+D+T+D+RL +CK S+L+ I+ R +EL +PN
Sbjct: 13 SSEALTTTLRNAIQALGRGFDVTSDVRLLYCKGAPGSRLVRIEEGQNRDLELSHGFLLPN 72
Query: 60 VPKSINCDKGDRMRLCSDVLSFQQMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAAN 119
VP I+C +G+ V SF +M+E+FN + G IP G FN+ F ++G WQ DAA+
Sbjct: 73 VPADIDCSRGNSGTQRISVCSFHEMAEEFNVRSGVKGNIPLGCFNAMFNYTGSWQVDAAS 132
Query: 120 TKSLAFDGVSITLYNIALDKTHVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIG 179
TKSLA G I LY++ L K +VL + ++RAVPSSWDPA+LA FIE YGTH V V IG
Sbjct: 133 TKSLALVGYFIPLYDVKLAKLTLVLHNEIRRAVPSSWDPASLASFIENYGTHIVTSVTIG 192
Query: 180 GTDIIYAKQQYSSPLQPSDVQKKLKDMADELFRGQAGQNNANDGTFNSKE 229
G D++Y +Q SSPL S+++ + DM F Q+ + K+
Sbjct: 193 GRDVVYIRQHQSSPLPVSEIENYVNDMIKHRFHEAESQSITGPLKYKDKD 242
>At1g61140 unknown protein
Length = 1287
Score = 30.8 bits (68), Expect = 2.3
Identities = 22/74 (29%), Positives = 35/74 (46%), Gaps = 6/74 (8%)
Query: 423 SSLPKTFQLKD------ETNRNVSDASSERKYYEKVQWKSFSHICTAPVESYDDNAVVTG 476
SS+P F ++D ETNR S ASS Y+ K +S++ D+ ++
Sbjct: 355 SSVPGEFSVRDDAYLSGETNRWWSGASSSAVSYQTDIEKGYSYMAPQTALPSQDSGKISS 414
Query: 477 AHFEVGETGLKKVL 490
HF +T L+ V+
Sbjct: 415 NHFYDSDTCLQYVV 428
>At3g04610 putative RNA-binding protein
Length = 577
Score = 30.4 bits (67), Expect = 3.0
Identities = 19/66 (28%), Positives = 30/66 (44%), Gaps = 4/66 (6%)
Query: 498 KVADATRVRAPEWDGSPGLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPP 557
K+ + TR R DG PG T+++ M +S + LPP + V+ + G
Sbjct: 211 KIVEETRARIKILDGPPGTTERAVM----VSGKEEPESSLPPSMDGLLRVHMRIVDGLDG 266
Query: 558 VPAQAP 563
+QAP
Sbjct: 267 EASQAP 272
>At5g55540 putative protein
Length = 1380
Score = 30.0 bits (66), Expect = 4.0
Identities = 14/46 (30%), Positives = 26/46 (56%)
Query: 422 LSSLPKTFQLKDETNRNVSDASSERKYYEKVQWKSFSHICTAPVES 467
L +P+ +QL ++ + +SD SE ++WK+F+ +C V S
Sbjct: 703 LQRVPRVYQLCNDIVQLLSDWRSENSNKPIMRWKAFADLCQFKVPS 748
>At5g23150 transcription factor-like protein (gb|AAD31171.1)
Length = 1392
Score = 29.6 bits (65), Expect = 5.2
Identities = 14/30 (46%), Positives = 17/30 (56%), Gaps = 6/30 (20%)
Query: 534 PQKLPPPQPSDVNVNSALYPGGPPVPAQAP 563
P LPPP P+ AL+P PP P+Q P
Sbjct: 1088 PSSLPPPPPA------ALFPPLPPPPSQPP 1111
>At4g33250 unknown protein
Length = 226
Score = 29.6 bits (65), Expect = 5.2
Identities = 33/120 (27%), Positives = 53/120 (43%), Gaps = 21/120 (17%)
Query: 83 QMSEQFNQEVSLSGKIPTGHFNSAFQFSGVWQRDAANTKSL--------AFDGVSITLYN 134
QM EQF + LS + TG F QF W A N L A + L +
Sbjct: 96 QMEEQFKSLIVLSHYLETGRFQ---QF---WDEAAKNRHILEAVPGFEQAIQAYASHLLS 149
Query: 135 IALDKT-HVVLSDHVKRAVPSSWDPAALARFIEKYGTHAVVGVKIGGTDIIYAKQQYSSP 193
++ K VL++ V + D A+L +FIE+ T++ V+ G I+ + +++ P
Sbjct: 150 LSYQKVPRSVLAEAV------NMDGASLDKFIEQQVTNSGWIVEKEGGSIVLPQNEFNHP 203
>At2g17770 bZIP transcription factor atbzip27
Length = 234
Score = 29.6 bits (65), Expect = 5.2
Identities = 18/43 (41%), Positives = 23/43 (52%), Gaps = 8/43 (18%)
Query: 538 PPPQPSDVNVNSALYPGGPPVPAQAPKL-------LKFVDTTE 573
PPP PS + +ALY G P+P A L +F+DTTE
Sbjct: 90 PPPPPSSSTIVTALY-GSLPLPPPATVLSLNSGVGFEFLDTTE 131
>At1g31810 hypothetical protein
Length = 1201
Score = 29.6 bits (65), Expect = 5.2
Identities = 20/62 (32%), Positives = 28/62 (44%), Gaps = 7/62 (11%)
Query: 528 STRFSGPQKLPPPQPSDVNVNSALY-PGGPPVPAQAPKLLKFVDTTEMTRG----PQDLP 582
+T FS Q PPP P + +++ + P PP P P L F TT + P LP
Sbjct: 496 TTSFSPSQPPPPPPPPPLFMSTTSFSPSQPPPPPPPPPL--FTSTTSFSPSQPPPPPPLP 553
Query: 583 GY 584
+
Sbjct: 554 SF 555
>At3g06870 unknown protein
Length = 207
Score = 29.3 bits (64), Expect = 6.8
Identities = 16/42 (38%), Positives = 24/42 (57%), Gaps = 5/42 (11%)
Query: 534 PQKLPPPQPSDVNVNSALYPGGPPVPAQAPKLLKFVDTTEMT 575
P+KL PQP+D+++N+ L PP P + FV +E T
Sbjct: 50 PEKLLLPQPTDLDLNTFLPNDEPPPPP-----IPFVGVSETT 86
>At3g05750 hypothetical protein
Length = 798
Score = 29.3 bits (64), Expect = 6.8
Identities = 15/46 (32%), Positives = 28/46 (60%), Gaps = 1/46 (2%)
Query: 184 IYAKQQYSSPLQPSDVQKKLKDMADELFRGQAGQN-NANDGTFNSK 228
+ AK ++SS S + K++D+ ++L Q GQ+ ++GT N+K
Sbjct: 224 VVAKTRFSSSDSSSSLPMKIRDLKEKLEASQKGQSPQISNGTCNNK 269
>At2g38250 putative GT-1-like transcription factor
Length = 289
Score = 29.3 bits (64), Expect = 6.8
Identities = 18/67 (26%), Positives = 34/67 (49%), Gaps = 12/67 (17%)
Query: 179 GGTDIIYAKQQYSSPLQPSDVQKKLKDMADE------------LFRGQAGQNNANDGTFN 226
GGT K++YSS + +V ++L D++++ +G + +N+N+G
Sbjct: 143 GGTSGAARKREYSSDEEEENVNEELVDVSNDPKILNPKKNIAKKRKGGSNSSNSNNGVRE 202
Query: 227 SKEKFMR 233
E+FMR
Sbjct: 203 VLEEFMR 209
>At1g32790 RNA-binding protein, putative
Length = 358
Score = 29.3 bits (64), Expect = 6.8
Identities = 18/72 (25%), Positives = 33/72 (45%), Gaps = 4/72 (5%)
Query: 169 GTHAVVGVKIGGTDIIYAKQQYSSPLQP-SDVQKKLKDMADELFR---GQAGQNNANDGT 224
G ++ +G + +D + + + ++ ++ KL MA+E + G N N G
Sbjct: 58 GLYSKIGSHVARSDGVDGGESFKRDMRELQELFSKLNPMAEEFVPPSLNKQGGNGVNGGF 117
Query: 225 FNSKEKFMRDNG 236
F S F R+NG
Sbjct: 118 FTSAGSFFRNNG 129
>At1g09970 leucine-rich repeat receptor-like kinase At1g09970
Length = 976
Score = 29.3 bits (64), Expect = 6.8
Identities = 17/47 (36%), Positives = 22/47 (46%)
Query: 454 WKSFSHICTAPVESYDDNAVVTGAHFEVGETGLKKVLFLRLHFCKVA 500
WKS + + V S DN A F V LKK+ +L L C +A
Sbjct: 162 WKSLRNATSLVVLSLGDNPFDATADFPVEVVSLKKLSWLYLSNCSIA 208
>At3g45770 nuclear receptor binding factor-like protein
Length = 375
Score = 28.9 bits (63), Expect = 8.9
Identities = 12/22 (54%), Positives = 14/22 (63%)
Query: 539 PPQPSDVNVNSALYPGGPPVPA 560
P PSD+N +YP PPVPA
Sbjct: 83 PINPSDINRIEGVYPVRPPVPA 104
>At2g47700 unknown protein
Length = 358
Score = 28.9 bits (63), Expect = 8.9
Identities = 19/45 (42%), Positives = 21/45 (46%), Gaps = 3/45 (6%)
Query: 515 GLTQKSGMISTFISTRFSGPQKLPPPQPSDVNVNSALYPGGPPVP 559
G T S +IS TR P PPP D NV +YPGG P
Sbjct: 284 GPTLASPLISM---TRRGLPPPPPPPPMPDQNVGFFIYPGGHHEP 325
>At1g33770 protein kinase, putative
Length = 614
Score = 28.9 bits (63), Expect = 8.9
Identities = 14/40 (35%), Positives = 22/40 (55%), Gaps = 1/40 (2%)
Query: 421 HLSSLPKTFQLKDETNRNVSDASSERKYYEKVQWKSFSHI 460
H+SSLPK + E + V D ++RK E V+W+ +
Sbjct: 433 HISSLPK-YPPSKELDAKVRDEEAKRKKAEAVKWRGHESV 471
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,966,721
Number of Sequences: 26719
Number of extensions: 624203
Number of successful extensions: 1683
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1635
Number of HSP's gapped (non-prelim): 37
length of query: 618
length of database: 11,318,596
effective HSP length: 105
effective length of query: 513
effective length of database: 8,513,101
effective search space: 4367220813
effective search space used: 4367220813
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148758.4


