

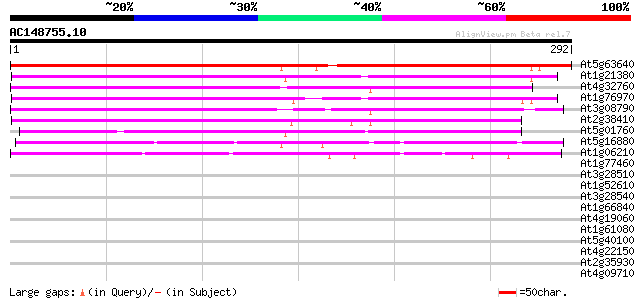
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.10 - phase: 0 /pseudo
(292 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g63640 unknown protein (At5g63640) 358 2e-99
At1g21380 unknown protein 199 1e-51
At4g32760 unknown protein 189 2e-48
At1g76970 unknown protein 184 4e-47
At3g08790 hypothetical protein 179 2e-45
At2g38410 unknown protein 160 1e-39
At5g01760 unknown protein 142 2e-34
At5g16880 TOM (target of myb1) -like protein 89 2e-18
At1g06210 unknown protein 70 1e-12
At1g77460 unknown protein 35 0.062
At3g28510 hypothetical protein 31 0.69
At1g52610 hypothetical protein 31 0.90
At3g28540 hypothetical protein 30 1.2
At1g66840 hypothetical protein 30 1.2
At4g19060 unknown protein (At4g19060) 30 1.5
At1g61080 hypothetical protein 30 2.0
At5g40100 disease resistence like - protein 29 2.6
At4g22150 unknown protein 29 2.6
At2g35930 unknown protein 29 2.6
At4g09710 RNA-directed DNA polymerase -like protein 29 3.4
>At5g63640 unknown protein (At5g63640)
Length = 447
Score = 358 bits (918), Expect = 2e-99
Identities = 201/339 (59%), Positives = 240/339 (70%), Gaps = 51/339 (15%)
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
MAAELV++ATSEKL+++DW KNIEI EL ARD+R+AKDV+KAIKKRLG+KNPN QLYAV
Sbjct: 1 MAAELVSSATSEKLADVDWAKNIEICELAARDERQAKDVIKAIKKRLGSKNPNTQLYAVQ 60
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKF 120
LLEMLMNNIG++I++QV+ V+P LVKIVKKKSDLPVRE+IFLLLDATQTSLGGASGKF
Sbjct: 61 LLEMLMNNIGENIHKQVIDTGVLPTLVKIVKKKSDLPVRERIFLLLDATQTSLGGASGKF 120
Query: 121 PQYYKAYYDLVSAGVQFPQR--AQVVQSNRPSLQPNTTNN--VPKREPSPLRRGRVAQKA 176
PQYY AYY+LV+AGV+F QR A V ++ NT N R P Q+
Sbjct: 121 PQYYTAYYELVNAGVKFTQRPNATPVVVTAQAVPRNTLNEQLASARNEGP----ATTQQR 176
Query: 177 ESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVM 236
ES +V S I+QKAS LE+LKEVLDAVD+++P+GA+DEFTLDLVEQCSFQK+RVMHLVM
Sbjct: 177 ESQSVSPSSILQKASTALEILKEVLDAVDSQNPEGAKDEFTLDLVEQCSFQKERVMHLVM 236
Query: 237 ASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDT-----TTVN----------------- 274
SRDE+ VS+AIE+NEQLQ++L RH+DLLS + T TT N
Sbjct: 237 TSRDEKAVSKAIELNEQLQRILNRHEDLLSGRITVPSRSTTSNGYHSNLEPVRPISNGDQ 296
Query: 275 ---------------------HFDHEEAEEEEEPEQLFR 292
H EE +EEEEPEQLFR
Sbjct: 297 KRELKASNANTESSSFISNRAHLKLEEEDEEEEPEQLFR 335
>At1g21380 unknown protein
Length = 506
Score = 199 bits (507), Expect = 1e-51
Identities = 107/296 (36%), Positives = 170/296 (57%), Gaps = 15/296 (5%)
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
AA AT++ L DW NIE+ +++ + +AK+ VK +KKRLG+KN Q+ A+
Sbjct: 5 AAACAERATNDMLIGPDWAINIELCDIINMEPSQAKEAVKVLKKRLGSKNSKVQILALYA 64
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFP 121
LE L N G+ + + +V +++P +VKIVKKK DL VRE+I LLD Q + GG+ G+FP
Sbjct: 65 LETLSKNCGESVYQLIVDRDILPDMVKIVKKKPDLTVREKILSLLDTWQEAFGGSGGRFP 124
Query: 122 QYYKAYYDLVSAGVQFPQRAQ--VVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAESN 179
QYY AY +L SAG++FP R + V P QP E + ++ + A +
Sbjct: 125 QYYNAYNELRSAGIEFPPRTESSVPFFTPPQTQPIVAQATASDEDAAIQASLQSDDASAL 184
Query: 180 TVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASR 239
++ E IQ A ++VL ++L A+D HP+G ++E +DLVEQC ++RVM LV +
Sbjct: 185 SMEE---IQSAQGSVDVLTDMLGALDPSHPEGLKEELIVDLVEQCRTYQRRVMALVNTTS 241
Query: 240 DERIVSRAIEVNEQLQKVLERHDDLLSSKDT----------TTVNHFDHEEAEEEE 285
DE ++ + + +N+ LQ+VL+ HDD ++NH D ++ +++
Sbjct: 242 DEELMCQGLALNDNLQRVLQHHDDKAKGNSVPATAPTPIPLVSINHDDDDDESDDD 297
>At4g32760 unknown protein
Length = 675
Score = 189 bits (479), Expect = 2e-48
Identities = 105/277 (37%), Positives = 160/277 (56%), Gaps = 8/277 (2%)
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
M +V ATSE L DW N+EI +++ D +AKDVVK IKKR+G++NP AQL A+
Sbjct: 1 MVNAMVERATSEMLIGPDWAMNLEICDMLNSDPAQAKDVVKGIKKRIGSRNPKAQLLALT 60
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKF 120
LLE ++ N GD ++ V VI +V+IVKKK D V+E+I +L+D Q + GG ++
Sbjct: 61 LLETIVKNCGDMVHMHVAEKGVIHEMVRIVKKKPDFHVKEKILVLIDTWQEAFGGPRARY 120
Query: 121 PQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAESNT 180
PQYY Y +L+ AG FPQR+ + + P P T + P+ G E +
Sbjct: 121 PQYYAGYQELLRAGAVFPQRS---ERSAPVFTPPQTQPLTSYPPNLRNAGPGNDVPEPSA 177
Query: 181 VPESRI-----IQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLV 235
PE IQ A +++VL E+L A++ + + + E +DLVEQC KQRV+HLV
Sbjct: 178 EPEFPTLSLSEIQNAKGIMDVLAEMLSALEPGNKEDLKQEVMVDLVEQCRTYKQRVVHLV 237
Query: 236 MASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTT 272
++ DE ++ + + +N+ LQ+VL ++ + S T+
Sbjct: 238 NSTSDESLLCQGLALNDDLQRVLTNYEAIASGLPGTS 274
>At1g76970 unknown protein
Length = 446
Score = 184 bits (468), Expect = 4e-47
Identities = 108/301 (35%), Positives = 170/301 (55%), Gaps = 28/301 (9%)
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
AA AT++ L DW NIE+ +L+ D +AK+ VK +KKRLG+KN Q+ A+
Sbjct: 5 AAACAERATNDMLIGPDWAINIELCDLINMDPSQAKEAVKVLKKRLGSKNSKVQILALYA 64
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFP 121
LE L N G+++ + ++ ++ +VKIVKKK +L VRE+I LLD Q + GG G++P
Sbjct: 65 LETLSKNCGENVYQLIIDRGLLNDMVKIVKKKPELNVREKILTLLDTWQEAFGGRGGRYP 124
Query: 122 QYYKAYYDLVSAGVQFPQRAQVVQS--NRPSLQPNTTNNVPKREPSPLRRGRVAQKAESN 179
QYY AY DL SAG++FP R + S P QP+ E + ++ A S
Sbjct: 125 QYYNAYNDLRSAGIEFPPRTESSLSFFTPPQTQPD--------EDAAIQASLQGDDASSL 176
Query: 180 TVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASR 239
++ E IQ A ++VL ++L A D +P+ ++E +DLVEQC ++RVM LV +
Sbjct: 177 SLEE---IQSAEGSVDVLMDMLGAHDPGNPESLKEEVIVDLVEQCRTYQRRVMTLVNTTT 233
Query: 240 DERIVSRAIEVNEQLQKVLERHDDLL-------SSKDT--------TTVNHFDHEEAEEE 284
DE ++ + + +N+ LQ VL+RHDD+ + ++T +NH D ++ ++
Sbjct: 234 DEELLCQGLALNDNLQHVLQRHDDIANVGSVPSNGRNTRAPPPVQIVDINHDDEDDESDD 293
Query: 285 E 285
E
Sbjct: 294 E 294
>At3g08790 hypothetical protein
Length = 607
Score = 179 bits (453), Expect = 2e-45
Identities = 101/296 (34%), Positives = 170/296 (57%), Gaps = 24/296 (8%)
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
M LV+ ATS+ L DW N+EI +++ + + ++VV IKKRL ++ QL A+
Sbjct: 1 MVHPLVDRATSDMLIGPDWAMNLEICDMLNHEPGQTREVVSGIKKRLTSRTSKVQLLALT 60
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKF 120
LLE ++ N G+ I+ QV +++ +VK+ K+K ++ V+E+I +L+D Q S G G+
Sbjct: 61 LLETIITNCGELIHMQVAEKDILHKMVKMAKRKPNIQVKEKILILIDTWQESFSGPQGRH 120
Query: 121 PQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTNNVPK-REPSPLRRGRVAQKAESN 179
PQYY AY +L+ AG+ FPQ RP + P++ N P R P + R A++ +
Sbjct: 121 PQYYAAYQELLRAGIVFPQ--------RPQITPSSGQNGPSTRYP---QNSRNARQEAID 169
Query: 180 TVPESRI-------IQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVM 232
T ES IQ A +++VL E+++A+D + +G + E +DLV QC KQRV+
Sbjct: 170 TSTESEFPTLSLTEIQNARGIMDVLAEMMNAIDGNNKEGLKQEVVVDLVSQCRTYKQRVV 229
Query: 233 HLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVNHFDHEEAEEEEEPE 288
HLV ++ DE ++ + + +N+ LQ++L +H+ + S + EE ++E P+
Sbjct: 230 HLVNSTSDESMLCQGLALNDDLQRLLAKHEAIASGN-----SMIKKEEKSKKEVPK 280
>At2g38410 unknown protein
Length = 671
Score = 160 bits (404), Expect = 1e-39
Identities = 99/310 (31%), Positives = 164/310 (51%), Gaps = 45/310 (14%)
Query: 2 AAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVML 61
A V+ ATS+ L DW N+EI + V +AKDVVKA+KKRL +K+ QL A+ L
Sbjct: 8 ATVAVDKATSDLLLGPDWTTNMEICDSVNSLHWQAKDVVKAVKKRLQHKSSRVQLLALTL 67
Query: 62 LEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFP 121
LE L+ N GD+++ QV ++ +VKIVKKK+D+ VR++I +++D+ Q + GG GK+P
Sbjct: 68 LETLVKNCGDYLHHQVAEKNILGEMVKIVKKKADMQVRDKILVMVDSWQQAFGGPEGKYP 127
Query: 122 QYYKAYYDLVSAGVQFPQRAQVVQ-------SNRPSLQPNTTNNVPKREPSPLRRGRVAQ 174
QYY AY +L +GV+FP+R+ S+ P QP VP + G
Sbjct: 128 QYYWAYDELRRSGVEFPRRSPDASPIITPPVSHPPLRQPQGGYGVPPAGYGVHQAGYGVP 187
Query: 175 KA----------------------------ESNTVPESRI----------IQKASNVLEV 196
+A S + E+ I+ +V+++
Sbjct: 188 QAGYGIPQAGYGVPQAGYGIPQVGYGMPSGSSRRLDEAMATEVEGLSLSSIESMRDVMDL 247
Query: 197 LKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQK 256
L ++L AVD + +DE +DLVE+C ++++M ++ ++ D+ ++ R +++N+ LQ
Sbjct: 248 LGDMLQAVDPSDREAVKDEVIVDLVERCRSNQKKLMQMLTSTGDDELLGRGLDLNDSLQI 307
Query: 257 VLERHDDLLS 266
+L +HD + S
Sbjct: 308 LLAKHDAIAS 317
>At5g01760 unknown protein
Length = 539
Score = 142 bits (359), Expect = 2e-34
Identities = 79/265 (29%), Positives = 148/265 (55%), Gaps = 8/265 (3%)
Query: 6 VNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEML 65
V+ ATSE L DW I I + + ++ + KD +KA+K+RL +K+ QL + L +
Sbjct: 26 VDKATSELLRTPDWTIIIAICDSLNSNRWQCKDAIKAVKRRLQHKSSRVQL---LTLTAM 82
Query: 66 MNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFPQYYK 125
+ N GD ++ + ++ +VK+V+KK D VR ++ +LLD + G + K P Y
Sbjct: 83 LKNCGDFVHSHIAEKHLLEDMVKLVRKKGDFEVRNKLLILLDTWNEAFSGVACKHPHYNW 142
Query: 126 AYYDLVSAGVQFPQRAQ----VVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAESNTV 181
AY +L GV+FPQR++ +++ P Q ++++++ RR E ++
Sbjct: 143 AYQELKRCGVKFPQRSKEAPLMLEPPPPVTQSSSSSSMNLMSIGSFRRLDETMATEIESL 202
Query: 182 PESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLVMASRDE 241
S ++ NV++++ +++ AV+ +DE +DLVEQC +++++ ++ + DE
Sbjct: 203 SLSS-LESMRNVMDLVNDMVQAVNPSDKSALKDELIVDLVEQCRSNQKKLIQMLTTTADE 261
Query: 242 RIVSRAIEVNEQLQKVLERHDDLLS 266
+++R +E+N+ LQ VL RHD + S
Sbjct: 262 DVLARGLELNDSLQVVLARHDAIAS 286
>At5g16880 TOM (target of myb1) -like protein
Length = 407
Score = 89.4 bits (220), Expect = 2e-18
Identities = 72/294 (24%), Positives = 144/294 (48%), Gaps = 17/294 (5%)
Query: 4 ELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVMLLE 63
++V AT+E L E DW N+EI +++ ++ + ++++ IKKR+ K P Q A++LLE
Sbjct: 50 KIVEDATTENLEEPDWDMNLEICDMINQETINSVELIRGIKKRIMMKQPRIQYLALVLLE 109
Query: 64 MLMNNIGDHINEQVVRAEVIPILVKIV-KKKSDLPVREQIFLLLDATQTSLGGASGKFPQ 122
+ N +E V V+ +VK++ ++ + R + +L++A S P
Sbjct: 110 TCVKNCEKAFSE-VAAERVLDEMVKLIDDPQTVVNNRNKALMLIEAWGESTSELR-YLPV 167
Query: 123 YYKAYYDLVSAGVQFPQR-----AQVVQSNRPSLQPNTTNNVPK--REPSPLRRGRVAQK 175
+ + Y L + G++FP R A + R + P ++P+ EP+ ++ +
Sbjct: 168 FEETYKSLKARGIRFPGRDNESLAPIFTPARSTPAPELNADLPQHVHEPAHIQYDVPVRS 227
Query: 176 AESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQKQRVMHLV 235
+ E+ I A N +E+L VL + + +D+ T LV+QC + V ++
Sbjct: 228 FTAEQTKEAFDI--ARNSIELLSTVLSS--SPQHDALQDDLTTTLVQQCRQSQTTVQRII 283
Query: 236 -MASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVNHFDHEEAEEEEEPE 288
A +E ++ A+ VN++L K L +++++ +K + + + EEP+
Sbjct: 284 ETAGENEALLFEALNVNDELVKTLSKYEEM--NKPSAPLTSHEPAMIPVAEEPD 335
>At1g06210 unknown protein
Length = 383
Score = 70.1 bits (170), Expect = 1e-12
Identities = 69/310 (22%), Positives = 141/310 (45%), Gaps = 29/310 (9%)
Query: 1 MAAELVNAATSEKLSEIDWMKNIEISELVARDQRKAKDVVKAIKKRLGNKNPNAQLYAVM 60
+ +++V+ AT E L E +W N+ I + D+ ++V+AIK+++ K+P +Q ++
Sbjct: 37 LESKMVDEATLETLEEPNWGMNMRICAQINNDEFNGTEIVRAIKRKISGKSPVSQRLSLE 96
Query: 61 LLEMLMNNIGDHINEQVVRAEVIPILVKIVKK-KSDLPVREQIFLLLDATQTSLGGASGK 119
LLE N + + +V +V+ +V ++K ++D R++ F L+ A S
Sbjct: 97 LLEACAMNC-EKVFSEVASEKVLDEMVWLIKNGEADSENRKRAFQLIRAWGQSQD--LTY 153
Query: 120 FPQYYKAYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTNNVPKREPS--PLRRGRVAQKAE 177
P +++ Y L + + + SL+ VP P P+ A +
Sbjct: 154 LPVFHQTYMSLEGENGLHARGEENSMPGQSSLESLMQRPVPVPPPGSYPVPNQEQALGDD 213
Query: 178 S---------NTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEFTLDLVEQCSFQK 228
+ + I+ N LE+L +L+ P D+ T+ L+E+C Q
Sbjct: 214 DGLDYNFGNLSIKDKKEQIEITRNSLELLSSMLNT--EGKPNHTEDDLTVSLMEKCK-QS 270
Query: 229 QRVMHLVMASR--DERIVSRAIEVNEQLQKVL---------ERHDDLLSSKDTTTVNHFD 277
Q ++ +++ S DE ++ A+ +N++LQ+VL E+ ++ + + + +
Sbjct: 271 QPLIQMIIESTTDDEGVLFEALHLNDELQQVLSSYKKPDETEKKASIVEQESSGSKDTGP 330
Query: 278 HEEAEEEEEP 287
+EE+EP
Sbjct: 331 KPTEQEEQEP 340
>At1g77460 unknown protein
Length = 2110
Score = 34.7 bits (78), Expect = 0.062
Identities = 20/69 (28%), Positives = 40/69 (56%), Gaps = 1/69 (1%)
Query: 40 VKAIKKRLGNKNPNAQLYAVMLLEMLMNNIGDHINEQVVRAEVIPILVKIVKKKSDLPVR 99
V + L + NPN+Q A LL L+ + D I ++++ + V+ L++++++K+D+ VR
Sbjct: 190 VDIVVSLLSSDNPNSQANAASLLARLVLSFCDSI-QKILNSGVVKSLIQLLEQKNDINVR 248
Query: 100 EQIFLLLDA 108
L+A
Sbjct: 249 ASAADALEA 257
>At3g28510 hypothetical protein
Length = 530
Score = 31.2 bits (69), Expect = 0.69
Identities = 33/141 (23%), Positives = 57/141 (40%), Gaps = 24/141 (17%)
Query: 155 TTNNVPKREPSPLRRGRVAQKAESNTVPESRIIQKASNVLEV-LKEVLDAVDAKHPQGAR 213
TTN V K +P+ +RRGR+ E + A N LE+ ++ ++ K
Sbjct: 369 TTNFVDKLDPALIRRGRMDNHIEMSYCKFEAFKVLAKNYLEIETHDLYGEIERK------ 422
Query: 214 DEFTLDLVEQCSFQKQRVMHLVMASRDE--------RIVSRAIEVNEQLQKVLERHDDLL 265
+E+ V +M DE R+V E E+ +K+ E +
Sbjct: 423 -------LEETDMSPADVAETLMPKSDEEDADICIKRLVKTLEEEKEKARKLAEEEEKKK 475
Query: 266 SSKDTTTVNHFDHEEAEEEEE 286
+ K+ + EEAEE+++
Sbjct: 476 AEKEAKKMK--KAEEAEEKKK 494
>At1g52610 hypothetical protein
Length = 792
Score = 30.8 bits (68), Expect = 0.90
Identities = 18/58 (31%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Query: 173 AQKAESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARDEF-TLDLVEQCSFQKQ 229
A ESN VPE +++++ + E + DA H G DE+ D E+ S +K+
Sbjct: 419 AMDEESNKVPEDTVVEESDDEDEFETQCCDAERPDHAMGTDDEWDAYDQAERVSSRKK 476
>At3g28540 hypothetical protein
Length = 510
Score = 30.4 bits (67), Expect = 1.2
Identities = 32/143 (22%), Positives = 55/143 (38%), Gaps = 20/143 (13%)
Query: 155 TTNNVPKREPSPLRRGRVAQKAESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGARD 214
TTN + K +P+ +RRGR+ E + A N LE+ L +
Sbjct: 368 TTNYLDKLDPALIRRGRMDNHIEMSYCRFEAFKVLAKNYLEIESHDLFGEIKR------- 420
Query: 215 EFTLDLVEQCSFQKQRVMHLVMASRDE--------RIVSRAIEVNEQLQKVLERHDDLLS 266
LVE+ V +M DE R+V E E+ +K+ E +
Sbjct: 421 -----LVEETDMSPADVAENLMPKSDEDDADICLTRLVKSLEEEKEKAKKLAEEEKMKKA 475
Query: 267 SKDTTTVNHFDHEEAEEEEEPEQ 289
++D + EE +++ + E+
Sbjct: 476 ARDARRIKKKAEEEHKKKNKVEE 498
>At1g66840 hypothetical protein
Length = 607
Score = 30.4 bits (67), Expect = 1.2
Identities = 34/135 (25%), Positives = 56/135 (41%), Gaps = 23/135 (17%)
Query: 171 RVAQKAESNTVPESRIIQKASNVLEVLKEVLDAVDAKHPQGA----------------RD 214
R Q+ ES + ++R ++ + L VLKEV +A +AK + A R
Sbjct: 248 RKVQRNESMSRSKNRAFERGKDNLSVLKEVTEATEAKKAELASINAELFCLVNTMDTLRK 307
Query: 215 EFTLDLVEQCSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHDDLLSSKDTTTVN 274
EF E K VM ER+ ++ + +QL+ V + + + D T +
Sbjct: 308 EFDHAKKETAWLDKMIQKDDVML---ERLNTKLLIAKDQLEAVSKAEERISYLADNLTTS 364
Query: 275 ----HFDHEEAEEEE 285
D E A++EE
Sbjct: 365 FEKLKSDREAAKKEE 379
>At4g19060 unknown protein (At4g19060)
Length = 383
Score = 30.0 bits (66), Expect = 1.5
Identities = 14/39 (35%), Positives = 20/39 (50%)
Query: 224 CSFQKQRVMHLVMASRDERIVSRAIEVNEQLQKVLERHD 262
C F K ++M SRDER+ + E LQ++ R D
Sbjct: 259 CGFPKGFGGKVIMTSRDERLAKAIVGEEENLQRLFPRSD 297
>At1g61080 hypothetical protein
Length = 907
Score = 29.6 bits (65), Expect = 2.0
Identities = 31/134 (23%), Positives = 65/134 (48%), Gaps = 15/134 (11%)
Query: 165 SPLRRGRVAQKAESNTVPESRI--IQKAS--NVLEVLKEVLDAVDAKHPQGARDEFTLDL 220
SP R G A A S+ + R+ ++K S +V + +L +A+ P + DE + +
Sbjct: 290 SPGRVGDFANSA-SHLLWNMRVQALEKLSPIDVKRLAIHILSQKEAQEPNESNDEDVISV 348
Query: 221 VEQCSFQKQRVMHLVMASRDERIVS---RAIEVNEQLQKVLERHDDLLSSKDTTTVNHFD 277
VE+ +K + + + E V+ ++ +N + +++ +SS ++T+ + +
Sbjct: 349 VEEIKQKKDEIESIDVKMETEESVNLDEESVVLNGEQDTIMK-----ISSLESTSESKLN 403
Query: 278 HEEAEEEEEPEQLF 291
H +E+ E QLF
Sbjct: 404 H--SEKYENSSQLF 415
>At5g40100 disease resistence like - protein
Length = 1017
Score = 29.3 bits (64), Expect = 2.6
Identities = 33/145 (22%), Positives = 60/145 (40%), Gaps = 22/145 (15%)
Query: 69 IGDHINEQVVRAEVIPILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFPQYYK--- 125
+GDHI+E++ RA V +V ++ R + L + + G G FP +Y+
Sbjct: 51 LGDHISEELQRAIEGSDFVVVVLSENYPTSRWCLMELQSIMELQMEGRLGVFPVFYRVEP 110
Query: 126 --AYYDLVSAGVQFPQRAQVVQSNRPSLQPNTTNNVPKREPSPLRRGRVAQKAESNTVPE 183
Y L S ++ QR P + VPK R + A+ + V
Sbjct: 111 SAVRYQLGSFDLEGYQR-----------DPQMADMVPK------WRQALKLIADLSGVAS 153
Query: 184 SRIIQKASNVLEVLKEVLDAVDAKH 208
+ I +A+ V ++++++ KH
Sbjct: 154 GQCIDEATMVRKIVEDISKRKTLKH 178
>At4g22150 unknown protein
Length = 302
Score = 29.3 bits (64), Expect = 2.6
Identities = 18/67 (26%), Positives = 31/67 (45%), Gaps = 2/67 (2%)
Query: 84 PILVKIVKKKSDLPVREQIFLLLDATQTSLGGASGKFPQYYKAYYDLVSAGVQFPQRAQV 143
P+ V ++++ P +E++ + +LGGAS D+ A VQ P ++ V
Sbjct: 164 PVHVNLMRRDEKCPEKEKLKVSFQGVGRTLGGASSSTASSQSNLTDV--AAVQSPLQSLV 221
Query: 144 VQSNRPS 150
V PS
Sbjct: 222 VDETLPS 228
>At2g35930 unknown protein
Length = 411
Score = 29.3 bits (64), Expect = 2.6
Identities = 17/77 (22%), Positives = 44/77 (57%), Gaps = 4/77 (5%)
Query: 17 IDWMKNIEISELVARD---QRKAKDVVKAIKKRLGNKNPNAQLYAVMLLEMLMNNIGDHI 73
++ + ++E SE V ++ +K ++VK++ K + +++YA +LL+ ++ + D +
Sbjct: 165 LNLLYHLETSETVLKNLLNNKKDNNIVKSLTKIMQRGMYESRVYATLLLKNIL-EVADPM 223
Query: 74 NEQVVRAEVIPILVKIV 90
++ EV +V+I+
Sbjct: 224 QSMTLKPEVFTEVVQIL 240
>At4g09710 RNA-directed DNA polymerase -like protein
Length = 1274
Score = 28.9 bits (63), Expect = 3.4
Identities = 24/92 (26%), Positives = 42/92 (45%), Gaps = 9/92 (9%)
Query: 76 QVVRAEVIPILV-----KIVKKKSDLPVREQIFLLLDATQTSLGGASGKFPQYYKAYYDL 130
QVV+ + PI+ +++K S L ++E +F + + G G ++ AY+D+
Sbjct: 347 QVVQEALSPIISSHCNEELIKISSLLEIKEALFSI---SADKAPGPDGFSASFFHAYWDI 403
Query: 131 VSAGVQFPQRAQVVQS-NRPSLQPNTTNNVPK 161
+ A V R+ V S P L +PK
Sbjct: 404 IEADVSRDIRSFFVDSCLSPRLNETHVTLIPK 435
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.130 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,842,059
Number of Sequences: 26719
Number of extensions: 230446
Number of successful extensions: 899
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 872
Number of HSP's gapped (non-prelim): 40
length of query: 292
length of database: 11,318,596
effective HSP length: 98
effective length of query: 194
effective length of database: 8,700,134
effective search space: 1687825996
effective search space used: 1687825996
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148755.10


