

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148757.5 + phase: 0
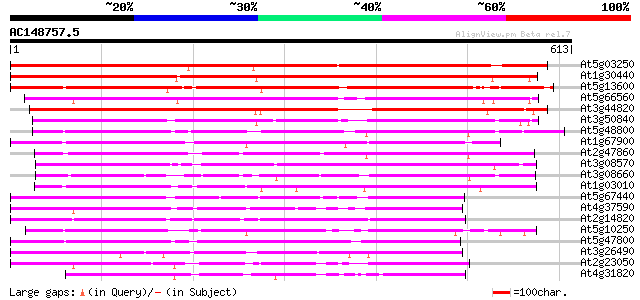
(613 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g03250 photoreceptor-interacting protein - like 617 e-177
At1g30440 non-phototropic hypocotyl, putative 584 e-167
At5g13600 photoreceptor-interacting protein-like; non-phototropi... 581 e-166
At5g66560 unknown protein 422 e-118
At3g44820 non-phototropic hypocotyl 3-like protein 409 e-114
At3g50840 putative protein 379 e-105
At5g48800 non-phototropic hypocotyl-like protein 353 2e-97
At1g67900 unknown protein 323 2e-88
At2g47860 unknown protein 309 3e-84
At3g08570 hypothetical protein 303 1e-82
At3g08660 putative non-phototropic hypocotyl 301 7e-82
At1g03010 hypothetical protein 300 2e-81
At5g67440 photoreceptor-interacting protein-like 295 4e-80
At4g37590 unknown protein 286 3e-77
At2g14820 hypothetical protein 280 2e-75
At5g10250 non-phototropic hypocotyl 3-like protein 280 2e-75
At5g47800 photoreceptor-interacting protein-like 270 1e-72
At3g26490 non-phototropic hypocotyl, putative 269 4e-72
At2g23050 unknown protein 263 2e-70
At4g31820 unknown protein 234 8e-62
>At5g03250 photoreceptor-interacting protein - like
Length = 592
Score = 617 bits (1592), Expect = e-177
Identities = 323/598 (54%), Positives = 425/598 (71%), Gaps = 24/598 (4%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDI 60
MA ++LGSKSE F+ GQ+WLC+TGL SDV IE+GD FHLHKFPL+SRS LER +++
Sbjct: 1 MAFMRLGSKSEAFHREGQTWLCTTGLVSDVTIEVGDMKFHLHKFPLLSRSGLLERLIEES 60
Query: 61 PSDSEKS-ILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEG 119
+D +L L ++PGG K F L+ +FCYGVK+ELT NVV LRCAAE+L+M+++YGEG
Sbjct: 61 STDDGSGCVLSLDEIPGGGKTFELVTKFCYGVKIELTAFNVVSLRCAAEYLEMTDNYGEG 120
Query: 120 NLMIQTENFLNHIFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKA-ADPTLAIL 178
NL+ TE FLN +FG WTD++KAL+TCE+V+ AE+LHI SRC+ SL +KA ADP+L
Sbjct: 121 NLVGMTETFLNEVFGNWTDSIKALQTCEEVIDYAEDLHIISRCVDSLAVKACADPSLFNW 180
Query: 179 PLSGPSSVQSPDNSE----MWNGISMS-LTSKETGEDWWFDDVSSLSLPLYKRFMQGAIA 233
P+ G + S N+E +WNGIS S + TGEDWWFDD S LSLPL+KR + A
Sbjct: 181 PVGGGKNATSGQNTEDESHLWNGISASGKMLQHTGEDWWFDDASFLSLPLFKRLITAIEA 240
Query: 234 RHMKPRRVSGSLVYYAKKHIPSLSSFQNGNSS---KSNLSEADQRNLIEEIVELLPNEKG 290
R MK ++ +++YY +KH+P ++ N + N SE DQ+ +EEIV LLP++KG
Sbjct: 241 RGMKLENIAMAVMYYTRKHVPLMNRQVNMDEQVIETPNPSEEDQKTCLEEIVGLLPSKKG 300
Query: 291 VTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDC 350
V TKFLLR L+TAM L+AS +LE+RIG QLD+A L DLLIPN+GYS ET++D++C
Sbjct: 301 VNPTKFLLRLLQTAMVLHASQSSRENLERRIGNQLDQAALVDLLIPNMGYS-ETLYDVEC 359
Query: 351 VQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPDVN 410
V RM++ F + + A + I+EE + TP T VA L+D YLAEVAPDVN
Sbjct: 360 VLRMIEQF-VSSTEQAGIVPSPCIIEEGHLVKDGADLLTPTTLVATLVDGYLAEVAPDVN 418
Query: 411 LKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEAS 470
LK KF+++AA IPD AR LDDG+Y AID+YLK+H W+T+SE+E ICRLMNCQKLSLEAS
Sbjct: 419 LKLAKFEAIAAAIPDYARPLDDGVYHAIDVYLKAHPWITDSEREHICRLMNCQKLSLEAS 478
Query: 471 THAAQNERLPLRVVVQVLFFEQLKLRTSVAGWFFASDTLENSTTLSG-NLALLRNDGNTT 529
THAAQNERLPLRV+VQVLFFEQL+LRTSV+GWFF S+ L+N G N LL+ G
Sbjct: 479 THAAQNERLPLRVIVQVLFFEQLRLRTSVSGWFFVSENLDNPDNQHGANGGLLKPRG--- 535
Query: 530 HNNPVVAFDHMKDRVSELEKECLSMKQDLEKMMKSKGSWNMLLKKLGCRLIPKPSNPK 587
+++++RVSELEKEC++MKQ+L K++++K SW +KL + + PK
Sbjct: 536 --------ENVRERVSELEKECMNMKQELHKLVRTKRSWKNFTRKLNFKKKSECCKPK 585
>At1g30440 non-phototropic hypocotyl, putative
Length = 665
Score = 584 bits (1506), Expect = e-167
Identities = 313/616 (50%), Positives = 415/616 (66%), Gaps = 42/616 (6%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDI 60
MAC+KLGSKS+ F GQ+W C+TGLPSD+++E+G+ SFHLHKFPL+SRS +ER + +
Sbjct: 1 MACMKLGSKSDAFQRQGQAWFCTTGLPSDIVVEVGEMSFHLHKFPLLSRSGVMERRIAEA 60
Query: 61 PSDSE-KSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEG 119
+ + K ++E+ DLPGG K F L+A+FCYGVK+ELT SNVV LRCAAE L+M+E++GEG
Sbjct: 61 SKEGDDKCLIEISDLPGGDKTFELVAKFCYGVKLELTASNVVYLRCAAEHLEMTEEHGEG 120
Query: 120 NLMIQTENFLNHI-FGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAA-DPTLAI 177
NL+ QTE F N + W D++KAL +C++VL A+EL+IT +CI SL ++A+ DP L
Sbjct: 121 NLISQTETFFNQVVLKSWKDSIKALHSCDEVLEYADELNITKKCIESLAMRASTDPNLFG 180
Query: 178 LPL---SGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIAR 234
P+ GP +QSP S +WNGIS K T DWW++D S LS PL+KR + +R
Sbjct: 181 WPVVEHGGP--MQSPGGSVLWNGISTGARPKHTSSDWWYEDASMLSFPLFKRLITVMESR 238
Query: 235 HMKPRRVSGSLVYYAKKHIPSLSSFQNGNSSKSN----------LSEADQRNLIEEIVEL 284
++ ++GSL YY +KH+P L + G S LSE +Q+NL+EEI EL
Sbjct: 239 GIREDIIAGSLTYYTRKHLPGLKRRRGGPESSGRFSTPLGSGNVLSEEEQKNLLEEIQEL 298
Query: 285 LPNEKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMET 344
L +KG+ TKF + LR A L AS C A+LEKRIG QLD+A LEDL++P+ ++MET
Sbjct: 299 LRMQKGLVPTKFFVDMLRIAKILKASPDCIANLEKRIGMQLDQAALEDLVMPSFSHTMET 358
Query: 345 IHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAE 404
++D+D VQR+LDHF+ D + ++ ++G+ Q TPMT VA L+D YLAE
Sbjct: 359 LYDVDSVQRILDHFLGTDQIMPGGVGSPCSSVDDGNLIGSPQSITPMTAVAKLIDGYLAE 418
Query: 405 VAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQK 464
VAPDVNLK PKFQ+LAA IP+ AR LDDG+YRAIDIYLK H W+ E+E+E +CRL++CQK
Sbjct: 419 VAPDVNLKLPKFQALAASIPEYARLLDDGLYRAIDIYLKHHPWLAETERENLCRLLDCQK 478
Query: 465 LSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAGWFFASDTLENST--TLSGNLALL 522
LSLEA THAAQNERLPLR++VQVLFFEQL+LRTSVAG F SD L+ + SG
Sbjct: 479 LSLEACTHAAQNERLPLRIIVQVLFFEQLQLRTSVAGCFLVSDNLDGGSRQLRSGGYVGG 538
Query: 523 RNDG--------NTTHNNPV--VAFDHMKDRVSELEKECLSMKQDLEKMMK-SKG----- 566
N+G N V V D M+ RV ELEKEC +M+Q++EK+ K +KG
Sbjct: 539 PNEGGGGGGGWATAVRENQVLKVGMDSMRMRVCELEKECSNMRQEIEKLGKTTKGGGSAS 598
Query: 567 ------SWNMLLKKLG 576
+W + KKLG
Sbjct: 599 NGVGSKTWENVSKKLG 614
>At5g13600 photoreceptor-interacting protein-like; non-phototropic
hypocotyl-like protein
Length = 591
Score = 581 bits (1498), Expect = e-166
Identities = 329/610 (53%), Positives = 425/610 (68%), Gaps = 48/610 (7%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDI 60
MA LKLGSKSE+F+L G +WLC TGL DV+I++ D SFHLHKFPL+SRS LE
Sbjct: 1 MASLKLGSKSEVFHLSGHTWLCKTGLKPDVMIQVVDESFHLHKFPLLSRSGYLETLFSK- 59
Query: 61 PSDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGN 120
+ + +LHD+PGG + FLL+A+FCYGV++E+TP N V LRCAAE+LQMSE+YG+ N
Sbjct: 60 -ASETTCVAQLHDIPGGPETFLLVAKFCYGVRIEVTPENAVSLRCAAEYLQMSENYGDAN 118
Query: 121 LMIQTENFLN-HIFGQWTDTLKAL-KTCED-VLPLAEELHITSRCIHSLVLKAA---DPT 174
L+ TE+FLN H+F W D++KAL K+CE VLPLAEELHI SRCI SL +KA + +
Sbjct: 119 LIYLTESFLNDHVFVNWEDSIKALEKSCEPKVLPLAEELHIVSRCIGSLAMKACAEDNTS 178
Query: 175 LAILPLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSS-LSLPLYKRFMQGAIA 233
P+S P + + WNGI +K T E+WWF+DVSS L LP+YKRF++ +
Sbjct: 179 FFNWPISLPEGTTT--TTIYWNGIQ----TKATSENWWFNDVSSFLDLPMYKRFIKTVES 232
Query: 234 RHMKPRRVSGSLVYYAKKHIPSLS-SFQNGNSSKSNLSEAD--------QRNLIEEIVEL 284
R + ++ S+ +YAK+++P L S ++G+ S+ + D QR+L+EEIVEL
Sbjct: 233 RGVNAGIIAASVTHYAKRNLPLLGCSRKSGSPSEEGTNYGDDMYYSHEEQRSLLEEIVEL 292
Query: 285 LPNEKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMET 344
LP +K VT TKFLLR LRT+M L+AS +LEKRIG QLDEA LEDLLIPN+ YS ET
Sbjct: 293 LPGKKCVTSTKFLLRLLRTSMVLHASQVTQETLEKRIGMQLDEAALEDLLIPNMKYSGET 352
Query: 345 IHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAE 404
++D D VQR+LDHFM+ T ++ IVEE ++++G+ +TKVA L+D YLAE
Sbjct: 353 LYDTDSVQRILDHFML--------TFDSSIVEE-KQMMGDSHPLKSITKVASLIDGYLAE 403
Query: 405 VAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQK 464
VA D NLK KFQ+L A+IP+ R +DDGIYRAIDIY+K+H W+TESE+EQ+C LMNCQK
Sbjct: 404 VASDENLKLSKFQALGALIPEDVRPMDDGIYRAIDIYIKAHPWLTESEREQLCLLMNCQK 463
Query: 465 LSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAGWFFASDTLENSTTLSGNLALLRN 524
LSLEA THAAQNERLPLRV+VQVLFFEQ++LRTS+AGW F S+ EN+ T SG L
Sbjct: 464 LSLEACTHAAQNERLPLRVIVQVLFFEQMRLRTSIAGWLFGSE--ENNDT-SGAL----- 515
Query: 525 DGNTTHNNPVVAFDHMKDRVSELEKECLSMKQDLEKMMKSKGSWNMLLKKLGCRLIPKPS 584
+GN N +V M++RV ELEKEC+SMKQDL+K++K+K N K G R K S
Sbjct: 516 EGNKNTNANMV-MHGMRERVFELEKECMSMKQDLDKLVKTKEGRNFFSKIFGSRSKTKTS 574
Query: 585 NPKASKPCRK 594
PC K
Sbjct: 575 ------PCGK 578
>At5g66560 unknown protein
Length = 668
Score = 422 bits (1086), Expect = e-118
Identities = 254/651 (39%), Positives = 377/651 (57%), Gaps = 102/651 (15%)
Query: 17 GQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSI-------- 68
GQ+W C+TGLPSD+ IE+ D +FHLHKFPL+S+S++L R + + + S S+
Sbjct: 10 GQAWFCTTGLPSDIEIEVDDMTFHLHKFPLMSKSRKLHRLITEQETRSSSSMALITVIDP 69
Query: 69 -----------------------------------LELHDLPGGAKAFLLIARFCYGVKM 93
++L D PG +++F ++A+FCYGVKM
Sbjct: 70 KVEETDKKGKGHEIEDDKEEEEVEEQEIEENGYPHIKLEDFPGSSESFEMVAKFCYGVKM 129
Query: 94 ELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPL 152
+L+ S VVPLRCAAE L+M+E+Y NL+ +TE FL+H ++ +++KALK CE V PL
Sbjct: 130 DLSASTVVPLRCAAEHLEMTEEYSPDNLISKTERFLSHSVYKSLRESIKALKACESVSPL 189
Query: 153 AEELHITSRCIHSLVLKA--ADPTLAILPLS---GPSSVQSPDNSEMWNGISMSLT--SK 205
A L IT +CI S+V +A ADP+L P++ G ++ + D + G + S S+
Sbjct: 190 AGSLGITEQCIDSIVSRASSADPSLFGWPVNDGGGRGNISATDLQLIPGGAAKSRKKPSR 249
Query: 206 ETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSL--SSFQNGN 263
++ + WF+D++ LSLP++K + + + + L+ YAKKHIP + S+ + +
Sbjct: 250 DSNMELWFEDLTQLSLPIFKTVILSMRSGDLSSDIIESCLICYAKKHIPGILRSNRKPPS 309
Query: 264 SSKSNLSEADQRNLIEEIVELLPNEKGV--TQTKFLLRSLRTAMALYASSCCCASLEKRI 321
SS + +SE +QR L+E I LP +K + T+FL LRTA+ L A+ C LE++I
Sbjct: 310 SSSTAVSENEQRELLETITSNLPLDKSSISSTTRFLFGLLRTAIILNAAEICRDLLERKI 369
Query: 322 GFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRI 381
G QL+ A L+DLL+P+ Y ET++D+D V+R+L HF+ D SN IVE + +
Sbjct: 370 GSQLERATLDDLLVPSYSYLNETLYDVDLVERILGHFL-----DTLEQSNTAIVEVDGK- 423
Query: 382 VGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIY 441
+ + V L+D +LAE+A D NLK KF +LA +PD AR DDG+YRA+D+Y
Sbjct: 424 ------SPSLMLVGKLIDGFLAEIASDANLKSDKFYNLAISLPDQARLYDDGLYRAVDVY 477
Query: 442 LKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAG 501
LK+H W++E+E+E+IC +M+CQKL+LEA THAAQNERLPLR VVQVLFFEQL+LR ++AG
Sbjct: 478 LKAHPWVSEAEREKICGVMDCQKLTLEACTHAAQNERLPLRAVVQVLFFEQLQLRHAIAG 537
Query: 502 WFFASDTLENSTTLS-------GNLALLRNDGN---------------TTHNNPVVA--F 537
A+ + S + NL + DG+ T N V+
Sbjct: 538 TLLAAQSPSTSQSTEPRPSAAIRNLTITEEDGDEAEGERQVDAGKWKKTVRENQVLRLDM 597
Query: 538 DHMKDRVSELEKECLSMKQDLEKMMKSKG----------SWNMLLKKLGCR 578
D M+ RV LE+EC +MK+ + K+ K SW+ + KK GC+
Sbjct: 598 DTMRTRVHRLERECSNMKKVIAKIDKEGSSPATTTDRPRSWS-ITKKFGCK 647
>At3g44820 non-phototropic hypocotyl 3-like protein
Length = 661
Score = 409 bits (1050), Expect = e-114
Identities = 236/600 (39%), Positives = 362/600 (60%), Gaps = 72/600 (12%)
Query: 22 CSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILE--LHDLPGGAK 79
C TGL SD+ + + D FHLHKFPL+S+ +L R +D S ++S+ L + PGGA
Sbjct: 62 CKTGLSSDITVVVDDVKFHLHKFPLVSKCGKLARMYEDSKSTDKQSLWTTVLEEFPGGAD 121
Query: 80 AFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLN-HIFGQWTD 138
FL++ARFCYG ++++T N+V + CAAE+L+M+ +YGE NL+ Q E FL+ H+ W D
Sbjct: 122 NFLIVARFCYGARVDITSKNLVSIHCAAEYLEMTNEYGEDNLISQVETFLHKHVLRNWKD 181
Query: 139 TLKALKTCEDVLPLAEELHITSRCIHSL-VLKAADPTLAILPLSGPSSVQSPDNSEMWNG 197
+ AL++ VL AE+L + + ++++ + DP+L P+ ++QSP S +WNG
Sbjct: 182 CILALQSSSPVLKSAEKLQMIPKLMNAVSTMVCTDPSLFGWPMMMYGTLQSPGGSILWNG 241
Query: 198 ISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLS 257
I+ + +G DWW++D+S LS+ L+KR ++ + ++ ++G+++YYA+K++P L
Sbjct: 242 INTGARMRSSGSDWWYEDISYLSVDLFKRLIKTMETKGIRAESLAGAMMYYARKYLPGLG 301
Query: 258 SFQNGNSSKS---------NLSEA---------DQRNLIEEIVELLPNEKGVTQTKFLLR 299
+Q+G S S NL++A DQ L+E I+ LLP ++G + KFLL
Sbjct: 302 RWQSGTSDSSKSRRRVVSFNLAKASSPSSMPPLDQIALLETILSLLPEKRGRSFCKFLLG 361
Query: 300 SLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFM 359
LR A L C LEKRIG QL+ A L++LLI N S ET++++DCV+R++ HF
Sbjct: 362 LLRVAFILGVDGNCVKKLEKRIGMQLELATLDNLLILNYSDS-ETLYNVDCVERIVRHFW 420
Query: 360 IVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKFQSL 419
L+DSY+AEVA DVNLK K +SL
Sbjct: 421 ------------------------------------RLVDSYMAEVASDVNLKPDKMRSL 444
Query: 420 AAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERL 479
AA +P+ +R L DG+YRA DIY K H W+++ +KEQ+C +M+ Q+LS++A HA+ N+RL
Sbjct: 445 AAALPESSRPLYDGLYRAFDIYFKEHPWLSDRDKEQLCNIMDYQRLSIDACAHASHNDRL 504
Query: 480 PLRVVVQVLFFEQLKLRTSVAGWFFASDT-LENSTTLSGNLA---LLRNDGNTT--HNNP 533
PLRVV+QVLFFEQ+ LRT++AG ++T ++ T+ G +++ DG T N
Sbjct: 505 PLRVVLQVLFFEQMHLRTALAGGLNVANTETAHAVTIPGGRTGQEIVQRDGWVTVVRQNQ 564
Query: 534 VVAFD--HMKDRVSELEKECLSMKQDLEKMMKSKGSWNM---LLKKLGCR-LIPKPSNPK 587
V+ D M+ RV ELE+E S+KQ+++K + SK S +M L KLGC+ L+P+ S+ K
Sbjct: 565 VLKVDMQKMRSRVGELEEEFQSIKQEMKKRV-SKSSSSMSSPRLVKLGCKFLLPRASDAK 623
>At3g50840 putative protein
Length = 567
Score = 379 bits (973), Expect = e-105
Identities = 226/568 (39%), Positives = 333/568 (57%), Gaps = 52/568 (9%)
Query: 26 LPSDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILELHDLPGGAKAFLLIA 85
LPSD+ IE+ D +FHLHKFPL+S+SK+L + + + S ++L + PGG++ F ++
Sbjct: 14 LPSDIEIEVDDITFHLHKFPLMSKSKKLHQLITEQEQSKVYSHIKLENFPGGSEIFEMVI 73
Query: 86 RFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNH-IFGQWTDTLKALK 144
+ YG K++++ S VPLRCAAE+L+M+E+Y NL+ +TE FL+ +F +++KALK
Sbjct: 74 KISYGFKVDISVSTAVPLRCAAEYLEMTEEYSPENLISKTEKFLSEFVFTNVQESIKALK 133
Query: 145 TCEDVLPLAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQS-PDNSEMWNGISMSLT 203
CE V LAE L IT +CI S+V +A+ + PSS P N+ +
Sbjct: 134 ACESVSSLAESLCITEQCIDSIVFQASS--------TDPSSFYGWPINNGGIFTVDRKKQ 185
Query: 204 SKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGN 263
SK++ + WF+D++ LS P+++R + + + P V SL+ YAKKHIP +S + +
Sbjct: 186 SKDSKTELWFEDLTELSFPIFRRVILSMKSSVLSPEIVERSLLTYAKKHIPGISRSSSAS 245
Query: 264 SSKSN-----LSEADQRNLIEEIVELLPNEKGVTQTKFLLRSLRTAMALYASSCCCASLE 318
SS S+ SE QR L+E I LP T T+ L LR A+ L AS C LE
Sbjct: 246 SSSSSSSTTIASENQQRELLETITSDLPLT--ATTTRSLFGLLRAAIILNASENCRKFLE 303
Query: 319 KRIGFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEE 378
K+IG L++A L+DLLIP+ Y ET++DID V+R+L F+ +N S+S
Sbjct: 304 KKIGSNLEKATLDDLLIPSYSYLNETLYDIDLVERLLRRFL--ENVAVSSSS-------- 353
Query: 379 RRIVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAI 438
+T V L+D L E+A D NLK +F +LA ++P AR DDG+YRA+
Sbjct: 354 ------------LTVVGRLIDGVLGEIASDANLKPEQFYNLAVLLPVQARVYDDGLYRAV 401
Query: 439 DIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTS 498
DIY K+H+W+ E EKE+IC +M+C+KL++E THAAQNERLPLR VVQVLF EQL+LR
Sbjct: 402 DIYFKTHSWILEEEKEKICSVMDCRKLTVEGCTHAAQNERLPLRAVVQVLFLEQLQLRQV 461
Query: 499 VAGWFFASDTLENSTTLSGNLALLRNDGNTTHNNPVVAFDHMKDRVSELEKECLSMKQ-- 556
+ G + + + G + + D M+ RV++LEKECL +K+
Sbjct: 462 ITGTLLTEEDGDKTVVDLGRWKEAVKENQVLR----LDMDTMRTRVNQLEKECLYLKKVI 517
Query: 557 ---DLEKMMKSK---GSWNMLLKKLGCR 578
D E ++K+K G W+ + KK GC+
Sbjct: 518 AKIDKESLLKAKHGAGKWS-IGKKFGCK 544
>At5g48800 non-phototropic hypocotyl-like protein
Length = 614
Score = 353 bits (906), Expect = 2e-97
Identities = 223/603 (36%), Positives = 333/603 (54%), Gaps = 58/603 (9%)
Query: 26 LPSDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILELHDLPGGAKAFLLIA 85
+PSD+ IE+ +F LHKFPL+SRS + R + + DS+ S +EL +LPGGA+ F L A
Sbjct: 41 VPSDITIEVNGGNFALHKFPLVSRSGRIRRIVAE-HRDSDISKVELLNLPGGAETFELAA 99
Query: 86 RFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNHIFGQWTDT-LKALK 144
+FCYG+ E+T SNV L C +++L+M+E+Y + NL +TE +L I + + ++ LK
Sbjct: 100 KFCYGINFEITSSNVAQLFCVSDYLEMTEEYSKDNLASRTEEYLESIVCKNLEMCVQVLK 159
Query: 145 TCEDVLPLAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTS 204
E +LPLA+EL+I RCI ++ KA +A S S ++ + + +S + S
Sbjct: 160 QSEILLPLADELNIIGRCIDAIASKACAEQIA----SSFSRLEYSSSGRLH--MSRQVKS 213
Query: 205 KETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGNS 264
G DWW +D+S L + LY+R M R ++P + SLV YA++ + S +
Sbjct: 214 SGDGGDWWIEDLSVLRIDLYQRVMNAMKCRGVRPESIGASLVSYAERELTKRSEHE---- 269
Query: 265 SKSNLSEADQRNLIEEIVELLPNEKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQ 324
+ ++E IV LLP E V FL LR A+ L S C LE+R+G Q
Sbjct: 270 ----------QTIVETIVTLLPVENLVVPISFLFGLLRRAVILDTSVSCRLDLERRLGSQ 319
Query: 325 LDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGN 384
LD A L+DLLIP+ ++ +T+ DID V R+L +F DD+ E+ V
Sbjct: 320 LDMATLDDLLIPSFRHAGDTLFDIDTVHRILVNFSQQGGDDS----------EDEESVFE 369
Query: 385 CQRA-----TPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAID 439
C T M KVA L+DSYLAE+APD NL KF +A +P ARTL DG+YRAID
Sbjct: 370 CDSPHSPSQTAMFKVAKLVDSYLAEIAPDANLDLSKFLLIAEALPPHARTLHDGLYRAID 429
Query: 440 IYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSV 499
+YLK+H +++S+K+++ +L++ QKLS EA HAAQNERLPL+ +VQVL+FEQLKLR+S+
Sbjct: 430 LYLKAHQGLSDSDKKKLSKLIDFQKLSQEAGAHAAQNERLPLQSIVQVLYFEQLKLRSSL 489
Query: 500 A----------------GWFFASDTLENSTTLSGNLALLRNDGNTTHNNPVVAFDHMKDR 543
W S L + + N A LR + + ++ R
Sbjct: 490 CSSYSDEEPKPKQQQQQSWRINSGALSATMSPKDNYASLRRENRELK----LELARLRMR 545
Query: 544 VSELEKECLSMKQDLEKMMKSKGSWNMLLKKLGCRLIPKPSNPKASKPCRKSKIAPDAVT 603
+++LEKE + MK+D+++ S+ + KK+G S+ + S K D+
Sbjct: 546 LNDLEKEHICMKRDMQR-SHSRKFMSSFSKKMGKLSFFGHSSSRGSSSPSKQSFRTDSKV 604
Query: 604 ELE 606
+E
Sbjct: 605 LME 607
>At1g67900 unknown protein
Length = 631
Score = 323 bits (828), Expect = 2e-88
Identities = 202/564 (35%), Positives = 309/564 (53%), Gaps = 47/564 (8%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDI 60
M +KLGS+ + FY S+ + SD IE+ + + LHKFPL+S+ L+R
Sbjct: 1 MKFMKLGSRPDTFYTSEDLRCVSSEVSSDFTIEVSGSRYLLHKFPLLSKCLRLQRMC--- 57
Query: 61 PSDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGN 120
S+S +SI++L + PGG +AF L A+FCYG+ + ++ N+V RCAAE+LQMSE+ +GN
Sbjct: 58 -SESPESIIQLPEFPGGVEAFELCAKFCYGITITISAYNIVAARCAAEYLQMSEEVEKGN 116
Query: 121 LMIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKA-ADPTLAIL 178
L+ + E F N I W D++ L+T + +E+L ITSRCI ++ K + P+ L
Sbjct: 117 LVYKLEVFFNSCILNGWRDSIVTLQTTKAFPLWSEDLAITSRCIEAIASKVLSHPSKVSL 176
Query: 179 PLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKP 238
S V+ D S S + + WW +D++ L + LY R M + P
Sbjct: 177 SHSHSRRVRDDDMS--------SNRAAASSRGWWAEDIAELGIDLYWRTMIAIKSGGKVP 228
Query: 239 RRVSG-SLVYYAKKHIPSLS-------SFQNGNSSKSNLSEADQRNLIEEIVELLPNEKG 290
+ G +L YA K +P+L ++ +S + + R L+E I+ LLP EKG
Sbjct: 229 ASLIGDALRVYASKWLPTLQRNRKVVKKKEDSDSDSDTDTSSKHRLLLESIISLLPAEKG 288
Query: 291 VTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDC 350
FLL+ L+ A L AS+ L +R+ QL+EA + DLLIP + Y E ++D+D
Sbjct: 289 AVSCSFLLKLLKAANILNASTSSKMELARRVALQLEEATVSDLLIPPMSYKSELLYDVDI 348
Query: 351 VQRMLDHFMIVDNDD------------------ADSTSNNDIVEEERRIVGNCQRATPMT 392
V +L+ FM+ + S N D+ +E R + ++ +
Sbjct: 349 VATILEQFMVQGQTSPPTSPLRGKKGMMDRRRRSRSAENIDLEFQESRRSSSASHSSKL- 407
Query: 393 KVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESE 452
KVA L+D YL ++A DVNL KF +LA +P+ +R D +YRAIDIYLK+H + +SE
Sbjct: 408 KVAKLVDGYLQQIARDVNLPLSKFVTLAESVPEFSRLDHDDLYRAIDIYLKAHKNLNKSE 467
Query: 453 KEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAGWFFASDTLENS 512
++++CR+++C+KLS+EA HAAQNE LPLRVVVQVLF+EQ + + ++ +N+
Sbjct: 468 RKRVCRVLDCKKLSMEACMHAAQNEMLPLRVVVQVLFYEQARAAAA------TNNGEKNT 521
Query: 513 TTLSGNLALLRNDGNTTHNNPVVA 536
T L N+ L N +NP A
Sbjct: 522 TELPSNIKALLAAHNIDPSNPNAA 545
>At2g47860 unknown protein
Length = 635
Score = 309 bits (792), Expect = 3e-84
Identities = 202/572 (35%), Positives = 321/572 (55%), Gaps = 51/572 (8%)
Query: 28 SDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILELHDLPGGAKAFLLIARF 87
SD+ IE+G A+F LHKFPL+SRS + + + + S+ + L L +PGG+++F L A+F
Sbjct: 39 SDLTIEVGSATFSLHKFPLVSRSGRIRKLVLE----SKDTNLNLAAVPGGSESFELAAKF 94
Query: 88 CYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFL-NHIFGQWTDTLKALKTC 146
CYGV ++ SN+ LRC A +L+M+ED E NL +TE +L + IF ++++ L +C
Sbjct: 95 CYGVGVQYNSSNIAALRCVAHYLEMTEDLSEKNLEARTEAYLKDSIFNDISNSITVLHSC 154
Query: 147 EDVLPLAEELHITSRCIHSLVLKAADPTLA--ILPLSGPSSVQSPDNSEMWNGISMSLTS 204
E +LP+AEE+++ R ++++ + A LA +L L S P+ ++
Sbjct: 155 ERLLPVAEEINLVGRLVNAIAVNACKEQLASGLLKLDQSFSCGVPETAKPC--------- 205
Query: 205 KETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGNS 264
DWW + L L ++R + ++ + +S L+ YA+K SL + N
Sbjct: 206 -----DWWGRSLPILKLDFFQRVLSAMKSKGLNHDIISDILMSYARK---SLQIIREPNL 257
Query: 265 SKSNLS-EADQRNLIEEIVELLPNE--KGVTQTKFLLRSLRTAMALYASSCCCASLEKRI 321
KS+ + QR ++E +V LLP + K FL L+TA+ S C + LE+RI
Sbjct: 258 VKSDSDLQRKQRIVLEAVVGLLPTQANKSSIPISFLSSLLKTAIGSGTSVSCRSDLERRI 317
Query: 322 GFQLDEADLEDLLIP-NIGYSMETIHDIDCVQRMLDHFMIVDNDDA--DSTSNNDIVEEE 378
LD+A LED+LIP NIG ++D D VQR+ F+ +D + D D V+E
Sbjct: 318 SHLLDQAILEDILIPANIG----AMYDTDSVQRIFSMFLNLDECEYRDDDDDEEDAVDES 373
Query: 379 RRIVGNCQRA-----TPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDG 433
+ + + A + + KV+ LMDSYLAEVA D +L KF +LA ++PD AR + DG
Sbjct: 374 EMAMYDFEGAESPKQSSIFKVSKLMDSYLAEVALDSSLPPSKFIALAELLPDHARVVCDG 433
Query: 434 IYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQL 493
+YRA+DI+LK H M +SE+ ++C+ ++C+KLS +AS+HAAQNERLP+++ VQVLF+EQ
Sbjct: 434 LYRAVDIFLKVHPHMKDSERYRLCKTVSCKKLSQDASSHAAQNERLPVQIAVQVLFYEQT 493
Query: 494 KLRTSVA----------GWFFASDTLENSTTLSGNLALLRNDGNTTHNNPVVAFD--HMK 541
+L+ ++ FF S SG ++ N + N + + M+
Sbjct: 494 RLKNAMTSGGGTGGSNQSQFFLFPNRSGSGMASGAISPRDNYASVRRENRELRLEVARMR 553
Query: 542 DRVSELEKECLSMKQDLEKMMKSKGSWNMLLK 573
R+++LEK+ +SMK+D K + + ML K
Sbjct: 554 MRLTDLEKDHVSMKRDFVKPQSRRRRYGMLRK 585
>At3g08570 hypothetical protein
Length = 612
Score = 303 bits (777), Expect = 1e-82
Identities = 186/557 (33%), Positives = 308/557 (54%), Gaps = 29/557 (5%)
Query: 29 DVIIEIGDASFHLHKFPLISRSKELERFMKDIP-SDSEKSILELHDLPGGAKAFLLIARF 87
D+ I + SF LHKFPL++R ++ + + ++ S S S EL D PGG+K F L +F
Sbjct: 32 DITIVVDGESFLLHKFPLVARCGKIRKMVAEMKESSSNLSHTELRDFPGGSKTFELAMKF 91
Query: 88 CYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNHI-FGQWTDTLKALKTC 146
CYG+ E+T SNVV +RCAA +L+M+ED+ E NL+ +TE +L + F +++ L +C
Sbjct: 92 CYGINFEITISNVVAIRCAAGYLEMTEDFKEENLIARTETYLEQVAFRSLEKSVEVLCSC 151
Query: 147 EDVLP--LAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTS 204
E + P +AE HI RC+ ++ + A L +L LS + + ++ E+ G S
Sbjct: 152 ETLYPQDIAETAHIPDRCVEAIAVNACREQL-VLGLSRLN--RGTESGELKRGDS----- 203
Query: 205 KETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGNS 264
+WW +D+S+L + Y R + ++ + SL++YA++ + + + +
Sbjct: 204 ----PEWWIEDLSALRIDYYARVVSAMARTGLRSESIITSLMHYAQESLKGIRNCKERTK 259
Query: 265 SKSNLSEADQRNLIEEIVELLPNEKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQ 324
S E +QRN++E IV L PN+ FL LR + + + C LE+RI Q
Sbjct: 260 LDSGTFENEQRNVLEAIVSLFPNDN--VPLSFLFGMLRVGITINVAISCRLELERRIAQQ 317
Query: 325 LDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGN 384
L+ L+DLLIP + ++++D+D V R+L F+ ++ + + E + G+
Sbjct: 318 LETVSLDDLLIPVVRDG-DSMYDVDTVHRILVCFLKKIEEEEEYDEDCCYENETENLTGS 376
Query: 385 CQRATPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKS 444
++ + KV +MD+YLAE+APD L KF +L ++PD AR +DDG+YRAID++LK
Sbjct: 377 MCHSS-LLKVGRIMDAYLAEIAPDPCLSLHKFMALIEILPDYARVMDDGLYRAIDMFLKG 435
Query: 445 HAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAGWFF 504
H + E E + +C+ ++ QKLS EA H AQN+RLP+++VV+VL+ EQL+++ ++G
Sbjct: 436 HPSLNEQECKSLCKFIDTQKLSQEACNHVAQNDRLPMQMVVRVLYSEQLRMKNVMSGESG 495
Query: 505 ASDTLENSTTLSGNLALLRNDGNT------THNNPVVAFDHMKDRVSELEKECLSMKQDL 558
L + S N + + +T + + ++ R++ELEKE + MKQ
Sbjct: 496 EGLLLSSQKHSSENPSRAVSPRDTYASLRRENRELKLEISRVRVRLTELEKEQILMKQG- 554
Query: 559 EKMMKSKGSWNMLLKKL 575
MM+ G LL L
Sbjct: 555 --MMEKSGHGGTLLTSL 569
>At3g08660 putative non-phototropic hypocotyl
Length = 582
Score = 301 bits (771), Expect = 7e-82
Identities = 188/561 (33%), Positives = 309/561 (54%), Gaps = 63/561 (11%)
Query: 29 DVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILELHDLPGGAKAFLLIARFC 88
D+I+ + SF LHKFPL++RS ++ + ++D+ S S++EL D PGG F L +FC
Sbjct: 37 DIIVVVDGESFLLHKFPLVARSGKMRKMVRDLKDSS--SMIELRDFPGGPSTFELTMKFC 94
Query: 89 YGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNHI-FGQWTDTLKALKTCE 147
YG+ ++T NVV LRCAA +L+M+EDY E NL+ + EN+L+ I F + +++ L +CE
Sbjct: 95 YGINFDITAFNVVSLRCAAGYLEMTEDYKEQNLIFRAENYLDQIVFRSFHESVLVLCSCE 154
Query: 148 DVLPLAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTSKET 207
+AE I RC+ ++ + A ++ +G+S L ++
Sbjct: 155 -TQEIAETYEIPDRCVEAIAMNAC-------------------RKQLVSGLSEELKGRDC 194
Query: 208 GEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGNSSKS 267
E W +++S+L + Y + + ++ + SLV+YAK SL + N
Sbjct: 195 LE-MWTEELSALGIDYYVQVVSAMARLSVRSESIVASLVHYAKT---SLKGIIDRNCQ-- 248
Query: 268 NLSEADQRNLIEEIVELLPN-EKG-----VTQTKFLLRSLRTAMALYASSCCCASLEKRI 321
+QR ++E +V LLPN EKG + FL L+ + C LE+RI
Sbjct: 249 -----EQRKIVEAMVNLLPNDEKGSYSLSIIPLGFLFGMLKVGTIIDIEISCRLELERRI 303
Query: 322 GFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLDHFMI-VDNDDADSTSNNDIVEEERR 380
G QL+ A L+DLLIP++ + ++++D+D V R+L F+ ++ +D + ++D +
Sbjct: 304 GHQLETASLDDLLIPSV-QNEDSMYDVDTVHRILTFFLERIEEEDDECGYDSDSTGQHSS 362
Query: 381 IVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDI 440
++ KV +MD+YL E+APD L KF ++ +P+ +R +DDGIYRAID+
Sbjct: 363 LL----------KVGRIMDAYLVEIAPDPYLSLHKFTAIIETLPEHSRIVDDGIYRAIDM 412
Query: 441 YLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLKLRTSVA 500
YLK+H +TE E++++C ++C+KLS EAS H AQN+RLP+++VV+VL+ EQL+L+ +++
Sbjct: 413 YLKAHPLLTEEERKKLCNFIDCKKLSQEASNHVAQNDRLPVQMVVRVLYTEQLRLKKALS 472
Query: 501 G------WFFASDTLENSTTLSGNLALLRNDGNTTHNNPVVAFDHMKDRVSELEKECLSM 554
G W S + + A LR + + M+ RVSELEKE M
Sbjct: 473 GDSEEGSWVLPSGVQSRAVSPRDTYAALRRENRELK----LEISRMRVRVSELEKEHNLM 528
Query: 555 KQD-LEKMMKSKGSWNMLLKK 574
K + +EK + G++ L K
Sbjct: 529 KHEMMEKSGNNGGTFLTSLSK 549
>At1g03010 hypothetical protein
Length = 634
Score = 300 bits (767), Expect = 2e-81
Identities = 192/577 (33%), Positives = 326/577 (56%), Gaps = 46/577 (7%)
Query: 28 SDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSILELHDLPGGAKAFLLIARF 87
SD+ +++G +SF LHKFPL+SRS ++ + + +D + S + L + PGG++AF L A+F
Sbjct: 38 SDLTVQVGSSSFCLHKFPLVSRSGKIRKLL----ADPKISNVCLSNAPGGSEAFELAAKF 93
Query: 88 CYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNH-IFGQWTDTLKALKTC 146
CYG+ +E+ N+ LRCA+ +L+M+ED+ E NL +TE+FL IF +++ L C
Sbjct: 94 CYGINIEINLLNIAKLRCASHYLEMTEDFSEENLASKTEHFLKETIFPSILNSIIVLHHC 153
Query: 147 EDVLPLAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTSKE 206
E ++P++E+L++ +R I ++ A L S + D S I +
Sbjct: 154 ETLIPVSEDLNLVNRLIIAVANNACKEQLT-------SGLLKLDYSFSGTNIE-----PQ 201
Query: 207 TGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPSLSSFQNGNSSK 266
T DWW ++ L+L ++R + ++ + +S L+ Y K + L ++ K
Sbjct: 202 TPLDWWGKSLAVLNLDFFQRVISAVKSKGLIQDVISKILISYTNKSLQGLI-VRDPKLEK 260
Query: 267 SNLSEAD----QRNLIEEIVELLPNE--KGVTQTKFLLRSLRTAMALYASSC---CCASL 317
+ +++ QR ++E IV LLP + + FL L+ +A +S+ C + L
Sbjct: 261 ERVLDSEGKKKQRLIVETIVRLLPTQGRRSSVPMAFLSSLLKMVIATSSSASTGSCRSDL 320
Query: 318 EKRIGFQLDEADLEDLLIP-NIGYSMETIHDIDCVQRMLDHFMIVDNDDADSTSNNDIVE 376
E+RIG QLD+A LED+LIP N+ + T++DID + R+ F+ +D DD + ++
Sbjct: 321 ERRIGLQLDQAILEDVLIPINLNGTNNTMYDIDSILRIFSIFLNLDEDDEEEEHHHLQFR 380
Query: 377 EERRIVGNCQ-----RATPMTKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLD 431
+E ++ + + + + KV+ LMD+YLAE+A D NL KF +LA ++PD AR +
Sbjct: 381 DETEMIYDFDSPGSPKQSSILKVSKLMDNYLAEIAMDPNLTTSKFIALAELLPDHARIIS 440
Query: 432 DGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFE 491
DG+YRA+DIYLK H + +SE+ ++C+ ++ QKLS EA +HAAQNERLP+++ VQVL+FE
Sbjct: 441 DGLYRAVDIYLKVHPNIKDSERYRLCKTIDSQKLSQEACSHAAQNERLPVQMAVQVLYFE 500
Query: 492 QLKLRTSVAGWFFASDTLENST-----------TLSGNLALLRNDGNTTHNNPVVAFD-- 538
Q++LR +++ + L NS SG ++ N + N + +
Sbjct: 501 QIRLRNAMSSSIGPTQFLFNSNCHQFPQRSGSGAGSGAISPRDNYASVRRENRELKLEVA 560
Query: 539 HMKDRVSELEKECLSMKQDLEKMMKSKGSWNMLLKKL 575
M+ R+++LEK+ +S+KQ+L K + KK+
Sbjct: 561 RMRMRLTDLEKDHISIKQELVKSNPGTKLFKSFAKKI 597
>At5g67440 photoreceptor-interacting protein-like
Length = 579
Score = 295 bits (756), Expect = 4e-80
Identities = 182/506 (35%), Positives = 282/506 (54%), Gaps = 36/506 (7%)
Query: 1 MACLKLGSKSEIFYLYGQSW-LCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKD 59
M +KLGSK + F +T L +DV++ +GD FHLHKFPL+S+S L++ +
Sbjct: 1 MKFMKLGSKPDSFQSDEDCVRYVATELATDVVVIVGDVKFHLHKFPLLSKSARLQKLIAT 60
Query: 60 IPSD--SEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYG 117
+D S+ + + D+PGG AF + A+FCYG+ + L NVV +RCAAE+L+M E
Sbjct: 61 TTTDEQSDDDEIRIPDIPGGPPAFEICAKFCYGMAVTLNAYNVVAVRCAAEYLEMYESIE 120
Query: 118 EGNLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPTLA 176
GNL+ + E FLN + W D++ L+T P +E++ + RC+ S+ LKAA
Sbjct: 121 NGNLVYKMEVFLNSSVLRSWKDSIIVLQTTRSFYPWSEDVKLDVRCLESIALKAA----- 175
Query: 177 ILPLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARH- 235
P+ V + + + DWW +D++ LS+ L+KR + I R
Sbjct: 176 ----MDPARVDWSYTYNRRKLLPPEMNNNSVPRDWWVEDLAELSIDLFKRVVS-TIRRKG 230
Query: 236 -MKPRRVSGSLVYYAKKHIPSLSSFQNGNSSKSNLSEADQRNLIEEIVELLPNEKGVTQT 294
+ P + +L YA K IP + N + ++ E QR+L+E +V +LP+EK
Sbjct: 231 GVLPEVIGEALEVYAAKRIPGFMIQNDDNDDEEDVME--QRSLLETLVSMLPSEKQSVSC 288
Query: 295 KFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLI--PNIGYSMETIHDIDCVQ 352
FL++ L+++++ L +RIG +L+EA++ DLLI P G ET++DID V+
Sbjct: 289 GFLIKLLKSSVSFECGEEERKELSRRIGEKLEEANVGDLLIRAPEGG---ETVYDIDIVE 345
Query: 353 RMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAP-DVNL 411
++D F + + D +D + + + VA L+D YLAE++ + NL
Sbjct: 346 TLIDEF-VTQTEKRDELDCSDDINDSSK-----------ANVAKLIDGYLAEISRIETNL 393
Query: 412 KFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEAST 471
KF ++A + R DG+YRAID++LK H +T+SEK+ +LM+C+KLS EA
Sbjct: 394 STTKFITIAEKVSTFPRQSHDGVYRAIDMFLKQHPGITKSEKKSSSKLMDCRKLSPEACA 453
Query: 472 HAAQNERLPLRVVVQVLFFEQLKLRT 497
HA QNERLPLRVVVQ+LFFEQ++ T
Sbjct: 454 HAVQNERLPLRVVVQILFFEQVRATT 479
>At4g37590 unknown protein
Length = 580
Score = 286 bits (731), Expect = 3e-77
Identities = 189/508 (37%), Positives = 276/508 (54%), Gaps = 57/508 (11%)
Query: 1 MACLKLGSKSEIFYLYGQSW-LCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKD 59
M +KLGSK + F G + S+ L +DVI+ IGD F+LHKFPL+S+S L++ +
Sbjct: 1 MKFMKLGSKPDSFQSEGDNVRYVSSELATDVIVIIGDVKFYLHKFPLLSKSARLQKLITT 60
Query: 60 IPSDSEKSI----------LELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEF 109
S S + +E+ ++PGG +F + A+FCYG+ + L NVV RCAAEF
Sbjct: 61 STSSSNEENQIHHHHHEDEIEIAEIPGGPASFEICAKFCYGMTVTLNAYNVVAARCAAEF 120
Query: 110 LQMSEDYGEGNLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVL 168
L+M E +GNL+ + E FLN I W D++ L+T + P +EEL +T RC+ S+
Sbjct: 121 LEMYETVEKGNLVYKIEVFLNSSILQSWKDSIIVLQTTRALSPYSEELKLTGRCLDSIAS 180
Query: 169 KAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFM 228
+A+ T + S + NG+ + DWW +D+ L + LYKR +
Sbjct: 181 RASIDTSKV-----EWSYTYSKKKNLDNGLRKP---QAVPRDWWVEDLCDLHIDLYKRAL 232
Query: 229 QGAIARHMKPRRVSGSLVY-YAKKHIPSLSSFQNGNSSKSNLSEADQRNLIEEIVELLPN 287
AR V G ++ YA K IP S +SS A R L + I+EL+P+
Sbjct: 233 ATIEARGNVSADVIGEALHAYAIKRIPGFSK----SSSVQVTDFAKYRALADSIIELIPD 288
Query: 288 EKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHD 347
EK + FL + LR ++ L A L+ R+G +LDEA+L D+L+ +D
Sbjct: 289 EKRSVSSSFLTKLLRASIFLGCDEV--AGLKNRVGERLDEANLGDVLL----------YD 336
Query: 348 IDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAP 407
++ +Q +++ F+ + D E++ T VA L+D YLAE +
Sbjct: 337 VELMQSLVEVFL----------KSRDPREDD---------VTAKASVAKLVDGYLAEKSR 377
Query: 408 DV-NLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLS 466
D NL KF SLA ++ R DG+YRAID++LK H M +SEK++ICRLM+C+KLS
Sbjct: 378 DSDNLPLQKFLSLAEMVSSFPRQSHDGVYRAIDMFLKEHPEMNKSEKKRICRLMDCRKLS 437
Query: 467 LEASTHAAQNERLPLRVVVQVLFFEQLK 494
EA HA QNERLP+RVVVQVLFFEQ++
Sbjct: 438 AEACAHAVQNERLPMRVVVQVLFFEQVR 465
>At2g14820 hypothetical protein
Length = 634
Score = 280 bits (716), Expect = 2e-75
Identities = 179/504 (35%), Positives = 284/504 (55%), Gaps = 18/504 (3%)
Query: 1 MACLKLGSKSEIFYLYGQSW-LCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKD 59
M +K+GSK + F G + L SD+ +++ + F LHKFPL+S+ L++ +
Sbjct: 1 MKFMKIGSKLDSFKTDGNNVRYVENELASDISVDVEGSRFCLHKFPLLSKCACLQKLLSS 60
Query: 60 IPSDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEG 119
++ I ++ +PGG AF A+FCYG+ + L+ NVV RCAAE+L M E +G
Sbjct: 61 TDKNNIDDI-DISGIPGGPTAFETCAKFCYGMTVTLSAYNVVATRCAAEYLGMHETVEKG 119
Query: 120 NLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPTLAIL 178
NL+ + + FL+ +F W D++ L+T + LPL+E+L + S CI ++ KA + +
Sbjct: 120 NLIYKIDVFLSSSLFRSWKDSIIVLQTTKPFLPLSEDLKLVSLCIDAIATKAC---VDVS 176
Query: 179 PLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKP 238
+ + +E NG S+ +++ DWW +D+ L + YKR + + +
Sbjct: 177 HVEWSYTYNKKKLAEENNGAD-SIKARDVPHDWWVEDLCELEIDYYKRVIMNIKTKCILG 235
Query: 239 RRVSG-SLVYYAKKHIPSLSSFQNGNSSKSNLSEADQRNLIEEIVELLPNEKGVTQTKFL 297
V G +L Y + LS F G + +L + + +IE +V LLP EK FL
Sbjct: 236 GEVIGEALKAYGYRR---LSGFNKGVMEQGDLVK--HKTIIETLVWLLPAEKNSVSCGFL 290
Query: 298 LRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLDH 357
L+ L+ + + L +RIG QL+EA + +LLI + S ET++D+D VQ+++
Sbjct: 291 LKLLKAVTMVNSGEVVKEQLVRRIGQQLEEASMAELLIKSHQGS-ETLYDVDLVQKIVME 349
Query: 358 FMIVD-NDDADSTSNND--IVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFP 414
FM D N + + + D V+E R++ G A+ + VA ++DSYL E+A D NL
Sbjct: 350 FMRRDKNSEIEVQDDEDGFEVQEVRKLPGILSEASKLM-VAKVIDSYLTEIAKDPNLPAS 408
Query: 415 KFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAA 474
KF +A + R D +YRAID++LK H +T+ EK+++C+LM+C+KLS+EA HA
Sbjct: 409 KFIDVAESVTSIPRPAHDALYRAIDMFLKEHPGITKGEKKRMCKLMDCRKLSVEACMHAV 468
Query: 475 QNERLPLRVVVQVLFFEQLKLRTS 498
QN+RLPLRVVVQVLFFEQ++ S
Sbjct: 469 QNDRLPLRVVVQVLFFEQVRAAAS 492
>At5g10250 non-phototropic hypocotyl 3-like protein
Length = 607
Score = 280 bits (715), Expect = 2e-75
Identities = 185/592 (31%), Positives = 302/592 (50%), Gaps = 96/592 (16%)
Query: 18 QSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDIPSDSEKSI-LELHDLPG 76
+SW + +P+D+ I++ D +F HKFPLIS+ + PS SE L+L + PG
Sbjct: 42 RSWYVKSQIPTDLSIQVNDITFKAHKFPLISKCGYISSIELK-PSTSENGYHLKLENFPG 100
Query: 77 GAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNLMIQTENFLNHI-FGQ 135
GA F I +FCY + ++L P NV PLRCA+E+L M+E++ GNL+ +TE F+ +
Sbjct: 101 GADTFETILKFCYNLPLDLNPLNVAPLRCASEYLYMTEEFEAGNLISKTEAFITFVVLAS 160
Query: 136 WTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPTLAILPLSGPSSVQSPDNSEMW 195
W DTL L++C ++ P AE L I RC L KA +
Sbjct: 161 WRDTLTVLRSCTNLSPWAENLQIVRRCCDLLAWKACND---------------------- 198
Query: 196 NGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPRRVSGSLVYYAKKHIPS 255
N I + + E ++D+++L + + R + AR KP+ ++ YA +P
Sbjct: 199 NNIPEDVVDRN--ERCLYNDIATLDIDHFMRVITTMKARRAKPQITGKIIMKYADNFLPV 256
Query: 256 LS------------------SFQNGNSSKSN-LSEADQRNLIEEIVELLPNEKGVTQTKF 296
++ S G +SN L + + IE +V +LP + G F
Sbjct: 257 INDDLEGIKGYGLGKNELQFSVNRGRMEESNSLGCQEHKETIESLVSVLPPQSGAVSCHF 316
Query: 297 LLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLD 356
LLR L+T++ AS + LEKR+G L++A++ DLLIPN + R+ +
Sbjct: 317 LLRMLKTSIVYSASPALISDLEKRVGMALEDANVCDLLIPNFKNEEQQER-----VRIFE 371
Query: 357 HFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKF 416
F++ E+++++G ++ L+D+YLAE+A D L KF
Sbjct: 372 FFLM---------------HEQQQVLGK-------PSISKLLDNYLAEIAKDPYLPITKF 409
Query: 417 QSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQN 476
Q LA ++P+ A DG+YRAID++LK+H +++ ++ ++C+ MNC+KLSL+A HAAQN
Sbjct: 410 QVLAEMLPENAWKCHDGLYRAIDMFLKTHPSLSDHDRRRLCKTMNCEKLSLDACLHAAQN 469
Query: 477 ERLPLRVVV----QVLFFEQLKLRTSVAGWFFASDTL-ENSTTLSGNLALLRNDGNTTHN 531
+RLPLR +V QVLF EQ+K+R D L E SG R D + +
Sbjct: 470 DRLPLRTIVQINTQVLFSEQVKMR------MMMQDKLPEKEEENSGG----REDKRMSRD 519
Query: 532 NPVV-----AFDHMKDRVSELEKECLSMKQDLEKM---MKSKGSWNMLLKKL 575
N ++ +++K ++SEL+ + ++Q+ E++ KS +W + +K+
Sbjct: 520 NEIIKTLKEELENVKKKMSELQSDYNELQQEYERLSSKQKSSHNWGLRWQKV 571
>At5g47800 photoreceptor-interacting protein-like
Length = 559
Score = 270 bits (691), Expect = 1e-72
Identities = 166/496 (33%), Positives = 273/496 (54%), Gaps = 26/496 (5%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFMKDI 60
M +K+G+K + FY S + T P+D++I I + ++HLH+ L+ + L R D+
Sbjct: 1 MKFMKIGTKPDTFYTQEASRILITDTPNDLVIRINNTTYHLHRSCLVPKCGLLRRLCTDL 60
Query: 61 PSDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGN 120
+S+ +EL+D+PGGA AF L A+FCY + + L+ N+V CA++FL+MS+ +GN
Sbjct: 61 -EESDTVTIELNDIPGGADAFELCAKFCYDITINLSAHNLVNALCASKFLRMSDSVDKGN 119
Query: 121 LMIQTENFLNHIFGQ-WTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPTLAILP 179
L+ + E F + Q W D++ L++ + E L I +CI S+V K PT +
Sbjct: 120 LLPKLEAFFHSCILQGWKDSIVTLQSTTKLPEWCENLGIVRKCIDSIVEKILTPTSEV-- 177
Query: 180 LSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIARHMKPR 239
S + P ++ + DWW +D+S L L L++ + A + P
Sbjct: 178 -SWSHTYTRPGYAKRQH--------HSVPRDWWTEDISDLDLDLFRCVITAARSTFTLPP 228
Query: 240 RVSGSLVY-YAKKHIPSL-SSFQNGNSSKSNLSEADQ-RNLIEEIVELLPNEKGVTQTKF 296
++ G ++ Y + +P S+ +G S K N + ++ R L+ +V ++P +KG F
Sbjct: 229 QLIGEALHVYTCRWLPYFKSNSHSGFSVKENEAALERHRRLVNTVVNMIPADKGSVSEGF 288
Query: 297 LLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDIDCVQRMLD 356
LLR + A + AS L ++ QL+EA LEDLL+P+ S +D D V +L+
Sbjct: 289 LLRLVSIASYVRASLTTKTELIRKSSLQLEEATLEDLLLPSHSSSHLHRYDTDLVATVLE 348
Query: 357 HFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPDVNLKFPKF 416
F+++ + + +++ N Q + KVA L+DSYL VA DV++ KF
Sbjct: 349 SFLMLWRRQSSAHLSSN----------NTQLLHSIRKVAKLIDSYLQAVAQDVHMPVSKF 398
Query: 417 QSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLEASTHAAQN 476
SL+ +PD AR D +Y+AI+I+LK H +++ EK+++CR ++CQKLS + HA +N
Sbjct: 399 VSLSEAVPDIARQSHDRLYKAINIFLKVHPEISKEEKKRLCRSLDCQKLSAQVRAHAVKN 458
Query: 477 ERLPLRVVVQVLFFEQ 492
ER+PLR VVQ LFF+Q
Sbjct: 459 ERMPLRTVVQALFFDQ 474
>At3g26490 non-phototropic hypocotyl, putative
Length = 588
Score = 269 bits (687), Expect = 4e-72
Identities = 179/523 (34%), Positives = 284/523 (54%), Gaps = 45/523 (8%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTGLPSDVIIEIGDASFHLHKFPLISRSKELERFM--K 58
M ++LG + + FY S+ L +D++I++ + LHKFP++S+ L+ + +
Sbjct: 1 MKFMELGFRPDTFYTVESVRSVSSDLLNDLVIQVKSTKYLLHKFPMLSKCLRLKNLVSSQ 60
Query: 59 DIPSDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGE 118
+ + E+ +++L D PG +AF L A+FCYG+ + L NVV +RCAAE+L M+E+
Sbjct: 61 ETETSQEQQVIQLVDFPGETEAFELCAKFCYGITITLCAHNVVAVRCAAEYLGMTEEVEL 120
Query: 119 G---NLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPL-AEELHITSRCIHSL-----VL 168
G NL+ + E FL +F W D+ L+T + VLPL +E+L IT+RCI ++ V
Sbjct: 121 GETENLVQRLELFLTTCVFKSWRDSYVTLQTTK-VLPLWSEDLGITNRCIEAIANGVTVS 179
Query: 169 KAADPTLAILPLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFM 228
D + + + + + + NG SK WW +D++ L L LY+R M
Sbjct: 180 PGEDFSTQLETGLLRNRSRIRRDEILCNG---GGGSKAESLRWWGEDLAELGLDLYRRTM 236
Query: 229 QGAIARHMK--PRRVSGSLVYYAKKHIPSLSSFQNGNSSKSNLSEADQRNLIEEIVELLP 286
+ H K PR + +L YA K +PS+ S AD ++E ++ LLP
Sbjct: 237 VAIKSSHRKISPRLIGNALRIYASKWLPSIQE-----------SSADSNLVLESVISLLP 285
Query: 287 NEKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIH 346
EK FLL+ L+ A + S L + G QLD+A + +LLIP + ++
Sbjct: 286 EEKSSVPCSFLLQLLKMANVMNVSHSSKMELAIKAGNQLDKATVSELLIP-LSDKSGMLY 344
Query: 347 DIDCVQRMLDHFMIVDNDDADST-----------SNNDIVEEERRIVGNCQRATP----M 391
D+D V+ M+ F+ + + T N +EE + + G+ ++ +
Sbjct: 345 DVDVVKMMVKQFLSHISPEIRPTRTRTEHRRSRSEENINLEEIQEVRGSLSTSSSPPPLL 404
Query: 392 TKVADLMDSYLAEVAPDVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTES 451
+KVA L+DSYL E+A DVNL KF LA IPD +R D +Y AID+YL+ H + +
Sbjct: 405 SKVAKLVDSYLQEIARDVNLTVSKFVELAETIPDTSRICHDDLYNAIDVYLQVHKKIEKC 464
Query: 452 EKEQICRLMNCQKLSLEASTHAAQNERLPLRVVVQVLFFEQLK 494
E++++CR+++C+KLS+EAS AAQNE LPLRV+VQ+LF EQ +
Sbjct: 465 ERKRLCRILDCKKLSVEASKKAAQNELLPLRVIVQILFVEQAR 507
>At2g23050 unknown protein
Length = 481
Score = 263 bits (672), Expect = 2e-70
Identities = 175/515 (33%), Positives = 272/515 (51%), Gaps = 70/515 (13%)
Query: 1 MACLKLGSKSEIFYLYGQSWLCSTG-LPSDVIIEIGDASFHLHKFPLISRSKELERFMKD 59
M +KLG+K + F G + T L +++II IG+ F+LHKFPL+S+S L++ +
Sbjct: 1 MKFMKLGTKPDSFLSKGDNVRYVTNELETEIIIIIGNVKFYLHKFPLLSKSGFLQKHIAT 60
Query: 60 IPSDSEKSI----LELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSED 115
++ EK +++ ++PGG+ AF + +FCYG+ + L NVV +RCAAEFL+M+E
Sbjct: 61 SKNEEEKKNQIDEIDISEIPGGSVAFEICVKFCYGITVTLNAYNVVAVRCAAEFLEMNET 120
Query: 116 YGEGNLMIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPT 174
+ + NL+ + + FLN IF W D++ L+T +D+L E + RC+ S+ A+ T
Sbjct: 121 FEKSNLVYKIDVFLNSTIFRSWKDSIIVLQTTKDLLSDDSE-ELVKRCLGSIASTASIDT 179
Query: 175 LAIL------PLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFM 228
+ V+ P++ +DWW +D+ L + LYK+ +
Sbjct: 180 SKVKWSYTYNRKKKLEKVRKPEDG--------------VPKDWWVEDLCELHIDLYKQAI 225
Query: 229 QGAIARHMKPRRVSGSLVY-YAKKHIPSLSSFQNGNSSKSNLSEADQRNLIEEIVELLPN 287
+ R P V G ++ YA + I S K ++ D R+LI I+ELLP+
Sbjct: 226 KAIKNRGKVPSNVIGEALHAYAIRRIAGFS--------KESMQLID-RSLINTIIELLPD 276
Query: 288 EKGVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHD 347
EKG + FL + R ++ L L+KR+ QL+E + D+L+ +D
Sbjct: 277 EKGNISSSFLTKLHRASIFLGCEETVKEKLKKRVSEQLEETTVNDILM----------YD 326
Query: 348 IDCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAP 407
+D VQ ++ FM N D + S VA L+D YLAE +
Sbjct: 327 LDMVQSLVKEFM---NRDPKTHSK--------------------VSVAKLIDGYLAEKSR 363
Query: 408 DVNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSL 467
D NL F SLA + R D +YRAID++LK H+ +++SEK+++C LM+C+KLS
Sbjct: 364 DPNLPLQNFLSLAETLSSFPRHSHDVLYRAIDMFLKEHSGISKSEKKRVCGLMDCRKLSA 423
Query: 468 EASTHAAQNERLPLRVVVQVLFFEQLKLRTSVAGW 502
EA HA QNERLP+RV+VQVLFFEQ++ S G+
Sbjct: 424 EACEHAVQNERLPMRVIVQVLFFEQIRANGSSTGY 458
>At4g31820 unknown protein
Length = 518
Score = 234 bits (598), Expect = 8e-62
Identities = 155/450 (34%), Positives = 242/450 (53%), Gaps = 52/450 (11%)
Query: 62 SDSEKSILELHDLPGGAKAFLLIARFCYGVKMELTPSNVVPLRCAAEFLQMSEDYGEGNL 121
S+ + + + D+PGG KAF + A+FCYG+ + L N+ +RCAAE+L+M+ED GNL
Sbjct: 9 SEEKTDEITILDMPGGYKAFEICAKFCYGMTVTLNAYNITAVRCAAEYLEMTEDADRGNL 68
Query: 122 MIQTENFLNH-IFGQWTDTLKALKTCEDVLPLAEELHITSRCIHSLVLKAADPTLAIL-- 178
+ + E FLN IF W D++ L+T +LP +E+L + RCI S+ K I
Sbjct: 69 IYKIEVFLNSGIFRSWKDSIIVLQTTRSLLPWSEDLKLVGRCIDSVSAKILVNPETITWS 128
Query: 179 -----PLSGPSSVQSPDNSEMWNGISMSLTSKETGEDWWFDDVSSLSLPLYKRFMQGAIA 233
LSGP + + + +DWW +DV L + ++KR + +
Sbjct: 129 YTFNRKLSGPDKIVEYHREKREENV--------IPKDWWVEDVCELEIDMFKRVISVVKS 180
Query: 234 R-HMKPRRVSGSLVYYAKKHIPSLSSFQNGNSSKSNLSEADQ-RNLIEEIVELLPNEK-- 289
M ++ +L YY + +P S +S SEA ++L+E +V LLP
Sbjct: 181 SGRMNNGVIAEALRYYVARWLPE--------SMESLTSEASSNKDLVETVVFLLPKVNRA 232
Query: 290 -GVTQTKFLLRSLRTAMALYASSCCCASLEKRIGFQLDEADLEDLLIPNIGYSMETIHDI 348
+ FLL+ L+ ++ + A L + + +L EA ++DLLI H++
Sbjct: 233 MSYSSCSFLLKLLKVSILVGADETVREDLVENVSLKLHEASVKDLLI----------HEV 282
Query: 349 DCVQRMLDHFMIVDNDDADSTSNNDIVEEERRIVGNCQRATPMTKVADLMDSYLAEVAPD 408
+ V R++D FM D S +D +E ++GN + V L+D+YLA +
Sbjct: 283 ELVHRIVDQFMA----DEKRVSEDDRYKEF--VLGN----GILLSVGRLIDAYLAL---N 329
Query: 409 VNLKFPKFQSLAAVIPDCARTLDDGIYRAIDIYLKSHAWMTESEKEQICRLMNCQKLSLE 468
L F L+ ++P+ AR + DG+Y+AID ++K H +T+SEK+++C LM+ +KL+ E
Sbjct: 330 SELTLSSFVELSELVPESARPIHDGLYKAIDTFMKEHPELTKSEKKRLCGLMDVRKLTNE 389
Query: 469 ASTHAAQNERLPLRVVVQVLFFEQLKLRTS 498
ASTHAAQNERLPLRVVVQVL+FEQL+ S
Sbjct: 390 ASTHAAQNERLPLRVVVQVLYFEQLRANHS 419
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,709,788
Number of Sequences: 26719
Number of extensions: 583286
Number of successful extensions: 2001
Number of sequences better than 10.0: 51
Number of HSP's better than 10.0 without gapping: 43
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1774
Number of HSP's gapped (non-prelim): 72
length of query: 613
length of database: 11,318,596
effective HSP length: 105
effective length of query: 508
effective length of database: 8,513,101
effective search space: 4324655308
effective search space used: 4324655308
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148757.5


