

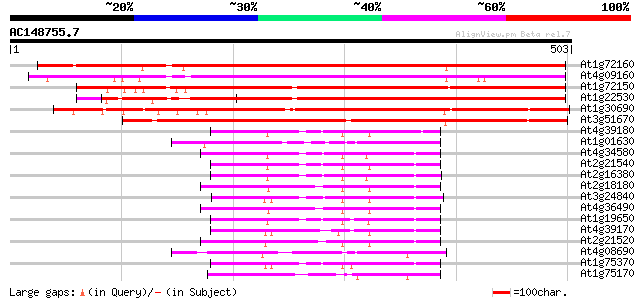
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.7 - phase: 0
(503 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g72160 cytosolic factor, putative 475 e-134
At4g09160 putative protein 445 e-125
At1g72150 putative cytosolic factor protein 429 e-120
At1g22530 unknown protein 415 e-116
At1g30690 putative phosphatidyl-inositol-transfer protein 368 e-102
At3g51670 unknown protein 340 1e-93
At4g39180 SEC14 - like protein 84 1e-16
At1g01630 polyphosphoinositide binding protein, putative 81 1e-15
At4g34580 putative protein 81 2e-15
At2g21540 putative phosphatidylinositol/phophatidylcholine trans... 80 2e-15
At2g16380 putative phosphatidylinositol/phosphatidylcholine tran... 78 1e-14
At2g18180 putative phosphatidylinositol/phophatidylcholine trans... 77 2e-14
At3g24840 phosphatidylinositol transfer protein like 76 4e-14
At4g36490 hypothetical protein 74 2e-13
At1g19650 sec14 cytosolic factor- like protein 74 2e-13
At4g39170 unknown protein 73 4e-13
At2g21520 putative phosphatidylinositol/phosphatidylcholine tran... 72 7e-13
At4g08690 putative phosphoglyceride transfer protein 70 3e-12
At1g75370 sec14 cytosolic factor, putative 70 3e-12
At1g75170 unknown protein 69 6e-12
>At1g72160 cytosolic factor, putative
Length = 490
Score = 475 bits (1223), Expect = e-134
Identities = 247/483 (51%), Positives = 325/483 (67%), Gaps = 17/483 (3%)
Query: 26 PQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNNENNSLQELQNLIQQAFNNHAFSAPP 85
P+K +P S +S+ + T + + + E N + + A PP
Sbjct: 13 PEKLPSPSLTPSEVSESTQDALPTETETLEKV--TETNPPETADTTTKPEEETAAEHHPP 70
Query: 86 LIKEQKQSTTTTVAAEPAQENKYQLEDKKENV---VSSVEDDGAKTVEAIEESIVAVSAS 142
+ E + ++T + K E+KK + + S +++ +K + ++
Sbjct: 71 TVTETETASTEKQEVKDEASQKEVAEEKKSMIPQNLGSFKEESSKLSDLSNSEKKSLD-- 128
Query: 143 VPPEQKPVVEKV--EASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMI 200
E K +V + PE+V I+GIPLL D+ SDV+LLKFLRAR+FKVK++F M+
Sbjct: 129 ---ELKHLVREALDNHQFTNTPEEVKIWGIPLLEDDRSDVVLLKFLRAREFKVKDSFAML 185
Query: 201 KNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSD 260
KNTI WRKEF I+EL++E L D+L+KVV+MHG D+EGHPVCYN+YGEFQNKELYNKTFSD
Sbjct: 186 KNTIKWRKEFKIDELVEEDLVDDLDKVVFMHGHDREGHPVCYNVYGEFQNKELYNKTFSD 245
Query: 261 EEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLF 320
EEKR +FL+ RIQFLE+SIR LDF+ GGV TI VND+K+SPG GK ELR ATKQA++L
Sbjct: 246 EEKRKHFLRTRIQFLERSIRKLDFSSGGVSTIFQVNDMKNSPGLGKKELRSATKQAVELL 305
Query: 321 QDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQL 380
QDNYPEFV KQ FINVPWWYL +I PF+T R+KSK VFAGPS+S ETL YI+PEQ+
Sbjct: 306 QDNYPEFVFKQAFINVPWWYLVFYTVIGPFMTPRSKSKLVFAGPSRSAETLFKYISPEQV 365
Query: 381 PVKYGGLSKD-----GEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRY 435
PV+YGGLS D +F DS +EIT++P +K TVE + EKC L WE+RV GWEV Y
Sbjct: 366 PVQYGGLSVDPCDCNPDFSLEDSASEITVKPGTKQTVEIIIYEKCELVWEIRVTGWEVSY 425
Query: 436 GAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRL 495
AEFVP + +YTV++QK RK+ S+E VL +SFK+NE GKV+LT+DN +S+KKKL+YR
Sbjct: 426 KAEFVPEEKDAYTVVIQKPRKMRPSDEPVLTHSFKVNELGKVLLTVDNPTSKKKKLVYRF 485
Query: 496 KTK 498
K
Sbjct: 486 NVK 488
>At4g09160 putative protein
Length = 723
Score = 445 bits (1144), Expect = e-125
Identities = 252/566 (44%), Positives = 341/566 (59%), Gaps = 95/566 (16%)
Query: 18 QPLYSHPQPQKKNNP----VTLQSHLSKPN-TEQEQTFNKPSDNIPNNENNSLQELQNLI 72
QP P P+ + + + + S +PN ++ + +PS + + E+++
Sbjct: 163 QPRGVTPTPETETSEADTSLLVTSETEEPNHAAEDYSETEPSQKLMLEQRRKYMEVEDWT 222
Query: 73 QQAFNNHA-FSAPPLIKEQKQ-----------STTTTVA----AEPAQENKYQLEDKKE- 115
+ + A A + E KQ +TT+TVA AE + ++E+K++
Sbjct: 223 EPELPDEAVLEAAASVPEPKQPEPQTPPPPPSTTTSTVASRSLAEMMNREEAEVEEKQKI 282
Query: 116 ---NVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIYGIPL 172
+ S +++ K + E + A+ E + +++ + S + SI+G+PL
Sbjct: 283 QIPRSLGSFKEETNKISDLSETELNALQ-----ELRHLLQVSQDS-----SKTSIWGVPL 332
Query: 173 LADETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHG 232
L D+ +DV+LLKFLRARDFK +EA++M+ T+ WR +F IEEL+DE LGD+L+KVV+M G
Sbjct: 333 LKDDRTDVVLLKFLRARDFKPQEAYSMLNKTLQWRIDFNIEELLDENLGDDLDKVVFMQG 392
Query: 233 FDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTI 292
DKE HPVCYN+YGEFQNK+LY KTFSDEEKR FL+WRIQFLEKSIRNLDF GGV TI
Sbjct: 393 QDKENHPVCYNVYGEFQNKDLYQKTFSDEEKRERFLRWRIQFLEKSIRNLDFVAGGVSTI 452
Query: 293 VHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLT 352
VNDLK+SPGPGK ELR ATKQAL L QDNYPEFV+KQ+FINVPWWYLA R+ISPF++
Sbjct: 453 CQVNDLKNSPGPGKTELRLATKQALHLLQDNYPEFVSKQIFINVPWWYLAFYRIISPFMS 512
Query: 353 QRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSKD-----GEFGNSDSVTEITIRPA 407
QR+KSK VFAGPS+S ETLL YI+PE +PV+YGGLS D +F + D TEIT++P
Sbjct: 513 QRSKSKLVFAGPSRSAETLLKYISPEHVPVQYGGLSVDNCECNSDFTHDDIATEITVKPT 572
Query: 408 SKHTVEFPVTEK-----------------CLLSW-------------------------- 424
+K TVE V E C+ W
Sbjct: 573 TKQTVEIIVYEVRPFCIKTFYTTFSNAHFCVCVWKITNCGFNTKYLRFSYVLKSSYICAS 632
Query: 425 ------------EVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKIN 472
E+RV+GWEV YGAEFVP N+ YTVI+QK RK+ + E V+ +SFK+
Sbjct: 633 KHFFLQKCTIVWEIRVVGWEVSYGAEFVPENKEGYTVIIQKPRKMTAKNELVVSHSFKVG 692
Query: 473 EPGKVVLTIDNTSSRKKKLLYRLKTK 498
E G+++LT+DN +S KK L+YR K K
Sbjct: 693 EVGRILLTVDNPTSTKKMLIYRFKVK 718
Score = 28.5 bits (62), Expect = 9.0
Identities = 24/101 (23%), Positives = 40/101 (38%), Gaps = 19/101 (18%)
Query: 10 TTPHVYVPQPLYSHPQPQKKNNPVTLQSHLSKPNTEQEQTFNKPSDNIPNN--------- 60
+ P P+P P P + V +S N E+ + K IP +
Sbjct: 237 SVPEPKQPEPQTPPPPPSTTTSTVASRSLAEMMNREEAEVEEKQKIQIPRSLGSFKEETN 296
Query: 61 --------ENNSLQELQNLIQ--QAFNNHAFSAPPLIKEQK 91
E N+LQEL++L+Q Q + + PL+K+ +
Sbjct: 297 KISDLSETELNALQELRHLLQVSQDSSKTSIWGVPLLKDDR 337
>At1g72150 putative cytosolic factor protein
Length = 573
Score = 429 bits (1104), Expect = e-120
Identities = 239/489 (48%), Positives = 319/489 (64%), Gaps = 59/489 (12%)
Query: 61 ENNSLQELQNLIQQAFNNHAFSAPPL---------------------IKEQKQSTTTTVA 99
+ +L+E + L+++A N F+AP +E+K+ TTT V
Sbjct: 93 QKKALEEFKELVREALNKREFTAPVTPVKEEKTEEKKTEEETKEEEKTEEKKEETTTEVK 152
Query: 100 AE------PAQENKYQLE------------DKKENVV----SSVEDDGAKTVEAIEESIV 137
E PA E + E ++K V SS E+DG KTVEAIEESIV
Sbjct: 153 VEEEKPAVPAAEEEKSSEAAPVETKSEEKPEEKAEVTTEKASSAEEDGTKTVEAIEESIV 212
Query: 138 AVSASVPPEQK--PVVEKVEA---SLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFK 192
+VS PPE PVV + A + P+ PE+VSI+G+PLL DE SDVIL KFLRARDFK
Sbjct: 213 SVS---PPESAVAPVVVETVAVAEAEPVEPEEVSIWGVPLLQDERSDVILTKFLRARDFK 269
Query: 193 VKEAFTMIKNTILWRKEFGIEELMDE-KLGDELEKVVYMHGFDKEGHPVCYNIYGEFQNK 251
VKEA TM+KNT+ WRKE I+EL++ + E EK+V+ HG DKEGH V Y+ YGEFQNK
Sbjct: 270 VKEALTMLKNTVQWRKENKIDELVESGEEVSEFEKMVFAHGVDKEGHVVIYSSYGEFQNK 329
Query: 252 ELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDF-NHGGVCTIVHVNDLKDSPGPGKWELR 310
EL FSD+EK + FL WRIQ EK +R +DF N + V V+D +++PG GK L
Sbjct: 330 EL----FSDKEKLNKFLSWRIQLQEKCVRAIDFSNPEAKSSFVFVSDFRNAPGLGKRALW 385
Query: 311 QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ-RTKSKFVFAGPSKSTE 369
Q ++A++ F+DNYPEF AK++FINVPWWY+ + +T RT+SK V AGPSKS +
Sbjct: 386 QFIRRAVKQFEDNYPEFAAKELFINVPWWYIPYYKTFGSIITSPRTRSKMVLAGPSKSAD 445
Query: 370 TLLSYIAPEQLPVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVI 429
T+ YIAPEQ+PVKYGGLSKD +++TE ++PA+ +T+E P +E C LSWE+RV+
Sbjct: 446 TIFKYIAPEQVPVKYGGLSKDTPL-TEETITEAIVKPAANYTIELPASEACTLSWELRVL 504
Query: 430 GWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKK 489
G +V YGA+F P+ EGSY VIV K RK+ S++E V+ +SFK+ EPGK+V+TIDN +S+KK
Sbjct: 505 GADVSYGAQFEPTTEGSYAVIVSKTRKIGSTDEPVITDSFKVGEPGKIVITIDNQTSKKK 564
Query: 490 KLLYRLKTK 498
K+LYR KT+
Sbjct: 565 KVLYRFKTQ 573
>At1g22530 unknown protein
Length = 683
Score = 415 bits (1067), Expect = e-116
Identities = 222/418 (53%), Positives = 290/418 (69%), Gaps = 21/418 (5%)
Query: 83 APPLIKEQKQSTTTTVAAEPAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSAS 142
A IK+ S TT+ E +E + E ++ +++ KTVEA+EESIV+++
Sbjct: 284 ASKFIKDIFVSVTTS---EKKKEEEKPAVVTIEKAFAADQEEETKTVEAVEESIVSITL- 339
Query: 143 VPPEQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKN 202
PE VE PE+VSI+GIPLL DE SDVILLKFLRARDFKVKEAFTM+KN
Sbjct: 340 --PETAAYVE---------PEEVSIWGIPLLEDERSDVILLKFLRARDFKVKEAFTMLKN 388
Query: 203 TILWRKEFGIEELMDEKL-GDELEKVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDE 261
T+ WRKE I++L+ E L G E EK+V+ HG DK+GH V Y+ YGEFQNKE+ FSD+
Sbjct: 389 TVQWRKENKIDDLVSEDLEGSEFEKLVFTHGVDKQGHVVIYSSYGEFQNKEI----FSDK 444
Query: 262 EKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQ 321
EK FLKWRIQF EK +R+LDF+ + V V+D +++PG G+ L Q K+A++ F+
Sbjct: 445 EKLSKFLKWRIQFQEKCVRSLDFSPEAKSSFVFVSDFRNAPGLGQRALWQFIKRAVKQFE 504
Query: 322 DNYPEFVAKQVFINVPWWYLAVNRMISPFLTQ-RTKSKFVFAGPSKSTETLLSYIAPEQL 380
DNYPEFVAK++FINVPWWY+ + +T RT+SK V +GPSKS ET+ Y+APE +
Sbjct: 505 DNYPEFVAKELFINVPWWYIPYYKTFGSIITSPRTRSKMVLSGPSKSAETIFKYVAPEVV 564
Query: 381 PVKYGGLSKDGEFGNSDSVTEITIRPASKHTVEFPVTEKCLLSWEVRVIGWEVRYGAEFV 440
PVKYGGLSKD F D VTE ++ SK+T++ P TE LSWE+RV+G +V YGA+F
Sbjct: 565 PVKYGGLSKDSPFTVEDGVTEAVVKSTSKYTIDLPATEGSTLSWELRVLGADVSYGAQFE 624
Query: 441 PSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKLLYRLKTK 498
PSNE SYTVIV K RKV ++E V+ +SFK +E GKVV+TIDN + +KKK+LYR KT+
Sbjct: 625 PSNEASYTVIVSKNRKVGLTDEPVITDSFKASEAGKVVITIDNQTFKKKKVLYRSKTQ 682
Score = 48.5 bits (114), Expect = 8e-06
Identities = 42/150 (28%), Positives = 69/150 (46%), Gaps = 14/150 (9%)
Query: 61 ENNSLQELQNLIQQAFNNHAFSAPP----LIKEQKQSTTTTVAAEPAQENKYQLEDKKEN 116
E N+L EL+ L+++A N F+APP +KE+K T E +E + + E+K
Sbjct: 94 EKNALAELKELVREALNKREFTAPPPPPAPVKEEKVEEKKTEETEEKKE-EVKTEEKSLE 152
Query: 117 VVSSVEDDGA--KTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLPPEQVSIY-GIPLL 173
+ E+ A TVE +E I+A A + V E + P+ P V P++
Sbjct: 153 AETKEEEKSAAPATVETKKEEILAAPAPI------VAETKKEETPVAPAPVETKPAAPVV 206
Query: 174 ADETSDVILLKFLRARDFKVKEAFTMIKNT 203
A+ + IL + KV+E ++ T
Sbjct: 207 AETKKEEILPAAPVTTETKVEEKVVPVETT 236
>At1g30690 putative phosphatidyl-inositol-transfer protein
Length = 540
Score = 368 bits (944), Expect = e-102
Identities = 213/507 (42%), Positives = 316/507 (62%), Gaps = 54/507 (10%)
Query: 40 SKPN-TEQEQTFNKPSD---NIPNNENNSLQELQNLIQQAFNNHAF-------SAPPLIK 88
SKP E+ +F + SD ++ +E +L +L++ +++A ++ S+P +K
Sbjct: 43 SKPEGVEKSASFKEESDFFADLKESEKKALSDLKSKLEEAIVDNTLLKTKKKESSP--MK 100
Query: 89 EQKQSTTTTVAA-----EPAQENKYQLEDKKENVVSSVEDDGAKTVEAI-------EESI 136
E+K+ A E A E K + E K E VV+ E A+TVEA+ +E +
Sbjct: 101 EKKEEVVKPEAEVEKKKEEAAEEKVEEEKKSEAVVTE-EAPKAETVEAVVTEEIIPKEEV 159
Query: 137 VAVSASVPPEQKP--------VVEKVEA-SLPLPPEQVSI------YGIPLLAD---ETS 178
V V E K V E+V+A ++ + E S+ +G+PLL E++
Sbjct: 160 TTVVEKVEEETKEEEKKTEDVVTEEVKAETIEVEDEDESVDKDIELWGVPLLPSKGAEST 219
Query: 179 DVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYMHGFDKEGH 238
DVILLKFLRARDFKV EAF M+K T+ WRK+ I+ ++ E+ G++L YM+G D+E H
Sbjct: 220 DVILLKFLRARDFKVNEAFEMLKKTLKWRKQNKIDSILGEEFGEDLATAAYMNGVDRESH 279
Query: 239 PVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDL 298
PVCYN++ E ELY +T E+ R FL+WR Q +EK I+ L+ GGV +++ ++DL
Sbjct: 280 PVCYNVHSE----ELY-QTIGSEKNREKFLRWRFQLMEKGIQKLNLKPGGVTSLLQIHDL 334
Query: 299 KDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSK 358
K++PG + E+ K+ ++ QDNYPEFV++ +FINVP+W+ A+ ++SPFLTQRTKSK
Sbjct: 335 KNAPGVSRTEIWVGIKKVIETLQDNYPEFVSRNIFINVPFWFYAMRAVLSPFLTQRTKSK 394
Query: 359 FVFAGPSKSTETLLSYIAPEQLPVKYGGLS--KDGEFGNSDSVTEITIRPASKHTVEFPV 416
FV A P+K ETLL YI ++LPV+YGG D EF N ++V+E+ ++P S T+E P
Sbjct: 395 FVVARPAKVRETLLKYIPADELPVQYGGFKTVDDTEFSN-ETVSEVVVKPGSSETIEIPA 453
Query: 417 TE-KCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKARKVASSEEAVLCNSFKINEPG 475
E + L W++ V+GWEV Y EFVP+ EG+YTVIVQK +K+ ++E + NSFK ++ G
Sbjct: 454 PETEGTLVWDIAVLGWEVNYKEEFVPTEEGAYTVIVQKVKKMGANEGPIR-NSFKNSQAG 512
Query: 476 KVVLTIDNTSSRKKKLLYRLKTKTSLS 502
K+VLT+DN S +KKK+LYR +TKT S
Sbjct: 513 KIVLTVDNVSGKKKKVLYRYRTKTESS 539
>At3g51670 unknown protein
Length = 409
Score = 340 bits (872), Expect = 1e-93
Identities = 186/406 (45%), Positives = 261/406 (63%), Gaps = 14/406 (3%)
Query: 102 PAQENKYQLEDKKENVVSSVEDDGAKTVEAIEESIVAVSASVPPEQKPVVEKVEASLPLP 161
P K Q + K++ ++S+ + I+E VS P EQK + E E
Sbjct: 7 PFDHQKTQNTEPKKSFITSLITLRSNN---IKEDTYFVSELKPTEQKSLQELKEKLSASS 63
Query: 162 PEQVSIYGIPLLA-DETSDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKL 220
+ S++G+ LL D+ +DVILLKFLRARDFKV ++ M++ + WR+EF E+L +E L
Sbjct: 64 SKASSMWGVSLLGGDDKADVILLKFLRARDFKVADSLRMLEKCLEWREEFKAEKLTEEDL 123
Query: 221 G-DELE-KVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKS 278
G +LE KV YM G+DKEGHPVCYN YG F+ KE+Y + F DEEK + FL+WR+Q LE+
Sbjct: 124 GFKDLEGKVAYMRGYDKEGHPVCYNAYGVFKEKEMYERVFGDEEKLNKFLRWRVQVLERG 183
Query: 279 IRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDNYPEFVAKQVFINVPW 338
++ L F GGV +I+ V DLKD P K ELR A+ Q L LFQDNYPE VA ++FINVPW
Sbjct: 184 VKMLHFKPGGVNSIIQVTDLKDMP---KRELRVASNQILSLFQDNYPELVATKIFINVPW 240
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK--DGEFGNS 396
++ + M SPFLTQRTKSKFV + + ETL +I PE +PV+YGGLS+ D + G
Sbjct: 241 YFSVIYSMFSPFLTQRTKSKFVMSKEGNAAETLYKFIRPEDIPVQYGGLSRPTDSQNGPP 300
Query: 397 DSVTEITIRPASKHTVEFPVTE-KCLLSWEVRVIGWEVRYGAEFVPSNEGSYTVIVQKAR 455
+E +I+ K ++ E ++W++ V GW++ Y AEFVP+ E SY ++V+K +
Sbjct: 301 KPASEFSIKGGEKVNIQIEGIEGGATITWDIVVGGWDLEYSAEFVPNAEESYAIVVEKPK 360
Query: 456 KVASSEEAVLCNSFKINEPGKVVLTIDNTSSRKKKL-LYRLKTKTS 500
K+ +++EAV CNSF E GK++L++DNT SRKKK+ YR + S
Sbjct: 361 KMKATDEAV-CNSFTTVEAGKLILSVDNTLSRKKKVAAYRYTVRKS 405
>At4g39180 SEC14 - like protein
Length = 554
Score = 84.3 bits (207), Expect = 1e-16
Identities = 64/218 (29%), Positives = 103/218 (46%), Gaps = 19/218 (8%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDK 235
++L+FLRAR F +++A M + + WRKE+G + +M++ E+E+VV HG DK
Sbjct: 96 MMLRFLRARKFDLEKAKQMWSDMLNWRKEYGADTIMEDFDFKEIEEVVKYYPQGYHGVDK 155
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
EG P+ G+ +L T D ++K+ ++ EK+ N+ F + H+
Sbjct: 156 EGRPIYIERLGQVDATKLMKVTTID-----RYVKYHVKEFEKTF-NVKFPACSIAAKRHI 209
Query: 296 ND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMIS 348
+ + D G G +A K LQ Q DNYPE + + IN + + +
Sbjct: 210 DQSTTILDVQGVGLSNFNKAAKDLLQSIQKIDNDNYPETLNRMFIINAGCGFRLLWNTVK 269
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T +K G T+ LL I +LP GG
Sbjct: 270 SFLDPKTTAKIHVLGNKYQTK-LLEIIDANELPEFLGG 306
>At1g01630 polyphosphoinositide binding protein, putative
Length = 255
Score = 81.3 bits (199), Expect = 1e-15
Identities = 69/253 (27%), Positives = 117/253 (45%), Gaps = 26/253 (10%)
Query: 146 EQKPVVEKVEASLPLPPEQVSIYGIPLL----------ADETSDVILLKFLRARDFKVKE 195
+Q+P + ++PL +++ + ++ E D+++ +FLRARD +++
Sbjct: 7 KQEPAAAAEQKTVPLIEDEIERSKVGIMRALCDRQDPETKEVDDLMIRRFLRARDLDIEK 66
Query: 196 AFTMIKNTILWRKEFGIE-ELMDEKLGDELE-KVVYMHGFDKEGHPVCYNIYGEFQNKEL 253
A TM N + W++ + + + ++ ++L + M G DK G P+ I N+
Sbjct: 67 ASTMFLNYLTWKRSMLPKGHIPEAEIANDLSHNKMCMQGHDKMGRPIAVAI----GNRHN 122
Query: 254 YNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQAT 313
+K DE KR + + LEK + G V + DL+ G ++R
Sbjct: 123 PSKGNPDEFKR-----FVVYTLEKICARMP---RGQEKFVAIGDLQGW-GYSNCDIR-GY 172
Query: 314 KQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLS 373
AL QD YPE + K ++ P+ ++ ++I PF+ TK K VF K T TLL
Sbjct: 173 LAALSTLQDCYPERLGKLYIVHAPYIFMTAWKVIYPFIDANTKKKIVFVENKKLTPTLLE 232
Query: 374 YIAPEQLPVKYGG 386
I QLP YGG
Sbjct: 233 DIDESQLPDIYGG 245
>At4g34580 putative protein
Length = 560
Score = 80.9 bits (198), Expect = 2e-15
Identities = 62/228 (27%), Positives = 111/228 (48%), Gaps = 20/228 (8%)
Query: 172 LLADETSDV-ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY- 229
LL + D+ ++L+FLRAR F +++A M + I WRK+FG + ++++ +E+++V+
Sbjct: 78 LLPSKLDDLHMMLRFLRARKFDIEKAKQMWSDMIQWRKDFGADTIIEDFDFEEIDEVMKH 137
Query: 230 ----MHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN 285
HG DKEG PV G+ +L T D ++K+ ++ EK+ + + F
Sbjct: 138 YPQGYHGVDKEGRPVYIERLGQIDANKLLQVTTMD-----RYVKYHVKEFEKTFK-VKFP 191
Query: 286 HGGVCTIVHVND---LKDSPGPGKWELRQATKQALQ----LFQDNYPEFVAKQVFINVPW 338
V H++ + D G G ++ ++ LQ + +NYPE + + IN
Sbjct: 192 SCSVAANKHIDQSTTILDVQGVGLKNFSKSARELLQRLCKIDNENYPETLNRMFIINAGS 251
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ + + FL +T +K G +K LL I +LP +GG
Sbjct: 252 GFRLLWSTVKSFLDPKTTAKIHVLG-NKYHSKLLEVIDASELPEFFGG 298
>At2g21540 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 371
Score = 80.5 bits (197), Expect = 2e-15
Identities = 62/218 (28%), Positives = 103/218 (46%), Gaps = 19/218 (8%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDK 235
++L+FLRAR F +++A M + I WRKEFG++ +M++ E+++V+ HG DK
Sbjct: 95 MMLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEIDEVLKYYPQGYHGVDK 154
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
+G PV G+ +L T D ++K+ ++ EK+ N+ + H+
Sbjct: 155 DGRPVYIERLGQVDATKLMQVTTID-----RYVKYHVREFEKTF-NIKLPACSIAAKKHI 208
Query: 296 ND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMIS 348
+ + D G G +A + LQ Q DNYPE + + IN + + +
Sbjct: 209 DQSTTILDVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSGFRLLWSTVK 268
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T +K G +K LL I +LP GG
Sbjct: 269 SFLDPKTTAKIHVLG-NKYQSKLLEIIDSNELPEFLGG 305
>At2g16380 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 582
Score = 77.8 bits (190), Expect = 1e-14
Identities = 60/219 (27%), Positives = 103/219 (46%), Gaps = 19/219 (8%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDK 235
++L+FLRAR F ++A M + + WR +FG++ ++++ +E+++V+ HG DK
Sbjct: 86 MMLRFLRARKFDKEKAKQMWSDMLQWRMDFGVDTIIEDFEFEEIDQVLKHYPQGYHGVDK 145
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
EG PV G+ +L T D + K+ ++ EK + + F H+
Sbjct: 146 EGRPVYIERLGQIDANKLLQATTMD-----RYEKYHVKEFEKMFK-IKFPSCSAAAKKHI 199
Query: 296 ND---LKDSPGPGKWELRQATKQALQ----LFQDNYPEFVAKQVFINVPWWYLAVNRMIS 348
+ + D G G ++ ++ LQ + DNYPE + + IN + + I
Sbjct: 200 DQSTTIFDVQGVGLKNFNKSARELLQRLLKIDNDNYPETLNRMFIINAGPGFRLLWAPIK 259
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGL 387
FL +T SK G +K LL I +LP +GGL
Sbjct: 260 KFLDPKTTSKIHVLG-NKYQPKLLEAIDASELPYFFGGL 297
>At2g18180 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 558
Score = 77.4 bits (189), Expect = 2e-14
Identities = 61/228 (26%), Positives = 104/228 (44%), Gaps = 20/228 (8%)
Query: 172 LLADETSDV-ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYM 230
LL D+ D ++L+FL+AR F +++ M + + WRKEFG + +M++ E+++V+
Sbjct: 71 LLPDKHDDYHMMLRFLKARKFDLEKTNQMWSDMLRWRKEFGADTVMEDFEFKEIDEVLKY 130
Query: 231 -----HGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN 285
HG DKEG PV G+ + +L T D ++ +++ F N+ F
Sbjct: 131 YPQGHHGVDKEGRPVYIERLGQVDSTKLMQVTTMDRYVNYHVMEFERTF------NVKFP 184
Query: 286 HGGVCTIVHVND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPW 338
+ H++ + D G G +A + + Q DNYPE + + IN
Sbjct: 185 ACSIAAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGS 244
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ + + FL +T +K G +K LL I +LP GG
Sbjct: 245 GFRMLWNTVKSFLDPKTTAKIHVLG-NKYQSKLLEIIDASELPEFLGG 291
>At3g24840 phosphatidylinositol transfer protein like
Length = 579
Score = 76.3 bits (186), Expect = 4e-14
Identities = 63/220 (28%), Positives = 100/220 (44%), Gaps = 19/220 (8%)
Query: 182 LLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKV--VYMHGF---DKE 236
+L+FL+AR F +++ M + + WRKE G++ ++ + + DE E+V Y HG+ D+E
Sbjct: 104 MLRFLKARRFDLEKTVQMWEEMLKWRKENGVDTIIQDFVYDEYEEVQQYYPHGYHGVDRE 163
Query: 237 GHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVN 296
G PV G+ +L T + FL++ +Q EK+ F + H+N
Sbjct: 164 GRPVYIERLGKIDPGKLMKVTTLE-----RFLRYHVQGFEKTFSE-KFPACSIAAKRHIN 217
Query: 297 D---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPWWYLAVNRMISP 349
+ D G R+ + + Q DNYPE + + IN + V +
Sbjct: 218 SSTTIIDVHGVSWMSFRKLAQDLVMRMQKIDGDNYPETLNQMYIINAGNGFKLVWNTVKG 277
Query: 350 FLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGGLSK 389
FL +T SK G +K LL I P +LP GG K
Sbjct: 278 FLDPKTTSKIHVLG-NKYRSHLLEIIDPSELPEFLGGNCK 316
>At4g36490 hypothetical protein
Length = 543
Score = 73.9 bits (180), Expect = 2e-13
Identities = 61/228 (26%), Positives = 102/228 (43%), Gaps = 20/228 (8%)
Query: 172 LLADETSDV-ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVYM 230
LL ++ D ++L+FL+AR F +++ M + WRKEFG + +M+E E+++V+
Sbjct: 68 LLPEKHDDYHMMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVMEEFDFKEIDEVLKY 127
Query: 231 -----HGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN 285
HG DKEG PV G + +L T D ++ +++ F N+ F
Sbjct: 128 YPQGHHGVDKEGRPVYIERLGLVDSTKLMQVTTMDRYVNYHVMEFERTF------NVKFP 181
Query: 286 HGGVCTIVHVND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPW 338
+ H++ + D G G +A + + Q DNYPE + + IN
Sbjct: 182 ACSIAAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGS 241
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ + + FL +T +K G +K LL I +LP GG
Sbjct: 242 GFRMLWNTVKSFLDPKTTAKIHVLG-NKYQSKLLEIIDESELPEFLGG 288
>At1g19650 sec14 cytosolic factor- like protein
Length = 608
Score = 73.6 bits (179), Expect = 2e-13
Identities = 65/221 (29%), Positives = 102/221 (45%), Gaps = 25/221 (11%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVVY-----MHGFDK 235
I+L+FL AR F + +A M N I WR++FG + ++++ EL++V+ HG DK
Sbjct: 104 IMLRFLFARKFDLGKAKLMWTNMIQWRRDFGTDTILEDFEFPELDEVLRYYPQGYHGVDK 163
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
EG PV G+ +L T + +L++ ++ EK+I + F + H+
Sbjct: 164 EGRPVYIERLGKVDASKLMQVTTLE-----RYLRYHVKEFEKTI-TVKFPACCIAAKRHI 217
Query: 296 ND---LKDSPGPGKWELRQATKQALQLF-------QDNYPEFVAKQVFINVPWWYLAVNR 345
+ + D G G L+ TK A L DNYPE + + IN + +
Sbjct: 218 DSSTTILDVQGLG---LKNFTKTARDLIIQLQKIDSDNYPETLHRMFIINAGSGFKLLWG 274
Query: 346 MISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ FL +T SK G +K LL I QLP +GG
Sbjct: 275 TVKSFLDPKTVSKIHVLG-NKYQNKLLEMIDASQLPDFFGG 314
>At4g39170 unknown protein
Length = 614
Score = 72.8 bits (177), Expect = 4e-13
Identities = 65/224 (29%), Positives = 100/224 (44%), Gaps = 31/224 (13%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV--YMHGF---DK 235
++L+FL+AR F +++A M + I WRKEFG + ++ + +E+++V+ Y HG+ DK
Sbjct: 108 MMLRFLKARKFDIEKAKHMWADMIQWRKEFGTDTIIQDFQFEEIDEVLKYYPHGYHSVDK 167
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIV-- 293
EG PV G+ +L T D R++ ++ F+ K CTI
Sbjct: 168 EGRPVYIERLGKVDPNKLMQVTTLDRYIRYHVKEFERSFMLKF---------PACTIAAK 218
Query: 294 ----HVNDLKDSPGPGKWELRQATKQALQLFQ-------DNYPEFVAKQVFINVPWWYLA 342
+ D G G L+ TK A +L DNYPE + + IN +
Sbjct: 219 KYIDSSTTILDVQGVG---LKNFTKSARELITRLQKIDGDNYPETLHQMFIINAGPGFRL 275
Query: 343 VNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ + FL +T SK G K LL I +LP GG
Sbjct: 276 LWSTVKSFLDPKTTSKIHVLG-CKYQSKLLEIIDSSELPEFLGG 318
>At2g21520 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 531
Score = 72.0 bits (175), Expect = 7e-13
Identities = 62/228 (27%), Positives = 100/228 (43%), Gaps = 20/228 (8%)
Query: 172 LLADETSDV-ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV-- 228
LL D D ++L+FL+AR F V++A M + I WRKEFG + ++ + +E+ +V+
Sbjct: 4 LLPDRHDDYHMMLRFLKARKFDVEKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEVLKH 63
Query: 229 ---YMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFN 285
HG DKEG P+ G+ L T D R++ ++ F+ K F
Sbjct: 64 YPQCYHGVDKEGRPIYIERLGKVDPNRLMQVTSMDRYVRYHVKEFERSFMIK------FP 117
Query: 286 HGGVCTIVHVND---LKDSPGPGKWELRQATKQALQLFQ----DNYPEFVAKQVFINVPW 338
+ H++ + D G G ++ + + Q DNYPE + + IN
Sbjct: 118 SCTISAKRHIDSSTTILDVQGVGLKNFNKSARDLITRLQKIDGDNYPETLHQMFIINAGP 177
Query: 339 WYLAVNRMISPFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ + + FL +T +K G K LL I +LP GG
Sbjct: 178 GFRLLWNTVKSFLDPKTSAKIHVLG-YKYLSKLLEVIDVNELPEFLGG 224
>At4g08690 putative phosphoglyceride transfer protein
Length = 301
Score = 70.1 bits (170), Expect = 3e-12
Identities = 60/250 (24%), Positives = 115/250 (46%), Gaps = 28/250 (11%)
Query: 146 EQKPVVEKVEASLPLPPEQVSIYGIPLLADETSDVILLKFLRARDFKVKEAFTMIKNTIL 205
E++ +E+V L PE++S + SD +L++LRAR++ VK+A M+K T+
Sbjct: 18 EEQAKIEEVRKLLGPLPEKLSSF--------CSDDAVLRYLRARNWHVKKATKMLKETLK 69
Query: 206 WRKEFGIEELMDEKLGDELE--KVVYMHGFDKEGHPVCYNIYGEFQNKELYNKTFSDEEK 263
WR ++ EE+ E++ E E K+ DK G PV + + + +
Sbjct: 70 WRVQYKPEEICWEEVAGEAETGKIYRSSCVDKLGRPVL-----------IMRPSVENSKS 118
Query: 264 RHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHVNDLKDSPGPGKWELRQATKQALQLFQDN 323
+++ + +E +++NL + ++ + LR TK+ + Q++
Sbjct: 119 VKGQIRYLVYCMENAVQNLPPGEEQMVWMIDFHGY----SLANVSLR-TTKETAHVLQEH 173
Query: 324 YPEFVAKQVFINVPWWYLAVNRMISPFLTQRT--KSKFVFAGPSKSTETLLSYIAPEQLP 381
YPE +A V N P ++ ++ PFL +T K KFV++ + + E++
Sbjct: 174 YPERLAFAVLYNPPKFFEPFWKVARPFLEPKTRNKVKFVYSDDPNTKVIMEENFDMEKME 233
Query: 382 VKYGGLSKDG 391
+ +GG G
Sbjct: 234 LAFGGNDDSG 243
>At1g75370 sec14 cytosolic factor, putative
Length = 612
Score = 70.1 bits (170), Expect = 3e-12
Identities = 61/218 (27%), Positives = 99/218 (44%), Gaps = 19/218 (8%)
Query: 181 ILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEELMDEKLGDELEKVV--YMHGF---DK 235
I+L+FL+AR F + + M N I WRK+FG + + ++ +E ++V+ Y HG+ DK
Sbjct: 113 IMLRFLKARKFDIGKTKLMWSNMIKWRKDFGTDTIFEDFEFEEFDEVLKYYPHGYHGVDK 172
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
EG PV G +L T + F+++ ++ EK++ N+ + H+
Sbjct: 173 EGRPVYIERLGLVDPAKLMQVTTVE-----RFIRYHVREFEKTV-NIKLPACCIAAKRHI 226
Query: 296 ND---LKDSPGPG----KWELRQATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMIS 348
+ + D G G R Q ++ DNYPE + + IN + V +
Sbjct: 227 DSSTTILDVQGVGFKNFSKPARDLIIQLQKIDNDNYPETLHRMFIINGGSGFKLVWATVK 286
Query: 349 PFLTQRTKSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
FL +T +K G +K LL I QLP GG
Sbjct: 287 QFLDPKTVTKIHVIG-NKYQNKLLEIIDASQLPDFLGG 323
>At1g75170 unknown protein
Length = 296
Score = 68.9 bits (167), Expect = 6e-12
Identities = 58/218 (26%), Positives = 102/218 (46%), Gaps = 29/218 (13%)
Query: 178 SDVILLKFLRARDFKVKEAFTMIKNTILWRKEFGIEEL-MDEKLGDELEKVVYMHGF-DK 235
SD L ++L AR++ V +A M++ T+ WR F EE+ +E G+ VY GF D+
Sbjct: 43 SDACLKRYLEARNWNVGKAKKMLEETLKWRSSFKPEEIRWNEVSGEGETGKVYKAGFHDR 102
Query: 236 EGHPVCYNIYGEFQNKELYNKTFSDEEKRHNFLKWRIQFLEKSIRNLDFNHGGVCTIVHV 295
G V G K L N+ +K + +E +I NL + + ++
Sbjct: 103 HGRTVLILRPGLQNTKSLENQ-----------MKHLVYLIENAILNLPEDQEQMSWLI-- 149
Query: 296 NDLKDSPGPGKWELR-----QATKQALQLFQDNYPEFVAKQVFINVPWWYLAVNRMISPF 350
D G W + ++ ++ + + Q++YPE +A N P + A +++ F
Sbjct: 150 ----DFTG---WSMSTSVPIKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKIVKYF 202
Query: 351 LTQRT--KSKFVFAGPSKSTETLLSYIAPEQLPVKYGG 386
+ +T K KFV+ S+S E + ++ E LP ++GG
Sbjct: 203 IDAKTFVKVKFVYPKNSESVELMSTFFDEENLPTEFGG 240
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.131 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,986,302
Number of Sequences: 26719
Number of extensions: 549319
Number of successful extensions: 1754
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 35
Number of HSP's successfully gapped in prelim test: 48
Number of HSP's that attempted gapping in prelim test: 1645
Number of HSP's gapped (non-prelim): 103
length of query: 503
length of database: 11,318,596
effective HSP length: 104
effective length of query: 399
effective length of database: 8,539,820
effective search space: 3407388180
effective search space used: 3407388180
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148755.7


