

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.9 + phase: 0
(527 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
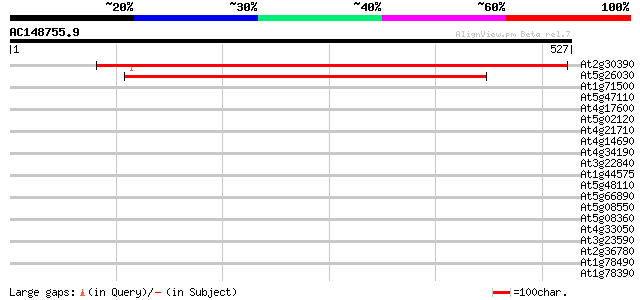
Score E
Sequences producing significant alignments: (bits) Value
At2g30390 putative ferrochelatase precusor 755 0.0
At5g26030 ferrochelatase-I 512 e-145
At1g71500 unknown protein 39 0.007
At5g47110 Lil3 protein 37 0.035
At4g17600 Lil3 protein (Lil3:1) 37 0.035
At5g02120 one helix protein (OHP) 33 0.30
At4g21710 DNA-directed RNA polymerase (EC 2.7.7.6) II second lar... 33 0.30
At4g14690 light induced protein like 33 0.39
At4g34190 unknown protein 32 0.66
At3g22840 early light-induced protein 31 1.9
At1g44575 Photosystem II chlorophyll-binding protein PsbS 31 1.9
At5g48110 terpene synthase 30 4.3
At5g66890 putative protein 29 5.6
At5g08550 putative protein 29 7.3
At5g08360 unknown protein 28 9.6
At4g33050 unknown protein 28 9.6
At3g23590 unknown protein 28 9.6
At2g36780 putative glucosyl transferase 28 9.6
At1g78490 unknown protein 28 9.6
At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase gb|AAF263... 28 9.6
>At2g30390 putative ferrochelatase precusor
Length = 512
Score = 755 bits (1949), Expect = 0.0
Identities = 382/455 (83%), Positives = 408/455 (88%), Gaps = 12/455 (2%)
Query: 82 SKQSLNRHLLPVEA-LVTSTTQDVSDTPLIGDD-----------KIGVLLLNLGGPETLD 129
S + L +H LP+ A LVTS ++S + +I D KIGVLLLNLGGPETLD
Sbjct: 56 SNRLLGKHSLPLRAALVTSNPLNISSSSVISDAISSSSVITDDAKIGVLLLNLGGPETLD 115
Query: 130 DVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPLRRMTDA 189
DVQPFLFNLFADPDIIRLP +F FLQKPLAQF+SV RAPKSKEGYASIGGGSPLR +TDA
Sbjct: 116 DVQPFLFNLFADPDIIRLPPVFQFLQKPLAQFISVARAPKSKEGYASIGGGSPLRHITDA 175
Query: 190 QAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRDGITKLVVLPLYPQFSISTSGS 249
QAEELRK L+EKNVPA VYVGMRYWHPFTEEAIE IKRDGITKLVVLPLYPQFSISTSGS
Sbjct: 176 QAEELRKCLWEKNVPAKVYVGMRYWHPFTEEAIEQIKRDGITKLVVLPLYPQFSISTSGS 235
Query: 250 SLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIEKELKGFDLPEKVMIFFSAH 309
SLRLLE IFREDEYLVNMQHTVIPSWYQREGYIKAMANLI+ EL F P +V+IFFSAH
Sbjct: 236 SLRLLERIFREDEYLVNMQHTVIPSWYQREGYIKAMANLIQSELGKFGSPNQVVIFFSAH 295
Query: 310 GVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETI 369
GVP+AYVEEAGDPYKAEMEECVDLIMEEL+KRKITNAYTLAYQSRVGPVEWLKPYT+E I
Sbjct: 296 GVPLAYVEEAGDPYKAEMEECVDLIMEELDKRKITNAYTLAYQSRVGPVEWLKPYTEEAI 355
Query: 370 IELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISD 429
ELGKKGV++LLAVPISFVSEHIETLEEIDVEYKELAL+SGI+NWGRVPALG EP FISD
Sbjct: 356 TELGKKGVENLLAVPISFVSEHIETLEEIDVEYKELALKSGIKNWGRVPALGTEPMFISD 415
Query: 430 LADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKS 489
LADAV+ESLPYVGAMAVSNLEARQSLVPLGSVEELLA YDSQRRELP P+ +WEWGWTKS
Sbjct: 416 LADAVVESLPYVGAMAVSNLEARQSLVPLGSVEELLATYDSQRRELPAPVTMWEWGWTKS 475
Query: 490 AETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGILP 524
AETWNGRAAM+AVL LL LEVTTG+GFLHQWGILP
Sbjct: 476 AETWNGRAAMLAVLALLVLEVTTGKGFLHQWGILP 510
>At5g26030 ferrochelatase-I
Length = 466
Score = 512 bits (1318), Expect = e-145
Identities = 244/340 (71%), Positives = 298/340 (86%)
Query: 109 LIGDDKIGVLLLNLGGPETLDDVQPFLFNLFADPDIIRLPRLFSFLQKPLAQFVSVLRAP 168
++ +DKIGVLLLNLGGPETL+DVQPFL+NLFADPDIIRLPR F FLQ +A+F+SV+RAP
Sbjct: 84 VVAEDKIGVLLLNLGGPETLNDVQPFLYNLFADPDIIRLPRPFQFLQGTIAKFISVVRAP 143
Query: 169 KSKEGYASIGGGSPLRRMTDAQAEELRKSLFEKNVPANVYVGMRYWHPFTEEAIELIKRD 228
KSKEGYA+IGGGSPLR++TD QA+ ++ SL KN+ ANVYVGMRYW+PFTEEA++ IK+D
Sbjct: 144 KSKEGYAAIGGGSPLRKITDEQADAIKMSLQAKNIAANVYVGMRYWYPFTEEAVQQIKKD 203
Query: 229 GITKLVVLPLYPQFSISTSGSSLRLLESIFREDEYLVNMQHTVIPSWYQREGYIKAMANL 288
IT+LVVLPLYPQ+SIST+GSS+R+L+ +FR+D YL + +I SWYQR GY+ +MA+L
Sbjct: 204 KITRLVVLPLYPQYSISTTGSSIRVLQDLFRKDPYLAGVPVAIIKSWYQRRGYVNSMADL 263
Query: 289 IEKELKGFDLPEKVMIFFSAHGVPVAYVEEAGDPYKAEMEECVDLIMEELEKRKITNAYT 348
IEKEL+ F P++VMIFFSAHGVPV+YVE AGDPY+ +MEEC+DLIMEEL+ R + N +
Sbjct: 264 IEKELQTFSDPKEVMIFFSAHGVPVSYVENAGDPYQKQMEECIDLIMEELKARGVLNDHK 323
Query: 349 LAYQSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALE 408
LAYQSRVGPV+WLKPYTDE +++LGK GVKSLLAVP+SFVSEHIETLEEID+EY+ELALE
Sbjct: 324 LAYQSRVGPVQWLKPYTDEVLVDLGKSGVKSLLAVPVSFVSEHIETLEEIDMEYRELALE 383
Query: 409 SGIENWGRVPALGCEPTFISDLADAVIESLPYVGAMAVSN 448
SG+ENWGRVPALG P+FI+DLADAVIESLP AM+ N
Sbjct: 384 SGVENWGRVPALGLTPSFITDLADAVIESLPSAEAMSNPN 423
>At1g71500 unknown protein
Length = 287
Score = 38.9 bits (89), Expect = 0.007
Identities = 20/40 (50%), Positives = 26/40 (65%)
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGIL 523
+G+TK E NG+AA+I LLLL E+ TG+G L G L
Sbjct: 237 FGFTKKNEVINGKAAVIGFLLLLDFELLTGKGLLKGTGFL 276
>At5g47110 Lil3 protein
Length = 258
Score = 36.6 bits (83), Expect = 0.035
Identities = 20/48 (41%), Positives = 25/48 (51%), Gaps = 6/48 (12%)
Query: 479 ILVWEWGWTK-----SAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWG 521
I+ W W W K AE NGRAAMI + F++ TG G + Q G
Sbjct: 155 IIPW-WAWMKRYHLPEAELLNGRAAMIGFFMAYFVDSLTGVGLVDQMG 201
>At4g17600 Lil3 protein (Lil3:1)
Length = 262
Score = 36.6 bits (83), Expect = 0.035
Identities = 20/48 (41%), Positives = 25/48 (51%), Gaps = 6/48 (12%)
Query: 479 ILVWEWGWTK-----SAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWG 521
I+ W W W K AE NGRAAMI + F++ TG G + Q G
Sbjct: 158 IIPW-WAWIKRYHLPEAELLNGRAAMIGFFMAYFVDSLTGVGLVDQMG 204
>At5g02120 one helix protein (OHP)
Length = 110
Score = 33.5 bits (75), Expect = 0.30
Identities = 14/38 (36%), Positives = 23/38 (59%)
Query: 485 GWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
G+T++AE WN RA MI ++ +E+ +G L G+
Sbjct: 63 GFTQTAEIWNSRACMIGLIGTFIVELILNKGILELIGV 100
>At4g21710 DNA-directed RNA polymerase (EC 2.7.7.6) II second
largest chain
Length = 1188
Score = 33.5 bits (75), Expect = 0.30
Identities = 37/150 (24%), Positives = 63/150 (41%), Gaps = 32/150 (21%)
Query: 386 SFVSEHIETLEEIDVEY--KELALESGIE------NWGRVPALGCEPTFISDLADAVIES 437
S+V + ++ +E+++++ K + SG++ NWG+ A G V+
Sbjct: 422 SYVQKCVDNGKEVNLQFAIKAKTITSGLKYSLATGNWGQANAAGTRAG-----VSQVLNR 476
Query: 438 LPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWGWTKSAETWNGRA 497
L Y A +S+L S P+G +L P + +WG AET G+A
Sbjct: 477 LTY--ASTLSHLRRLNS--PIGREGKLAK---------PRQLHNSQWGMMCPAETPEGQA 523
Query: 498 AMIAVLLLLFLEVTTGEG------FLHQWG 521
+ L L + +T G FL +WG
Sbjct: 524 CGLVKNLALMVYITVGSAAYPILEFLEEWG 553
>At4g14690 light induced protein like
Length = 193
Score = 33.1 bits (74), Expect = 0.39
Identities = 14/27 (51%), Positives = 17/27 (62%)
Query: 487 TKSAETWNGRAAMIAVLLLLFLEVTTG 513
T AE WNGR AM+ ++ L F E TG
Sbjct: 163 TSDAELWNGRFAMLGLVALAFTEYVTG 189
>At4g34190 unknown protein
Length = 146
Score = 32.3 bits (72), Expect = 0.66
Identities = 11/32 (34%), Positives = 21/32 (65%)
Query: 491 ETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+ W GR AM+ + + +E++TG+G L +G+
Sbjct: 85 DIWLGRGAMVGFAVAITVEISTGKGLLENFGV 116
>At3g22840 early light-induced protein
Length = 195
Score = 30.8 bits (68), Expect = 1.9
Identities = 13/27 (48%), Positives = 16/27 (59%)
Query: 487 TKSAETWNGRAAMIAVLLLLFLEVTTG 513
T AE WNGR AM+ ++ L F E G
Sbjct: 165 TSDAELWNGRFAMLGLVALAFTEFVKG 191
>At1g44575 Photosystem II chlorophyll-binding protein PsbS
Length = 265
Score = 30.8 bits (68), Expect = 1.9
Identities = 31/100 (31%), Positives = 42/100 (42%), Gaps = 10/100 (10%)
Query: 428 SDLADAVIESLPYVGAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEW--- 484
S LA + L G V+ RQS VPL + A + ++ P V +
Sbjct: 21 SPLAQPKVHHLFLSGNSPVALPSRRQSFVPLALFKPKTKA--APKKVEKPKSKVEDGIFG 78
Query: 485 -----GWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQ 519
G+TK+ E + GR AMI L E TG+G L Q
Sbjct: 79 TSGGIGFTKANELFVGRVAMIGFAASLLGEALTGKGILAQ 118
Score = 30.4 bits (67), Expect = 2.5
Identities = 15/39 (38%), Positives = 23/39 (58%)
Query: 484 WGWTKSAETWNGRAAMIAVLLLLFLEVTTGEGFLHQWGI 522
+G+TK+ E + GR A + + L E+ TG+G L Q I
Sbjct: 187 FGFTKANELFVGRLAQLGIAFSLIGEIITGKGALAQLNI 225
>At5g48110 terpene synthase
Length = 575
Score = 29.6 bits (65), Expect = 4.3
Identities = 19/57 (33%), Positives = 30/57 (52%), Gaps = 7/57 (12%)
Query: 366 DETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGC 422
DET+++ K K + +++ L+ I +KEL LES I N+ RV A+ C
Sbjct: 252 DETLLKFAKINFKFMQL-------HYVQELQTIVKWWKELDLESKIPNYYRVRAVEC 301
>At5g66890 putative protein
Length = 415
Score = 29.3 bits (64), Expect = 5.6
Identities = 22/84 (26%), Positives = 43/84 (51%), Gaps = 14/84 (16%)
Query: 331 VDLIMEELEKRKITNAYTLAYQSRVGPVEWLKPYTDETIIELGKKGVKSLLAVPISF--- 387
V +I LE K+TN L+ + + + K +++++ K G+KSL + + F
Sbjct: 183 VIIINHGLEPAKLTNLSCLSSLPNLKRIRFEK--VSISLLDIPKLGLKSLEKLSLWFCHV 240
Query: 388 ---------VSEHIETLEEIDVEY 402
VSE +++L+EI+++Y
Sbjct: 241 VDALNELEDVSETLQSLQEIEIDY 264
>At5g08550 putative protein
Length = 908
Score = 28.9 bits (63), Expect = 7.3
Identities = 30/110 (27%), Positives = 47/110 (42%), Gaps = 10/110 (9%)
Query: 377 VKSLLAVPISFVSEHIETLEEIDVEYKELALESGIENWGRVPALGCEPTFISDLADAVIE 436
++ ++V F+ +EEI+ + KEL AL I+D D +IE
Sbjct: 415 LRDFISVICDFMQNKGSLIEEIEDQMKEL---------NEKHALSILERRIADNNDEMIE 465
Query: 437 SLPYV-GAMAVSNLEARQSLVPLGSVEELLAAYDSQRRELPPPILVWEWG 485
V AM V N S V + LAA S R+++ P+ + E+G
Sbjct: 466 LGAAVKAAMTVLNKHGSSSSVIAAATGAALAASTSIRQQMNQPVKLDEFG 515
>At5g08360 unknown protein
Length = 186
Score = 28.5 bits (62), Expect = 9.6
Identities = 19/69 (27%), Positives = 32/69 (45%), Gaps = 3/69 (4%)
Query: 59 VNPLKTCIVGK---FSPGWSEAQPLVSKQSLNRHLLPVEALVTSTTQDVSDTPLIGDDKI 115
+N L+T + + FS WS A + S+Q+LN++ L +L + + +
Sbjct: 58 LNTLRTSDLSENSWFSITWSPATQIPSRQTLNQYFLTYHSLTPVIPETIPKGTEVELPAF 117
Query: 116 GVLLLNLGG 124
GVL L G
Sbjct: 118 GVLTTKLSG 126
>At4g33050 unknown protein
Length = 488
Score = 28.5 bits (62), Expect = 9.6
Identities = 34/120 (28%), Positives = 52/120 (43%), Gaps = 23/120 (19%)
Query: 132 QPFLFNLFA----DPDIIRLPRLFSFLQKPLAQFVSVLRAPKSKEGYASIGGGSPL---R 184
QPF + L D ++ + PR S LQK +++ P +E Y I L +
Sbjct: 222 QPFFYWLDIGDGKDVNLEKHPR--SVLQKQCIRYLG----PMEREAYEVIVEDGRLMYKQ 275
Query: 185 RMTDAQAEELRKSLFEKNVPANVYVGMRYWHPF----------TEEAIELIKRDGITKLV 234
MT + E KS+F + N+YVG++ F T A L+ RDGI + +
Sbjct: 276 GMTLINSTEEAKSIFVLSTTRNLYVGIKKKGLFQHSSFLSGGATTAAGRLVARDGILEAI 335
>At3g23590 unknown protein
Length = 1309
Score = 28.5 bits (62), Expect = 9.6
Identities = 11/28 (39%), Positives = 20/28 (71%)
Query: 111 GDDKIGVLLLNLGGPETLDDVQPFLFNL 138
G D++ ++LL+LGG +T+D F+ +L
Sbjct: 1280 GKDELAIVLLSLGGLKTMDYAADFIIHL 1307
>At2g36780 putative glucosyl transferase
Length = 496
Score = 28.5 bits (62), Expect = 9.6
Identities = 23/89 (25%), Positives = 45/89 (49%), Gaps = 16/89 (17%)
Query: 250 SLRLLESIFREDEYLVNMQHTVIPSWYQREGY------IKAMANLIEKELKGFDLPEKVM 303
+L +LE++ ++EY + +PS+ R + +KA A+ KE+ + E V
Sbjct: 167 NLEILENVKSDEEYFL------VPSFPDRVEFTKLQLPVKANASGDWKEI----MDEMVK 216
Query: 304 IFFSAHGVPVAYVEEAGDPYKAEMEECVD 332
++++GV V +E PY + +E +D
Sbjct: 217 AEYTSYGVIVNTFQELEPPYVKDYKEAMD 245
>At1g78490 unknown protein
Length = 479
Score = 28.5 bits (62), Expect = 9.6
Identities = 25/119 (21%), Positives = 49/119 (41%), Gaps = 9/119 (7%)
Query: 374 KKGVKSLLAVPISFVSEHIETLEEIDVEYKELA-----LESGIENWGRVPALGCEPTFIS 428
++G S A+ + F+S+ + L E+ E+K + E+G+ +W +S
Sbjct: 287 REGTSSCTALAVKFISKDPKVLAELKREHKAIVDNRKDKEAGV-SWEEYRHNMTFTNMVS 345
Query: 429 DLADAVIESLPYVGAMAVSNLEARQSLVPLG---SVEELLAAYDSQRRELPPPILVWEW 484
+ + + P + AV ++E + +P G +V +D E P W W
Sbjct: 346 NEVLRLANTTPLLFRKAVQDVEIKGYTIPAGWIVAVAPSAVHFDPAIYENPFEFNPWRW 404
>At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase
gb|AAF26356.1
Length = 657
Score = 28.5 bits (62), Expect = 9.6
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 7/51 (13%)
Query: 360 WLKPYTDETIIELGKKGVKSLLAVPISFVSEHIETLEEIDVEYKELALESG 410
W +P TDE ++ + S + P S +EH ETL+ + E + L L++G
Sbjct: 476 WEEPETDEVVV------IGSCMTPPDSIFNEHDETLQSVLSEIR-LNLKTG 519
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.137 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,961,825
Number of Sequences: 26719
Number of extensions: 514732
Number of successful extensions: 1320
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 1304
Number of HSP's gapped (non-prelim): 24
length of query: 527
length of database: 11,318,596
effective HSP length: 104
effective length of query: 423
effective length of database: 8,539,820
effective search space: 3612343860
effective search space used: 3612343860
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148755.9


