

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.3 - phase: 0
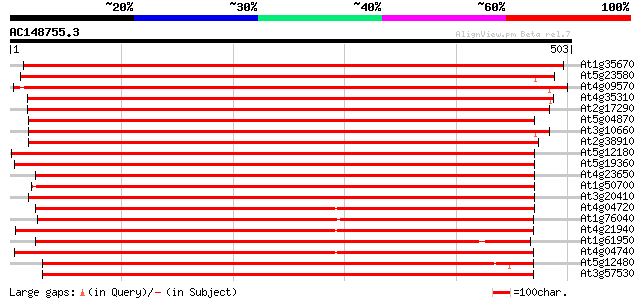
(503 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g35670 calcium-dependent protein kinase 760 0.0
At5g23580 calcium-dependent protein kinase (pir||S71196) 760 0.0
At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4) 759 0.0
At4g35310 calmodulin-domain protein kinase CDPK isoform 5 (CPK5) 686 0.0
At2g17290 calmodulin-domain protein kinase CDPK isoform 6 (CPK6) 683 0.0
At5g04870 calcium-dependent protein kinase 678 0.0
At3g10660 calmodulin-domain protein kinase CDPK isoform 2 678 0.0
At2g38910 putative calcium-dependent protein kinase 657 0.0
At5g12180 calcium-dependent protein kinase 619 e-177
At5g19360 calcium-dependent protein kinase - like 617 e-177
At4g23650 calcium-dependent protein kinase (CDPK6) 597 e-171
At1g50700 hypothetical protein 587 e-168
At3g20410 calmodulin-domain protein kinase CDPK isoform 9 586 e-167
At4g04720 putative calcium dependent protein kinase 583 e-166
At1g76040 calcium-dependent protein kinase (CPK29) 575 e-164
At4g21940 calcium-dependent protein kinase like protein 573 e-164
At1g61950 hypothetical protein 548 e-156
At4g04740 putative calcium dependent protein kinase 544 e-155
At5g12480 calcium-dependent protein kinase - like protein 535 e-152
At3g57530 calcium-dependent protein kinase 532 e-151
>At1g35670 calcium-dependent protein kinase
Length = 495
Score = 760 bits (1963), Expect = 0.0
Identities = 370/486 (76%), Positives = 427/486 (87%), Gaps = 2/486 (0%)
Query: 13 KPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDY 72
+P VLPY+T ++ + Y +G+KLGQGQFGTTYLCT KST+ +ACKSIPKRKL C+EDY
Sbjct: 9 RPSNTVLPYQTPRLRDHYLLGKKLGQGQFGTTYLCTEKSTSANYACKSIPKRKLVCREDY 68
Query: 73 EDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAA 132
EDVWREIQIMHHLSEHP+VVRI+GTYEDS VHIVME+CEGGELFDRIV KGH+SER+A
Sbjct: 69 EDVWREIQIMHHLSEHPNVVRIKGTYEDSVFVHIVMEVCEGGELFDRIVSKGHFSEREAV 128
Query: 133 GLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVV 192
LIKTI+ VVE+CHSLGVMHRDLKPENFLFD+ +DAKLKATDFGLSVFYKPG+ DVV
Sbjct: 129 KLIKTILGVVEACHSLGVMHRDLKPENFLFDSPKDDAKLKATDFGLSVFYKPGQYLYDVV 188
Query: 193 GSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSE 252
GSPYYVAPEVL+K YGPE DVWSAGVILYILLSGVPPFWAETE GIFRQIL GKLDF+S+
Sbjct: 189 GSPYYVAPEVLKKCYGPEIDVWSAGVILYILLSGVPPFWAETESGIFRQILQGKLDFKSD 248
Query: 253 PWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSA 312
PWP+IS++AKDLI KML+++P+ R++AHE L HPWIVD+ APDKP+D AVLSRLKQFS
Sbjct: 249 PWPTISEAAKDLIYKMLERSPKKRISAHEALCHPWIVDEQAAPDKPLDPAVLSRLKQFSQ 308
Query: 313 MNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEI 372
MNK+KKMALRVIAERLSEEEIGGLKELFKMIDTD+SGTITF+ELK GLKRVGSELMESEI
Sbjct: 309 MNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRVGSELMESEI 368
Query: 373 QDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQAC 432
+ LMDAAD+D SGTIDYGEF+AAT+H+NK+EREENL++AF+YFDKD SGYITIDE+ AC
Sbjct: 369 KSLMDAADIDNSGTIDYGEFLAATLHMNKMEREENLVAAFSYFDKDGSGYITIDELQSAC 428
Query: 433 KDFGLDDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGNG-GIGRRTMRN-TLNLRDALGL 490
+FGL D +D+MIKEID DNDG+ID+SEF AMMRKG+G G R M+N N+ DA G+
Sbjct: 429 TEFGLCDTPLDDMIKEIDLDNDGKIDFSEFTAMMRKGDGVGRSRTMMKNLNFNIADAFGV 488
Query: 491 VGNGSN 496
G S+
Sbjct: 489 DGEKSD 494
>At5g23580 calcium-dependent protein kinase (pir||S71196)
Length = 490
Score = 760 bits (1962), Expect = 0.0
Identities = 372/483 (77%), Positives = 429/483 (88%), Gaps = 4/483 (0%)
Query: 10 IALKPVT-WVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFC 68
+A KP T WVLPY+T+ V + Y +G+ LGQGQFGTT+LCTHK T +K ACKSIPKRKL C
Sbjct: 1 MANKPRTRWVLPYKTKNVEDNYFLGQVLGQGQFGTTFLCTHKQTGQKLACKSIPKRKLLC 60
Query: 69 KEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSE 128
+EDY+DV REIQIMHHLSE+P+VVRIE YED+ VH+VMELCEGGELFDRIVK+GHYSE
Sbjct: 61 QEDYDDVLREIQIMHHLSEYPNVVRIESAYEDTKNVHLVMELCEGGELFDRIVKRGHYSE 120
Query: 129 RQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESF 188
R+AA LIKTIV VVE+CHSLGV+HRDLKPENFLF + DEDA LK+TDFGLSVF PGE+F
Sbjct: 121 REAAKLIKTIVGVVEACHSLGVVHRDLKPENFLFSSSDEDASLKSTDFGLSVFCTPGEAF 180
Query: 189 SDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLD 248
S++VGS YYVAPEVL K YGPE DVWSAGVILYILL G PPFWAE+E GIFR+IL GKL+
Sbjct: 181 SELVGSAYYVAPEVLHKHYGPECDVWSAGVILYILLCGFPPFWAESEIGIFRKILQGKLE 240
Query: 249 FQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLK 308
F+ PWPSIS+SAKDLI+KML+ NP+ RLTAH+VL HPWIVDD +APDKP+D AV+SRLK
Sbjct: 241 FEINPWPSISESAKDLIKKMLESNPKKRLTAHQVLCHPWIVDDKVAPDKPLDCAVVSRLK 300
Query: 309 QFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELM 368
+FSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTD SGTITF+ELKD ++RVGSELM
Sbjct: 301 KFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDKSGTITFEELKDSMRRVGSELM 360
Query: 369 ESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEI 428
ESEIQ+L+ AADVD+SGTIDYGEF+AAT+HLNKLEREENL++AF++FDKDASGYITI+E+
Sbjct: 361 ESEIQELLRAADVDESGTIDYGEFLAATIHLNKLEREENLVAAFSFFDKDASGYITIEEL 420
Query: 429 SQACKDFGLDDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN---GGIGRRTMRNTLNLR 485
QA K+FG++D ++DEMIK+IDQDNDGQIDY EF AMMRKGN GGIGRRTMRN+LN
Sbjct: 421 QQAWKEFGINDSNLDEMIKDIDQDNDGQIDYGEFVAMMRKGNGTGGGIGRRTMRNSLNFG 480
Query: 486 DAL 488
L
Sbjct: 481 TTL 483
>At4g09570 calmodulin-domain protein kinase CDPK isoform 4 (CPK4)
Length = 501
Score = 759 bits (1960), Expect = 0.0
Identities = 373/499 (74%), Positives = 429/499 (85%), Gaps = 5/499 (1%)
Query: 4 ENPTPTIALKPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPK 63
E P P +P VLPY T ++ + Y +G+KLGQGQFGTTYLCT KS++ +ACKSIPK
Sbjct: 2 EKPNPR---RPSNSVLPYETPRLRDHYLLGKKLGQGQFGTTYLCTEKSSSANYACKSIPK 58
Query: 64 RKLFCKEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKK 123
RKL C+EDYEDVWREIQIMHHLSEHP+VVRI+GTYEDS VHIVME+CEGGELFDRIV K
Sbjct: 59 RKLVCREDYEDVWREIQIMHHLSEHPNVVRIKGTYEDSVFVHIVMEVCEGGELFDRIVSK 118
Query: 124 GHYSERQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYK 183
G +SER+AA LIKTI+ VVE+CHSLGVMHRDLKPENFLFD+ +DAKLKATDFGLSVFYK
Sbjct: 119 GCFSEREAAKLIKTILGVVEACHSLGVMHRDLKPENFLFDSPSDDAKLKATDFGLSVFYK 178
Query: 184 PGESFSDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQIL 243
PG+ DVVGSPYYVAPEVL+K YGPE DVWSAGVILYILLSGVPPFWAETE GIFRQIL
Sbjct: 179 PGQYLYDVVGSPYYVAPEVLKKCYGPEIDVWSAGVILYILLSGVPPFWAETESGIFRQIL 238
Query: 244 LGKLDFQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAV 303
GK+DF+S+PWP+IS+ AKDLI KMLD++P+ R++AHE L HPWIVD++ APDKP+D AV
Sbjct: 239 QGKIDFKSDPWPTISEGAKDLIYKMLDRSPKKRISAHEALCHPWIVDEHAAPDKPLDPAV 298
Query: 304 LSRLKQFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRV 363
LSRLKQFS MNK+KKMALRVIAERLSEEEIGGLKELFKMIDTD+SGTITF+ELK GLKRV
Sbjct: 299 LSRLKQFSQMNKIKKMALRVIAERLSEEEIGGLKELFKMIDTDNSGTITFEELKAGLKRV 358
Query: 364 GSELMESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYI 423
GSELMESEI+ LMDAAD+D SGTIDYGEF+AAT+H+NK+EREENL+ AF+YFDKD SGYI
Sbjct: 359 GSELMESEIKSLMDAADIDNSGTIDYGEFLAATLHINKMEREENLVVAFSYFDKDGSGYI 418
Query: 424 TIDEISQACKDFGLDDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGNGGIGRRTMRNTL- 482
TIDE+ QAC +FGL D +D+MIKEID DNDG+ID+SEF AMM+KG+G RTMRN L
Sbjct: 419 TIDELQQACTEFGLCDTPLDDMIKEIDLDNDGKIDFSEFTAMMKKGDGVGRSRTMRNNLN 478
Query: 483 -NLRDALGLVGNGSNQVID 500
N+ +A G+ S D
Sbjct: 479 FNIAEAFGVEDTSSTAKSD 497
>At4g35310 calmodulin-domain protein kinase CDPK isoform 5 (CPK5)
Length = 556
Score = 686 bits (1769), Expect = 0.0
Identities = 332/473 (70%), Positives = 396/473 (83%), Gaps = 2/473 (0%)
Query: 17 WVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVW 76
+VL ++T + +IY + RKLGQGQFGTTYLCT ++ +ACKSI KRKL KED EDV
Sbjct: 84 YVLGHKTPNIRDIYTLSRKLGQGQFGTTYLCTEIASGVDYACKSISKRKLISKEDVEDVR 143
Query: 77 REIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIK 136
REIQIMHHL+ H +V I+G YEDS VHIVMELC GGELFDRI+++GHYSER+AA L K
Sbjct: 144 REIQIMHHLAGHGSIVTIKGAYEDSLYVHIVMELCAGGELFDRIIQRGHYSERKAAELTK 203
Query: 137 TIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPY 196
IV VVE+CHSLGVMHRDLKPENFL D+D LKA DFGLSVF+KPG+ F+DVVGSPY
Sbjct: 204 IIVGVVEACHSLGVMHRDLKPENFLLVNKDDDFSLKAIDFGLSVFFKPGQIFTDVVGSPY 263
Query: 197 YVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPS 256
YVAPEVL K YGPE+DVW+AGVILYILLSGVPPFWAET+ GIF +L G +DF+S+PWP
Sbjct: 264 YVAPEVLLKRYGPEADVWTAGVILYILLSGVPPFWAETQQGIFDAVLKGYIDFESDPWPV 323
Query: 257 ISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKL 316
ISDSAKDLIR+ML P RLTAHEVLRHPWI ++ +APD+ +D AVLSRLKQFSAMNKL
Sbjct: 324 ISDSAKDLIRRMLSSKPAERLTAHEVLRHPWICENGVAPDRALDPAVLSRLKQFSAMNKL 383
Query: 317 KKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLM 376
KKMAL+VIAE LSEEEI GL+E+F+ +DTD+SG ITFDELK GL++ GS L ++EI DLM
Sbjct: 384 KKMALKVIAESLSEEEIAGLREMFQAMDTDNSGAITFDELKAGLRKYGSTLKDTEIHDLM 443
Query: 377 DAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFG 436
DAADVD SGTIDY EFIAAT+HLNKLEREE+L++AF YFDKD SG+ITIDE+ QAC + G
Sbjct: 444 DAADVDNSGTIDYSEFIAATIHLNKLEREEHLVAAFQYFDKDGSGFITIDELQQACVEHG 503
Query: 437 LDDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGNGGIGRRTMRNTLN--LRDA 487
+ D+ ++++IKE+DQ+NDG+IDY EF MM+KGN G+GRRTMRN+LN +RDA
Sbjct: 504 MADVFLEDIIKEVDQNNDGKIDYGEFVEMMQKGNAGVGRRTMRNSLNISMRDA 556
>At2g17290 calmodulin-domain protein kinase CDPK isoform 6 (CPK6)
Length = 544
Score = 683 bits (1763), Expect = 0.0
Identities = 329/468 (70%), Positives = 392/468 (83%)
Query: 17 WVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVW 76
+VL ++T + ++Y + RKLGQGQFGTTYLCT +T +ACKSI KRKL KED EDV
Sbjct: 72 YVLGHKTPNIRDLYTLSRKLGQGQFGTTYLCTDIATGVDYACKSISKRKLISKEDVEDVR 131
Query: 77 REIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIK 136
REIQIMHHL+ H ++V I+G YED VHIVMELC GGELFDRI+ +GHYSER+AA L K
Sbjct: 132 REIQIMHHLAGHKNIVTIKGAYEDPLYVHIVMELCAGGELFDRIIHRGHYSERKAAELTK 191
Query: 137 TIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPY 196
IV VVE+CHSLGVMHRDLKPENFL D+D LKA DFGLSVF+KPG+ F DVVGSPY
Sbjct: 192 IIVGVVEACHSLGVMHRDLKPENFLLVNKDDDFSLKAIDFGLSVFFKPGQIFKDVVGSPY 251
Query: 197 YVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPS 256
YVAPEVL K YGPE+DVW+AGVILYILLSGVPPFWAET+ GIF +L G +DF ++PWP
Sbjct: 252 YVAPEVLLKHYGPEADVWTAGVILYILLSGVPPFWAETQQGIFDAVLKGYIDFDTDPWPV 311
Query: 257 ISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKL 316
ISDSAKDLIRKML +P RLTAHEVLRHPWI ++ +APD+ +D AVLSRLKQFSAMNKL
Sbjct: 312 ISDSAKDLIRKMLCSSPSERLTAHEVLRHPWICENGVAPDRALDPAVLSRLKQFSAMNKL 371
Query: 317 KKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLM 376
KKMAL+VIAE LSEEEI GL+ +F+ +DTD+SG ITFDELK GL+R GS L ++EI+DLM
Sbjct: 372 KKMALKVIAESLSEEEIAGLRAMFEAMDTDNSGAITFDELKAGLRRYGSTLKDTEIRDLM 431
Query: 377 DAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFG 436
+AADVD SGTIDY EFIAAT+HLNKLEREE+L+SAF YFDKD SGYITIDE+ Q+C + G
Sbjct: 432 EAADVDNSGTIDYSEFIAATIHLNKLEREEHLVSAFQYFDKDGSGYITIDELQQSCIEHG 491
Query: 437 LDDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGNGGIGRRTMRNTLNL 484
+ D+ ++++IKE+DQDNDG+IDY EF AMM+KGN G+GRRTM+N+LN+
Sbjct: 492 MTDVFLEDIIKEVDQDNDGRIDYEEFVAMMQKGNAGVGRRTMKNSLNI 539
>At5g04870 calcium-dependent protein kinase
Length = 610
Score = 678 bits (1750), Expect = 0.0
Identities = 325/453 (71%), Positives = 386/453 (84%)
Query: 18 VLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWR 77
VL +TE E Y +GRKLGQGQFGTT+LC K+T K+FACKSI KRKL ED EDV R
Sbjct: 138 VLQRKTENFKEFYSLGRKLGQGQFGTTFLCVEKTTGKEFACKSIAKRKLLTDEDVEDVRR 197
Query: 78 EIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKT 137
EIQIMHHL+ HP+V+ I+G YED AVH+VME C GGELFDRI+++GHY+ER+AA L +T
Sbjct: 198 EIQIMHHLAGHPNVISIKGAYEDVVAVHLVMECCAGGELFDRIIQRGHYTERKAAELTRT 257
Query: 138 IVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYY 197
IV VVE+CHSLGVMHRDLKPENFLF + ED+ LK DFGLS+F+KP + F+DVVGSPYY
Sbjct: 258 IVGVVEACHSLGVMHRDLKPENFLFVSKHEDSLLKTIDFGLSMFFKPDDVFTDVVGSPYY 317
Query: 198 VAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSI 257
VAPEVLRK YGPE+DVWSAGVI+YILLSGVPPFWAETE GIF Q+L G LDF S+PWPSI
Sbjct: 318 VAPEVLRKRYGPEADVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSI 377
Query: 258 SDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLK 317
S+SAKDL+RKML ++P+ RLTAH+VL HPW+ D +APDKP+DSAVLSR+KQFSAMNK K
Sbjct: 378 SESAKDLVRKMLVRDPKKRLTAHQVLCHPWVQVDGVAPDKPLDSAVLSRMKQFSAMNKFK 437
Query: 318 KMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMD 377
KMALRVIAE LSEEEI GLKE+F MID D SG ITF+ELK GLKRVG+ L ESEI DLM
Sbjct: 438 KMALRVIAESLSEEEIAGLKEMFNMIDADKSGQITFEELKAGLKRVGANLKESEILDLMQ 497
Query: 378 AADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL 437
AADVD SGTIDY EFIAAT+HLNK+ERE++L +AF YFDKD SGYIT DE+ QAC++FG+
Sbjct: 498 AADVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFTYFDKDGSGYITPDELQQACEEFGV 557
Query: 438 DDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
+D+ I+E+++++DQDNDG+IDY+EF AMM+KG+
Sbjct: 558 EDVRIEELMRDVDQDNDGRIDYNEFVAMMQKGS 590
>At3g10660 calmodulin-domain protein kinase CDPK isoform 2
Length = 646
Score = 678 bits (1749), Expect = 0.0
Identities = 327/469 (69%), Positives = 396/469 (83%), Gaps = 2/469 (0%)
Query: 18 VLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWR 77
VL +TE E Y +GRKLGQGQFGTT+LC K T ++ACKSI KRKL ED EDV R
Sbjct: 174 VLQRKTENFKEFYSLGRKLGQGQFGTTFLCLEKGTGNEYACKSISKRKLLTDEDVEDVRR 233
Query: 78 EIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKT 137
EIQIMHHL+ HP+V+ I+G YED AVH+VMELC GGELFDRI+++GHY+ER+AA L +T
Sbjct: 234 EIQIMHHLAGHPNVISIKGAYEDVVAVHLVMELCSGGELFDRIIQRGHYTERKAAELART 293
Query: 138 IVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYY 197
IV V+E+CHSLGVMHRDLKPENFLF + +ED+ LK DFGLS+F+KP E F+DVVGSPYY
Sbjct: 294 IVGVLEACHSLGVMHRDLKPENFLFVSREEDSLLKTIDFGLSMFFKPDEVFTDVVGSPYY 353
Query: 198 VAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSI 257
VAPEVLRK YGPESDVWSAGVI+YILLSGVPPFWAETE GIF Q+L G LDF S+PWPSI
Sbjct: 354 VAPEVLRKRYGPESDVWSAGVIVYILLSGVPPFWAETEQGIFEQVLHGDLDFSSDPWPSI 413
Query: 258 SDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLK 317
S+SAKDL+RKML ++P+ RLTAH+VL HPW+ D +APDKP+DSAVLSR+KQFSAMNK K
Sbjct: 414 SESAKDLVRKMLVRDPKRRLTAHQVLCHPWVQIDGVAPDKPLDSAVLSRMKQFSAMNKFK 473
Query: 318 KMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMD 377
KMALRVIAE LSEEEI GLK++FKMID D+SG ITF+ELK GLKRVG+ L ESEI DLM
Sbjct: 474 KMALRVIAESLSEEEIAGLKQMFKMIDADNSGQITFEELKAGLKRVGANLKESEILDLMQ 533
Query: 378 AADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL 437
AADVD SGTIDY EFIAAT+HLNK+ERE++L +AF+YFDKD SG+IT DE+ QAC++FG+
Sbjct: 534 AADVDNSGTIDYKEFIAATLHLNKIEREDHLFAAFSYFDKDESGFITPDELQQACEEFGV 593
Query: 438 DDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN--GGIGRRTMRNTLNL 484
+D I+EM++++DQD DG+IDY+EF AMM+KG+ GG + + N++++
Sbjct: 594 EDARIEEMMRDVDQDKDGRIDYNEFVAMMQKGSIMGGPVKMGLENSISI 642
>At2g38910 putative calcium-dependent protein kinase
Length = 583
Score = 657 bits (1696), Expect = 0.0
Identities = 317/458 (69%), Positives = 386/458 (84%), Gaps = 1/458 (0%)
Query: 18 VLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWR 77
VL +TE + +IY +GRKLGQGQFGTT+LC K T K+FACK+I KRKL ED EDV R
Sbjct: 122 VLGRKTENLKDIYSVGRKLGQGQFGTTFLCVDKKTGKEFACKTIAKRKLTTPEDVEDVRR 181
Query: 78 EIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKT 137
EIQIMHHLS HP+V++I G YED+ AVH+VME+C GGELFDRI+++GHY+E++AA L +
Sbjct: 182 EIQIMHHLSGHPNVIQIVGAYEDAVAVHVVMEICAGGELFDRIIQRGHYTEKKAAELARI 241
Query: 138 IVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYY 197
IV V+E+CHSLGVMHRDLKPENFLF + DE+A LK DFGLSVF+KPGE+F+DVVGSPYY
Sbjct: 242 IVGVIEACHSLGVMHRDLKPENFLFVSGDEEAALKTIDFGLSVFFKPGETFTDVVGSPYY 301
Query: 198 VAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSI 257
VAPEVLRK Y E DVWSAGVI+YILLSGVPPFW ETE GIF Q+L G LDF SEPWPS+
Sbjct: 302 VAPEVLRKHYSHECDVWSAGVIIYILLSGVPPFWDETEQGIFEQVLKGDLDFISEPWPSV 361
Query: 258 SDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLK 317
S+SAKDL+R+ML ++P+ R+T HEVL HPW D +A DKP+DSAVLSRL+QFSAMNKLK
Sbjct: 362 SESAKDLVRRMLIRDPKKRMTTHEVLCHPWARVDGVALDKPLDSAVLSRLQQFSAMNKLK 421
Query: 318 KMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMD 377
K+A++VIAE LSEEEI GLKE+FKMIDTD+SG IT +ELK GL RVG++L +SEI LM
Sbjct: 422 KIAIKVIAESLSEEEIAGLKEMFKMIDTDNSGHITLEELKKGLDRVGADLKDSEILGLMQ 481
Query: 378 AADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL 437
AAD+D SGTIDYGEFIAA VHLNK+E+E++L +AF+YFD+D SGYIT DE+ QACK FGL
Sbjct: 482 AADIDNSGTIDYGEFIAAMVHLNKIEKEDHLFTAFSYFDQDGSGYITRDELQQACKQFGL 541
Query: 438 DDIHIDEMIKEIDQDNDGQIDYSEFAAMMR-KGNGGIG 474
D+H+D++++E+D+DNDG+IDYSEF MM+ G G +G
Sbjct: 542 ADVHLDDILREVDKDNDGRIDYSEFVDMMQDTGFGKMG 579
>At5g12180 calcium-dependent protein kinase
Length = 528
Score = 619 bits (1596), Expect = e-177
Identities = 299/470 (63%), Positives = 374/470 (78%), Gaps = 1/470 (0%)
Query: 2 AKENPTPTIALKPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSI 61
A +P P P+ VL E V Y +G++LG+GQFG T+LCT K+T +FACK+I
Sbjct: 45 APPSPPPATKQGPIGPVLGRPMEDVKASYSLGKELGRGQFGVTHLCTQKATGHQFACKTI 104
Query: 62 PKRKLFCKEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIV 121
KRKL KED EDV RE+QIMHHL+ P++V ++G YED +VH+VMELC GGELFDRI+
Sbjct: 105 AKRKLVNKEDIEDVRREVQIMHHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRII 164
Query: 122 KKGHYSERQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVF 181
KGHYSER AA L++TIV++V +CHS+GV+HRDLKPENFL DE++ LKATDFGLSVF
Sbjct: 165 AKGHYSERAAASLLRTIVQIVHTCHSMGVIHRDLKPENFLLLNKDENSPLKATDFGLSVF 224
Query: 182 YKPGESFSDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQ 241
YKPGE F D+VGS YY+APEVL++ YGPE+D+WS GV+LYILL GVPPFWAE+E GIF
Sbjct: 225 YKPGEVFKDIVGSAYYIAPEVLKRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNA 284
Query: 242 ILLGKLDFQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDS 301
IL G +DF S+PWPSIS AKDL++KML+ +P+ RLTA +VL HPWI +D APD P+D+
Sbjct: 285 ILRGHVDFSSDPWPSISPQAKDLVKKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDN 344
Query: 302 AVLSRLKQFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLK 361
AV+SRLKQF AMN KK+ALRVIA LSEEEI GLKE+FK +DTDSSGTIT +EL+ GL
Sbjct: 345 AVMSRLKQFKAMNNFKKVALRVIAGCLSEEEIMGLKEMFKGMDTDSSGTITLEELRQGLA 404
Query: 362 RVGSELMESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASG 421
+ G+ L E E+Q LM+AAD D +GTIDYGEFIAAT+H+N+L+REE+L SAF +FDKD SG
Sbjct: 405 KQGTRLSEYEVQQLMEAADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSG 464
Query: 422 YITIDEISQACKDFGLDD-IHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
YIT++E+ QA ++FG++D I E+I E+D DNDG+I+Y EF AMMRKGN
Sbjct: 465 YITMEELEQALREFGMNDGRDIKEIISEVDGDNDGRINYDEFVAMMRKGN 514
>At5g19360 calcium-dependent protein kinase - like
Length = 523
Score = 617 bits (1591), Expect = e-177
Identities = 297/467 (63%), Positives = 373/467 (79%), Gaps = 1/467 (0%)
Query: 5 NPTPTIALKPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKR 64
+P P P+ VL E V Y +G++LG+GQFG T+LCT K+T +FACK+I KR
Sbjct: 43 SPPPATKQGPIGPVLGRPMEDVKSSYTLGKELGRGQFGVTHLCTQKATGLQFACKTIAKR 102
Query: 65 KLFCKEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKG 124
KL KED EDV RE+QIMHHL+ P++V ++G YED +VH+VMELC GGELFDRI+ KG
Sbjct: 103 KLVNKEDIEDVRREVQIMHHLTGQPNIVELKGAYEDKHSVHLVMELCAGGELFDRIIAKG 162
Query: 125 HYSERQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKP 184
HYSER AA L++TIV+++ +CHS+GV+HRDLKPENFL + DE++ LKATDFGLSVFYKP
Sbjct: 163 HYSERAAASLLRTIVQIIHTCHSMGVIHRDLKPENFLLLSKDENSPLKATDFGLSVFYKP 222
Query: 185 GESFSDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILL 244
GE F D+VGS YY+APEVLR+ YGPE+D+WS GV+LYILL GVPPFWAE+E GIF IL
Sbjct: 223 GEVFKDIVGSAYYIAPEVLRRKYGPEADIWSIGVMLYILLCGVPPFWAESENGIFNAILS 282
Query: 245 GKLDFQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVL 304
G++DF S+PWP IS AKDL+RKML+ +P+ RLTA +VL HPWI +D APD P+D+AV+
Sbjct: 283 GQVDFSSDPWPVISPQAKDLVRKMLNSDPKQRLTAAQVLNHPWIKEDGEAPDVPLDNAVM 342
Query: 305 SRLKQFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVG 364
SRLKQF AMN KK+ALRVIA LSEEEI GLKE+FK +DTD+SGTIT +EL+ GL + G
Sbjct: 343 SRLKQFKAMNNFKKVALRVIAGCLSEEEIMGLKEMFKGMDTDNSGTITLEELRQGLAKQG 402
Query: 365 SELMESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYIT 424
+ L E E+Q LM+AAD D +GTIDYGEFIAAT+H+N+L+REE+L SAF +FDKD SGYIT
Sbjct: 403 TRLSEYEVQQLMEAADADGNGTIDYGEFIAATMHINRLDREEHLYSAFQHFDKDNSGYIT 462
Query: 425 IDEISQACKDFGLDD-IHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
+E+ QA ++FG++D I E+I E+D DNDG+I+Y EF AMMRKGN
Sbjct: 463 TEELEQALREFGMNDGRDIKEIISEVDGDNDGRINYEEFVAMMRKGN 509
>At4g23650 calcium-dependent protein kinase (CDPK6)
Length = 529
Score = 597 bits (1539), Expect = e-171
Identities = 286/448 (63%), Positives = 364/448 (80%), Gaps = 1/448 (0%)
Query: 24 EKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMH 83
E+V Y GR+LG+GQFG TYL THK T ++ ACKSIP R+L K+D EDV RE+QIMH
Sbjct: 72 EEVRRTYEFGRELGRGQFGVTYLVTHKETKQQVACKSIPTRRLVHKDDIEDVRREVQIMH 131
Query: 84 HLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVE 143
HLS H ++V ++G YED +V+++MELCEGGELFDRI+ KG YSER AA L + +V VV
Sbjct: 132 HLSGHRNIVDLKGAYEDRHSVNLIMELCEGGELFDRIISKGLYSERAAADLCRQMVMVVH 191
Query: 144 SCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEVL 203
SCHS+GVMHRDLKPENFLF + DE++ LKATDFGLSVF+KPG+ F D+VGS YYVAPEVL
Sbjct: 192 SCHSMGVMHRDLKPENFLFLSKDENSPLKATDFGLSVFFKPGDKFKDLVGSAYYVAPEVL 251
Query: 204 RKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAKD 263
++ YGPE+D+WSAGVILYILLSGVPPFW E E GIF IL G+LDF ++PWP++SD AKD
Sbjct: 252 KRNYGPEADIWSAGVILYILLSGVPPFWGENETGIFDAILQGQLDFSADPWPALSDGAKD 311
Query: 264 LIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALRV 323
L+RKML +P+ RLTA EVL HPWI +D A DKP+D+AVLSR+KQF AMNKLKKMAL+V
Sbjct: 312 LVRKMLKYDPKDRLTAAEVLNHPWIREDGEASDKPLDNAVLSRMKQFRAMNKLKKMALKV 371
Query: 324 IAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVDK 383
IAE LSEEEI GLKE+FK +DTD++G +T +EL+ GL ++GS++ E+EI+ LM+AAD+D
Sbjct: 372 IAENLSEEEIIGLKEMFKSLDTDNNGIVTLEELRTGLPKLGSKISEAEIRQLMEAADMDG 431
Query: 384 SGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL-DDIHI 442
G+IDY EFI+AT+H+N++ERE++L +AF +FD D SGYIT++E+ A K + + DD I
Sbjct: 432 DGSIDYLEFISATMHMNRIEREDHLYTAFQFFDNDNSGYITMEELELAMKKYNMGDDKSI 491
Query: 443 DEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
E+I E+D D DG+I+Y EF AMM+KGN
Sbjct: 492 KEIIAEVDTDRDGKINYEEFVAMMKKGN 519
>At1g50700 hypothetical protein
Length = 521
Score = 587 bits (1512), Expect = e-168
Identities = 286/452 (63%), Positives = 356/452 (78%), Gaps = 3/452 (0%)
Query: 20 PYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREI 79
PY E V Y + ++LG+GQFG TYLCT KST K+FACKSI K+KL K D ED+ REI
Sbjct: 65 PY--EDVKLFYTLSKELGRGQFGVTYLCTEKSTGKRFACKSISKKKLVTKGDKEDMRREI 122
Query: 80 QIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIV 139
QIM HLS P++V +G YED AV++VMELC GGELFDRI+ KGHYSER AA + + IV
Sbjct: 123 QIMQHLSGQPNIVEFKGAYEDEKAVNLVMELCAGGELFDRILAKGHYSERAAASVCRQIV 182
Query: 140 EVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVA 199
VV CH +GVMHRDLKPENFL + DE A +KATDFGLSVF + G + D+VGS YYVA
Sbjct: 183 NVVNICHFMGVMHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGRVYKDIVGSAYYVA 242
Query: 200 PEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISD 259
PEVL++ YG E D+WSAG+ILYILLSGVPPFWAETE GIF IL G++DF+S+PWPSIS+
Sbjct: 243 PEVLKRRYGKEIDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGEIDFESQPWPSISN 302
Query: 260 SAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKM 319
SAKDL+R+ML Q+P+ R++A EVL+HPW+ + A DKPIDSAVLSR+KQF AMNKLKK+
Sbjct: 303 SAKDLVRRMLTQDPKRRISAAEVLKHPWLREGGEASDKPIDSAVLSRMKQFRAMNKLKKL 362
Query: 320 ALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAA 379
AL+VIAE + EEI GLK +F IDTD+SGTIT++ELK+GL ++GS L E+E++ LMDAA
Sbjct: 363 ALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSRLTEAEVKQLMDAA 422
Query: 380 DVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL-D 438
DVD +G+IDY EFI AT+H ++LE EN+ AF +FDKD SGYIT DE+ A K++G+ D
Sbjct: 423 DVDGNGSIDYIEFITATMHRHRLESNENVYKAFQHFDKDGSGYITTDELEAALKEYGMGD 482
Query: 439 DIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
D I E++ ++D DNDG+I+Y EF AMMR GN
Sbjct: 483 DATIKEILSDVDADNDGRINYDEFCAMMRSGN 514
Score = 31.2 bits (69), Expect = 1.4
Identities = 21/57 (36%), Positives = 27/57 (46%), Gaps = 5/57 (8%)
Query: 445 MIKEIDQDNDGQIDYSEFAAMMRKGNGGIGRR-TMRNTLNLRDALGLVGNGSNQVID 500
M ID DN G I Y E +++G +G R T L DA + GNGS I+
Sbjct: 382 MFANIDTDNSGTITYEE----LKEGLAKLGSRLTEAEVKQLMDAADVDGNGSIDYIE 434
>At3g20410 calmodulin-domain protein kinase CDPK isoform 9
Length = 541
Score = 586 bits (1510), Expect = e-167
Identities = 283/454 (62%), Positives = 358/454 (78%), Gaps = 1/454 (0%)
Query: 18 VLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWR 77
+L E V Y +G++LG+GQFG TYLCT ST KK+ACKSI K+KL K D +D+ R
Sbjct: 79 ILENAFEDVKLFYTLGKELGRGQFGVTYLCTENSTGKKYACKSISKKKLVTKADKDDMRR 138
Query: 78 EIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKT 137
EIQIM HLS P++V +G YED AV++VMELC GGELFDRI+ KGHY+ER AA + +
Sbjct: 139 EIQIMQHLSGQPNIVEFKGAYEDEKAVNLVMELCAGGELFDRIIAKGHYTERAAASVCRQ 198
Query: 138 IVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYY 197
IV VV+ CH +GV+HRDLKPENFL + DE A +KATDFGLSVF + G+ + D+VGS YY
Sbjct: 199 IVNVVKICHFMGVLHRDLKPENFLLSSKDEKALIKATDFGLSVFIEEGKVYRDIVGSAYY 258
Query: 198 VAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSI 257
VAPEVLR+ YG E D+WSAG+ILYILLSGVPPFWAETE GIF IL G +DF+S+PWPSI
Sbjct: 259 VAPEVLRRRYGKEVDIWSAGIILYILLSGVPPFWAETEKGIFDAILEGHIDFESQPWPSI 318
Query: 258 SDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLK 317
S SAKDL+R+ML +P+ R++A +VL+HPW+ + A DKPIDSAVLSR+KQF AMNKLK
Sbjct: 319 SSSAKDLVRRMLTADPKRRISAADVLQHPWLREGGEASDKPIDSAVLSRMKQFRAMNKLK 378
Query: 318 KMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMD 377
K+AL+VIAE + EEI GLK +F IDTD+SGTIT++ELK+GL ++GS+L E+E++ LMD
Sbjct: 379 KLALKVIAENIDTEEIQGLKAMFANIDTDNSGTITYEELKEGLAKLGSKLTEAEVKQLMD 438
Query: 378 AADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL 437
AADVD +G+IDY EFI AT+H ++LE ENL AF +FDKD+SGYITIDE+ A K++G+
Sbjct: 439 AADVDGNGSIDYIEFITATMHRHRLESNENLYKAFQHFDKDSSGYITIDELESALKEYGM 498
Query: 438 -DDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
DD I E++ ++D DNDG+I+Y EF AMMR GN
Sbjct: 499 GDDATIKEVLSDVDSDNDGRINYEEFCAMMRSGN 532
Score = 30.0 bits (66), Expect = 3.1
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 5/57 (8%)
Query: 445 MIKEIDQDNDGQIDYSEFAAMMRKGNGGIGRR-TMRNTLNLRDALGLVGNGSNQVID 500
M ID DN G I Y E +++G +G + T L DA + GNGS I+
Sbjct: 400 MFANIDTDNSGTITYEE----LKEGLAKLGSKLTEAEVKQLMDAADVDGNGSIDYIE 452
>At4g04720 putative calcium dependent protein kinase
Length = 531
Score = 583 bits (1502), Expect = e-166
Identities = 283/448 (63%), Positives = 359/448 (79%), Gaps = 2/448 (0%)
Query: 24 EKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMH 83
E + + Y +G++LG+GQFG TY+C T +ACKSI KRKL K+D EDV REIQIM
Sbjct: 74 EDIRKFYSLGKELGRGQFGITYMCKEIGTGNTYACKSILKRKLISKQDKEDVKREIQIMQ 133
Query: 84 HLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVE 143
+LS P++V I+G YED ++H+VMELC GGELFDRI+ +GHYSER AAG+I++IV VV+
Sbjct: 134 YLSGQPNIVEIKGAYEDRQSIHLVMELCAGGELFDRIIAQGHYSERAAAGIIRSIVNVVQ 193
Query: 144 SCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEVL 203
CH +GV+HRDLKPENFL + +E+A LKATDFGLSVF + G+ + D+VGS YYVAPEVL
Sbjct: 194 ICHFMGVVHRDLKPENFLLSSKEENAMLKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVL 253
Query: 204 RKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAKD 263
R+ YG E D+WSAGVILYILLSGVPPFWAE E GIF +++ G++DF SEPWPSIS+SAKD
Sbjct: 254 RRSYGKEIDIWSAGVILYILLSGVPPFWAENEKGIFDEVIKGEIDFVSEPWPSISESAKD 313
Query: 264 LIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALRV 323
L+RKML ++P+ R+TA +VL HPWI APDKPIDSAVLSR+KQF AMNKLKK+AL+V
Sbjct: 314 LVRKMLTKDPKRRITAAQVLEHPWIKGGE-APDKPIDSAVLSRMKQFRAMNKLKKLALKV 372
Query: 324 IAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVDK 383
IAE LSEEEI GLK +F IDTD SGTIT++ELK GL R+GS L E+E++ LM+AADVD
Sbjct: 373 IAESLSEEEIKGLKTMFANIDTDKSGTITYEELKTGLTRLGSRLSETEVKQLMEAADVDG 432
Query: 384 SGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL-DDIHI 442
+GTIDY EFI+AT+H KL+R+E++ AF +FDKD SG+IT DE+ A K++G+ D+ I
Sbjct: 433 NGTIDYYEFISATMHRYKLDRDEHVYKAFQHFDKDNSGHITRDELESAMKEYGMGDEASI 492
Query: 443 DEMIKEIDQDNDGQIDYSEFAAMMRKGN 470
E+I E+D DNDG+I++ EF AMMR G+
Sbjct: 493 KEVISEVDTDNDGRINFEEFCAMMRSGS 520
>At1g76040 calcium-dependent protein kinase (CPK29)
Length = 561
Score = 575 bits (1482), Expect = e-164
Identities = 279/445 (62%), Positives = 354/445 (78%), Gaps = 2/445 (0%)
Query: 26 VTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMHHL 85
++ +Y + ++LG+GQFG TY CT KS +++ACKSI KRKL ++D EDV RE+ I+ HL
Sbjct: 108 LSALYDLHKELGRGQFGITYKCTDKSNGREYACKSISKRKLIRRKDIEDVRREVMILQHL 167
Query: 86 SEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVESC 145
+ P++V G YED +H+VMELC GGELFDRI+KKG YSE++AA + + IV VV C
Sbjct: 168 TGQPNIVEFRGAYEDKDNLHLVMELCSGGELFDRIIKKGSYSEKEAANIFRQIVNVVHVC 227
Query: 146 HSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEVLRK 205
H +GV+HRDLKPENFL + +ED+ +KATDFGLSVF + G+ + D+VGS YYVAPEVL +
Sbjct: 228 HFMGVVHRDLKPENFLLVSNEEDSPIKATDFGLSVFIEEGKVYRDIVGSAYYVAPEVLHR 287
Query: 206 LYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAKDLI 265
YG E DVWSAGV+LYILLSGVPPFW ETE IF IL GKLD ++ PWP+IS+SAKDLI
Sbjct: 288 NYGKEIDVWSAGVMLYILLSGVPPFWGETEKTIFEAILEGKLDLETSPWPTISESAKDLI 347
Query: 266 RKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALRVIA 325
RKML ++P+ R+TA E L HPW+ D I+ DKPI+SAVL R+KQF AMNKLKK+AL+VIA
Sbjct: 348 RKMLIRDPKKRITAAEALEHPWMTDTKIS-DKPINSAVLVRMKQFRAMNKLKKLALKVIA 406
Query: 326 ERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVDKSG 385
E LSEEEI GLK+ FK +DTD SGTITFDEL++GL R+GS+L ESEI+ LM+AADVDKSG
Sbjct: 407 ENLSEEEIKGLKQTFKNMDTDESGTITFDELRNGLHRLGSKLTESEIKQLMEAADVDKSG 466
Query: 386 TIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL-DDIHIDE 444
TIDY EF+ AT+H ++LE+EENL+ AF YFDKD SG+IT DE+ + ++G+ DD IDE
Sbjct: 467 TIDYIEFVTATMHRHRLEKEENLIEAFKYFDKDRSGFITRDELKHSMTEYGMGDDATIDE 526
Query: 445 MIKEIDQDNDGQIDYSEFAAMMRKG 469
+I ++D DNDG+I+Y EF AMMRKG
Sbjct: 527 VINDVDTDNDGRINYEEFVAMMRKG 551
>At4g21940 calcium-dependent protein kinase like protein
Length = 554
Score = 573 bits (1478), Expect = e-164
Identities = 275/465 (59%), Positives = 364/465 (78%), Gaps = 2/465 (0%)
Query: 6 PTPTIALKPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRK 65
P I + +L E++ ++Y +G++LG+GQFG TY C ST +ACKSI KRK
Sbjct: 78 PLKPIVFRETETILGKPFEEIRKLYTLGKELGRGQFGITYTCKENSTGNTYACKSILKRK 137
Query: 66 LFCKEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGH 125
L K+D +DV REIQIM +LS ++V I+G YED ++H+VMELC G ELFDRI+ +GH
Sbjct: 138 LTRKQDIDDVKREIQIMQYLSGQENIVEIKGAYEDRQSIHLVMELCGGSELFDRIIAQGH 197
Query: 126 YSERQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPG 185
YSE+ AAG+I++++ VV+ CH +GV+HRDLKPENFL + DE+A LKATDFGLSVF + G
Sbjct: 198 YSEKAAAGVIRSVLNVVQICHFMGVIHRDLKPENFLLASTDENAMLKATDFGLSVFIEEG 257
Query: 186 ESFSDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLG 245
+ + D+VGS YYVAPEVLR+ YG E D+WSAG+ILYILL GVPPFW+ETE GIF +I+ G
Sbjct: 258 KVYRDIVGSAYYVAPEVLRRSYGKEIDIWSAGIILYILLCGVPPFWSETEKGIFNEIIKG 317
Query: 246 KLDFQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLS 305
++DF S+PWPSIS+SAKDL+RK+L ++P+ R++A + L HPWI APDKPIDSAVLS
Sbjct: 318 EIDFDSQPWPSISESAKDLVRKLLTKDPKQRISAAQALEHPWIRGGE-APDKPIDSAVLS 376
Query: 306 RLKQFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGS 365
R+KQF AMNKLKK+AL+VIAE LSEEEI GLK +F +DTD SGTIT++ELK+GL ++GS
Sbjct: 377 RMKQFRAMNKLKKLALKVIAESLSEEEIKGLKTMFANMDTDKSGTITYEELKNGLAKLGS 436
Query: 366 ELMESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITI 425
+L E+E++ LM+AADVD +GTIDY EFI+AT+H + +R+E++ AF YFDKD SG+IT+
Sbjct: 437 KLTEAEVKQLMEAADVDGNGTIDYIEFISATMHRYRFDRDEHVFKAFQYFDKDNSGFITM 496
Query: 426 DEISQACKDFGL-DDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKG 469
DE+ A K++G+ D+ I E+I E+D DNDG+I+Y EF AMMR G
Sbjct: 497 DELESAMKEYGMGDEASIKEVIAEVDTDNDGRINYEEFCAMMRSG 541
>At1g61950 hypothetical protein
Length = 547
Score = 548 bits (1411), Expect = e-156
Identities = 269/446 (60%), Positives = 346/446 (77%), Gaps = 6/446 (1%)
Query: 24 EKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMH 83
E + E Y +GR+LG+GQFG TY+CT S+ K FACKSI KRKL +D EDV REIQIMH
Sbjct: 92 EDIKEKYSLGRELGRGQFGITYICTEISSGKNFACKSILKRKLIRTKDREDVRREIQIMH 151
Query: 84 HLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVE 143
+LS P++V I+G YED +VH+VMELCEGGELFD+I K+GHYSE+ AA +I+++V+VV+
Sbjct: 152 YLSGQPNIVEIKGAYEDRQSVHLVMELCEGGELFDKITKRGHYSEKAAAEIIRSVVKVVQ 211
Query: 144 SCHSLGVMHRDLKPENFLFDTVDE-DAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEV 202
CH +GV+HRDLKPENFL + DE + LKATDFG+SVF + G+ + D+VGS YYVAPEV
Sbjct: 212 ICHFMGVIHRDLKPENFLLSSKDEASSMLKATDFGVSVFIEEGKVYEDIVGSAYYVAPEV 271
Query: 203 LRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAK 262
L++ YG D+WSAGVILYILL G PPFWAET+ GIF +IL G++DF+SEPWPSIS+SAK
Sbjct: 272 LKRNYGKAIDIWSAGVILYILLCGNPPFWAETDKGIFEEILRGEIDFESEPWPSISESAK 331
Query: 263 DLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALR 322
DL+R ML +P+ R TA +VL HPWI + A DKPIDSAVLSR+KQ AMNKLKK+A +
Sbjct: 332 DLVRNMLKYDPKKRFTAAQVLEHPWIREGGEASDKPIDSAVLSRMKQLRAMNKLKKLAFK 391
Query: 323 VIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVD 382
IA+ L EEE+ GLK +F +DTD SGTIT+DELK GL+++GS L E+E++ L++ ADVD
Sbjct: 392 FIAQNLKEEELKGLKTMFANMDTDKSGTITYDELKSGLEKLGSRLTETEVKQLLEDADVD 451
Query: 383 KSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGL-DDIH 441
+GTIDY EFI+AT++ ++ERE+NL AF +FDKD SG E+ A K++ + DDI
Sbjct: 452 GNGTIDYIEFISATMNRFRVEREDNLFKAFQHFDKDNSG----QELETAMKEYNMGDDIM 507
Query: 442 IDEMIKEIDQDNDGQIDYSEFAAMMR 467
I E+I E+D DNDG I+Y EF MM+
Sbjct: 508 IKEIISEVDADNDGSINYQEFCNMMK 533
>At4g04740 putative calcium dependent protein kinase
Length = 520
Score = 544 bits (1402), Expect = e-155
Identities = 273/466 (58%), Positives = 351/466 (74%), Gaps = 2/466 (0%)
Query: 5 NPTPTIALKPVTWVLPYRTEKVTEIYRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKR 64
+P+ I+++ +L E + + Y +GR+LG+G G TY+C T +ACKSI KR
Sbjct: 44 SPSIPISVRDPETILGKPFEDIRKFYSLGRELGRGGLGITYMCKEIGTGNIYACKSILKR 103
Query: 65 KLFCKEDYEDVWREIQIMHHLSEHPHVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKG 124
KL + EDV EIQIM HLS P+VV I+G+YED +VH+VMELC GGELFDRI+ +G
Sbjct: 104 KLISELGREDVKTEIQIMQHLSGQPNVVEIKGSYEDRHSVHLVMELCAGGELFDRIIAQG 163
Query: 125 HYSERQAAGLIKTIVEVVESCHSLGVMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKP 184
HYSER AAG IK+IV+VV+ CH GV+HRDLKPENFLF + +E+A LK TDFGLS F +
Sbjct: 164 HYSERAAAGTIKSIVDVVQICHLNGVIHRDLKPENFLFSSKEENAMLKVTDFGLSAFIEE 223
Query: 185 GESFSDVVGSPYYVAPEVLRKLYGPESDVWSAGVILYILLSGVPPFWAETEPGIFRQILL 244
G+ + DVVGSPYYVAPEVLR+ YG E D+WSAGVILYILL GVPPFWA+ E G+F +IL
Sbjct: 224 GKIYKDVVGSPYYVAPEVLRQSYGKEIDIWSAGVILYILLCGVPPFWADNEEGVFVEILK 283
Query: 245 GKLDFQSEPWPSISDSAKDLIRKMLDQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVL 304
K+DF EPWPSISDSAKDL+ KML ++P+ R+TA +VL HPWI AP+KPIDS VL
Sbjct: 284 CKIDFVREPWPSISDSAKDLVEKMLTEDPKRRITAAQVLEHPWIKGGE-APEKPIDSTVL 342
Query: 305 SRLKQFSAMNKLKKMALRVIAERLSEEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVG 364
SR+KQF AMNKLKK+AL+V A LSEEEI GLK LF +DT+ SGTIT+++L+ GL R+
Sbjct: 343 SRMKQFRAMNKLKKLALKVSAVSLSEEEIKGLKTLFANMDTNRSGTITYEQLQTGLSRLR 402
Query: 365 SELMESEIQDLMDAADVDKSGTIDYGEFIAATVHLNKLEREENLLSAFAYFDKDASGYIT 424
S L E+E+Q L++A+DVD +GTIDY EFI+AT+H KL +E++ AF + DKD +G+IT
Sbjct: 403 SRLSETEVQQLVEASDVDGNGTIDYYEFISATMHRYKLHHDEHVHKAFQHLDKDKNGHIT 462
Query: 425 IDEISQACKDFGL-DDIHIDEMIKEIDQDNDGQIDYSEFAAMMRKG 469
DE+ A K++G+ D+ I E+I E+D DNDG+I++ EF AMMR G
Sbjct: 463 RDELESAMKEYGMGDEASIKEVISEVDTDNDGKINFEEFRAMMRCG 508
>At5g12480 calcium-dependent protein kinase - like protein
Length = 535
Score = 535 bits (1377), Expect = e-152
Identities = 258/444 (58%), Positives = 338/444 (76%), Gaps = 5/444 (1%)
Query: 30 YRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMHHLSEHP 89
Y +GR++G+G+FG TYLCT K T +K+ACKSI K+KL D EDV RE++IM H+ +HP
Sbjct: 59 YDLGREVGRGEFGITYLCTDKETGEKYACKSISKKKLRTAVDIEDVRREVEIMKHMPKHP 118
Query: 90 HVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVESCHSLG 149
+VV ++ ++ED AVHIVMELCEGGELFDRIV +GHY+ER AA ++KTIVEVV+ CH G
Sbjct: 119 NVVSLKDSFEDDDAVHIVMELCEGGELFDRIVARGHYTERAAAAVMKTIVEVVQICHKQG 178
Query: 150 VMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEVLRKLYGP 209
VMHRDLKPENFLF E + LKA DFGLSVF+KPGE F+++VGSPYY+APEVLR+ YGP
Sbjct: 179 VMHRDLKPENFLFANKKETSALKAIDFGLSVFFKPGEQFNEIVGSPYYMAPEVLRRNYGP 238
Query: 210 ESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAKDLIRKML 269
E DVWSAGVILYILL GVPPFWAETE G+ + I+ +DF+ +PWP +SDSAKDL+RKML
Sbjct: 239 EIDVWSAGVILYILLCGVPPFWAETEQGVAQAIIRSVIDFKRDPWPRVSDSAKDLVRKML 298
Query: 270 DQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALRVIAERLS 329
+ +P+ RLTA +VL H WI++ AP+ + V +RLKQFS MNKLKK ALRVIAE LS
Sbjct: 299 EPDPKKRLTAAQVLEHTWILNAKKAPNVSLGETVKARLKQFSVMNKLKKRALRVIAEHLS 358
Query: 330 EEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVDKSGTIDY 389
EE G+KE F+M+D + G I +ELK GL++ G ++ ++++Q LM+A DVD GT++Y
Sbjct: 359 VEEAAGIKEAFEMMDVNKRGKINLEELKYGLQKAGQQIADTDLQILMEATDVDGDGTLNY 418
Query: 390 GEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKDFGLDDIHIDEMI--- 446
EF+A +VHL K+ +E+L AF +FD++ SGYI IDE+ +A D LD+ +E+I
Sbjct: 419 SEFVAVSVHLKKMANDEHLHKAFNFFDQNQSGYIEIDELREALND-ELDNTSSEEVIAAI 477
Query: 447 -KEIDQDNDGQIDYSEFAAMMRKG 469
+++D D DG+I Y EF AMM+ G
Sbjct: 478 MQDVDTDKDGRISYEEFVAMMKAG 501
>At3g57530 calcium-dependent protein kinase
Length = 538
Score = 532 bits (1371), Expect = e-151
Identities = 255/441 (57%), Positives = 333/441 (74%), Gaps = 1/441 (0%)
Query: 30 YRMGRKLGQGQFGTTYLCTHKSTNKKFACKSIPKRKLFCKEDYEDVWREIQIMHHLSEHP 89
Y +GR+LG+G+FG TYLCT K T+ FACKSI K+KL D EDV RE++IM H+ EHP
Sbjct: 63 YTLGRELGRGEFGVTYLCTDKETDDVFACKSILKKKLRTAVDIEDVRREVEIMRHMPEHP 122
Query: 90 HVVRIEGTYEDSTAVHIVMELCEGGELFDRIVKKGHYSERQAAGLIKTIVEVVESCHSLG 149
+VV ++ TYED AVH+VMELCEGGELFDRIV +GHY+ER AA + KTI+EVV+ CH G
Sbjct: 123 NVVTLKETYEDEHAVHLVMELCEGGELFDRIVARGHYTERAAAAVTKTIMEVVQVCHKHG 182
Query: 150 VMHRDLKPENFLFDTVDEDAKLKATDFGLSVFYKPGESFSDVVGSPYYVAPEVLRKLYGP 209
VMHRDLKPENFLF E A LKA DFGLSVF+KPGE F+++VGSPYY+APEVL++ YGP
Sbjct: 183 VMHRDLKPENFLFGNKKETAPLKAIDFGLSVFFKPGERFNEIVGSPYYMAPEVLKRNYGP 242
Query: 210 ESDVWSAGVILYILLSGVPPFWAETEPGIFRQILLGKLDFQSEPWPSISDSAKDLIRKML 269
E D+WSAGVILYILL GVPPFWAETE G+ + I+ LDF+ +PWP +S++AKDLIRKML
Sbjct: 243 EVDIWSAGVILYILLCGVPPFWAETEQGVAQAIIRSVLDFRRDPWPKVSENAKDLIRKML 302
Query: 270 DQNPRTRLTAHEVLRHPWIVDDNIAPDKPIDSAVLSRLKQFSAMNKLKKMALRVIAERLS 329
D + + RLTA +VL HPW+ + AP+ + V +RLKQF+ MNKLKK ALRVIAE LS
Sbjct: 303 DPDQKRRLTAQQVLDHPWLQNAKTAPNVSLGETVRARLKQFTVMNKLKKRALRVIAEHLS 362
Query: 330 EEEIGGLKELFKMIDTDSSGTITFDELKDGLKRVGSELMESEIQDLMDAADVDKSGTIDY 389
+EE G++E F+++DT G I DELK GL+++G + + ++Q LMDA D+D+ G +D
Sbjct: 363 DEEASGIREGFQIMDTSQRGKINIDELKIGLQKLGHAIPQDDLQILMDAGDIDRDGYLDC 422
Query: 390 GEFIAATVHLNKLEREENLLSAFAYFDKDASGYITIDEISQACKD-FGLDDIHIDEMIKE 448
EFIA +VHL K+ +E+L AFA+FD++ +GYI I+E+ +A D G + +D +I++
Sbjct: 423 DEFIAISVHLRKMGNDEHLKKAFAFFDQNNNGYIEIEELREALSDELGTSEEVVDAIIRD 482
Query: 449 IDQDNDGQIDYSEFAAMMRKG 469
+D D DG+I Y EF MM+ G
Sbjct: 483 VDTDKDGRISYEEFVTMMKTG 503
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,669,151
Number of Sequences: 26719
Number of extensions: 532020
Number of successful extensions: 4397
Number of sequences better than 10.0: 1049
Number of HSP's better than 10.0 without gapping: 369
Number of HSP's successfully gapped in prelim test: 680
Number of HSP's that attempted gapping in prelim test: 2080
Number of HSP's gapped (non-prelim): 1332
length of query: 503
length of database: 11,318,596
effective HSP length: 104
effective length of query: 399
effective length of database: 8,539,820
effective search space: 3407388180
effective search space used: 3407388180
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148755.3


