

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148755.1 + phase: 0
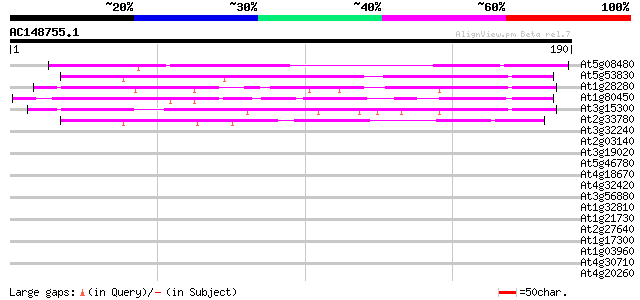
(190 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g08480 unknown protein 134 3e-32
At5g53830 unknown protein 77 5e-15
At1g28280 unknown protein 68 3e-12
At1g80450 unknown protein 61 3e-10
At3g15300 unknown protein 59 2e-09
At2g33780 hypothetical protein 54 5e-08
At3g32240 hypothetical protein 34 0.053
At2g03140 similar to late embryogenesis abundant proteins 34 0.053
At3g19020 hypothetical protein 33 0.091
At5g46780 unknown protein 33 0.12
At4g18670 extensin-like protein 33 0.12
At4g32420 cyclophilin (AtCYP95) 32 0.26
At3g56880 unknown protein 31 0.35
At1g32810 hypothetical protein 31 0.45
At1g21730 kinesin-related protein (MKRP1) 31 0.45
At2g27640 unknown protein 30 0.59
At1g17300 unknown protein 30 0.59
At1g03960 protein phosphatase 2A like protein 30 0.59
At4g30710 putative protein 30 0.77
At4g20260 endomembrane-associated protein 30 0.77
>At5g08480 unknown protein
Length = 142
Score = 134 bits (337), Expect = 3e-32
Identities = 84/179 (46%), Positives = 94/179 (51%), Gaps = 53/179 (29%)
Query: 14 CKPLTTFVHTNTGAFREVVQRLTGPSEGN---ATKEEQTAKVAPITKRTPPKLHERRKCM 70
CKP+TTFV T+T FRE+VQRLTGP+E N AT E K A I KR KLHERR+CM
Sbjct: 13 CKPVTTFVQTDTNTFREIVQRLTGPTENNAAAATPEATVIKTA-IQKRPTSKLHERRQCM 71
Query: 71 KPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIM 130
+PKLEIVKP L + G S +
Sbjct: 72 RPKLEIVKPPLSFKPTEGEPDSCTT----------------------------------- 96
Query: 131 EDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASPKACEK 189
NIEEEEKAIKERRFYLHPSPRSK G+ EPELL LFPL SP + K
Sbjct: 97 -------------NIEEEEKAIKERRFYLHPSPRSK-PGYTEPELLTLFPLTSPNSSGK 141
>At5g53830 unknown protein
Length = 243
Score = 77.0 bits (188), Expect = 5e-15
Identities = 63/198 (31%), Positives = 87/198 (43%), Gaps = 38/198 (19%)
Query: 18 TTFVHTNTGAFREVVQRLTG-------------PSEGNATKEEQTAKVAPITKRTPPKLH 64
TTFV +T F++VVQ LTG PS N + + + PI K KL+
Sbjct: 49 TTFVQADTSTFKQVVQMLTGSSTDTTTGKHHEAPSPVNNNNKGSSFSIPPIKKTNSFKLY 108
Query: 65 ERRKCMK------------------PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVS 106
ERR+ +L N +H+ P SP + S + + SP
Sbjct: 109 ERRQNNNNMFAKNDLMINTLRLQNSQRLMFTGGNSSHHQSPRFSPRNSSSSENILLSPSM 168
Query: 107 GSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSK 166
L SP TP L +D + S ++ N EE+KAI ++ FYLHPSP S
Sbjct: 169 LDFPKLGLNSPVTP------LRSNDDPFNKSSPLSLGNSSEEDKAIADKGFYLHPSPVST 222
Query: 167 ASGFNEPELLNLFPLASP 184
++P LL LFP+ASP
Sbjct: 223 PRD-SQPLLLPLFPVASP 239
>At1g28280 unknown protein
Length = 247
Score = 68.2 bits (165), Expect = 3e-12
Identities = 70/209 (33%), Positives = 94/209 (44%), Gaps = 52/209 (24%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEG---------NATKEEQTAKVAPITKRT 59
S +G+ P TTFV +T +F++VVQ LTG +E N T + + P +
Sbjct: 49 SESGNPYP-TTFVQADTSSFKQVVQMLTGSAERPKHGSSLKPNPTHHQPDPRSTPSSFSI 107
Query: 60 PP-----------------KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSF-P 101
PP +L+ERR MK NL+ + +P NS+F P
Sbjct: 108 PPIKAVPNKKQSSSSASGFRLYERRNSMK--------NLKINP---LNPVFNPVNSAFSP 156
Query: 102 SSPVSGSGS----SSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEIN-IEEEEKAIKERR 156
P S S SL+ SP TP I + + GS N + EEKA+KER
Sbjct: 157 RKPEILSPSILDFPSLVLSPVTP-------LIPDPFDRSGSSNQSPNELAAEEKAMKERG 209
Query: 157 FYLHPSPRSKASGFNEPELLNLFPLASPK 185
FYLHPSP + EP LL LFP+ SP+
Sbjct: 210 FYLHPSPATTPMD-PEPRLLPLFPVTSPR 237
>At1g80450 unknown protein
Length = 177
Score = 61.2 bits (147), Expect = 3e-10
Identities = 62/188 (32%), Positives = 85/188 (44%), Gaps = 34/188 (18%)
Query: 2 EKPPVHGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVA----PITK 57
++PP + + +P T FV + FR +VQ+LTG ++ A P+T
Sbjct: 4 QQPPSYAT-----EPNTMFVQADPSNFRNIVQKLTGAPPDISSSSFSAVSAAHQKLPLTP 58
Query: 58 RTPP-KLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPS 116
+ P KLHERR+ K K+E+ N+ P + S R F SPVS S
Sbjct: 59 KKPAFKLHERRQSSK-KMELKVNNIT---NPNDAFSHFHRG--FLVSPVSHLDPFWARVS 112
Query: 117 PTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELL 176
P + E+ Q +EE+KAI E+ FY PSPRS + PELL
Sbjct: 113 PHSAR---------EEHHAQPD-------KEEQKAIAEKGFYFLPSPRSGSE--PAPELL 154
Query: 177 NLFPLASP 184
LFPL SP
Sbjct: 155 PLFPLRSP 162
>At3g15300 unknown protein
Length = 219
Score = 58.5 bits (140), Expect = 2e-09
Identities = 59/200 (29%), Positives = 94/200 (46%), Gaps = 33/200 (16%)
Query: 7 HGSTTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHER 66
H T D P TTFV ++ +F++VVQ LTG S +P + R P +
Sbjct: 27 HIITRSDHYP-TTFVQADSSSFKQVVQMLTGSSSPR----------SPDSPRPPTTPSGK 75
Query: 67 RKCMKPKLEIVKP-----NLQYHKQPGASPSSKSRNSSFPSS-PVSGSGSSSLLPSP--- 117
+ P ++ +P N Y ++ ++ +NS ++ + G G+ S SP
Sbjct: 76 GNFVIPPIKTAQPKKHSGNKLYERRSHGGFNNNLKNSLMINTLMIGGGGAGSPRFSPRNQ 135
Query: 118 --TTPSTL-FSRLTIME--DEKKQGSVINEIN-------IEEEEKAIKERRFYLHPSPRS 165
+PS L F +L + KQG+ NE + + EEE+ I ++ +YLH SP S
Sbjct: 136 EILSPSCLDFPKLALNSPVTPLKQGTNGNEGDPFDKMSPLSEEERGIADKGYYLHRSPIS 195
Query: 166 KASGFNEPELLNLFPLASPK 185
+EP+LL LFP+ SP+
Sbjct: 196 TPRD-SEPQLLPLFPVTSPR 214
>At2g33780 hypothetical protein
Length = 204
Score = 53.9 bits (128), Expect = 5e-08
Identities = 55/184 (29%), Positives = 76/184 (40%), Gaps = 48/184 (26%)
Query: 18 TTFVHTNTGAFREVVQRLTG----PSEGNATKEEQTAKVAPITKRTPPK------LHERR 67
TTF+ T+ +F++VVQ LTG P+ + + PI T K L ERR
Sbjct: 36 TTFIRTDPSSFKQVVQLLTGIPKNPTHQPDPRFPPFHSIPPIKAVTNKKQSSSFRLSERR 95
Query: 68 KCMKPKL----------EIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSP 117
MK L EI+ P + SP ++ S P GS S PS
Sbjct: 96 NSMKHYLNINPTHSGPPEILTPTILNFPALDLSP-----DTPLMSDPFYRPGSFSQSPSD 150
Query: 118 TTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLN 177
+ PS +++E++IKE+ FYL PSP S EP LL+
Sbjct: 151 SKPSF----------------------DDDQERSIKEKGFYLRPSP-STTPRDTEPRLLS 187
Query: 178 LFPL 181
LFP+
Sbjct: 188 LFPM 191
>At3g32240 hypothetical protein
Length = 487
Score = 33.9 bits (76), Expect = 0.053
Identities = 33/127 (25%), Positives = 51/127 (39%), Gaps = 12/127 (9%)
Query: 36 TGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKS 95
+ P G+AT T+ V + P H RR P + P Q P S S +
Sbjct: 17 SNPVGGDATSPGNTSAVQDNRQAHPR--HSRRHSGSPAMSTGSP-AQSTGSPAQSTGSPA 73
Query: 96 RNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEE-EEKAIKE 154
S PSSP +G SP+TP++ + + S +N + ++E ++E
Sbjct: 74 LFSGSPSSPATG--------SPSTPASGSHSSPANQTSQPPSSRLNNLTLDELLVSPVRE 125
Query: 155 RRFYLHP 161
LHP
Sbjct: 126 HLKKLHP 132
>At2g03140 similar to late embryogenesis abundant proteins
Length = 1805
Score = 33.9 bits (76), Expect = 0.053
Identities = 26/118 (22%), Positives = 55/118 (46%), Gaps = 10/118 (8%)
Query: 39 SEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNS 98
+EG+A K + P+ K + K M+P + KP +Q QP N
Sbjct: 892 NEGDAGKSSASQ---PVEKDESNDQSKETKVMQPVSDQTKPAIQEPNQPNF-------NV 941
Query: 99 SFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVINEINIEEEEKAIKERR 156
S ++G S+ + + L + ++ +++EKK+G+ +++ ++EK +K+ +
Sbjct: 942 SQAFEALTGMDDSTQVAVNSVFGVLENMISQLDEEKKEGNEVSDEKNLKDEKNLKDAK 999
>At3g19020 hypothetical protein
Length = 951
Score = 33.1 bits (74), Expect = 0.091
Identities = 23/83 (27%), Positives = 31/83 (36%), Gaps = 2/83 (2%)
Query: 38 PSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRN 97
P + ++ +E + +P K PPK E K PK E KP QP + K
Sbjct: 502 PKQESSKQEPPKPEESP--KPEPPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEE 559
Query: 98 SSFPSSPVSGSGSSSLLPSPTTP 120
S P P + P P P
Sbjct: 560 SPKPQPPKQETPKPEESPKPQPP 582
Score = 33.1 bits (74), Expect = 0.091
Identities = 28/101 (27%), Positives = 36/101 (34%), Gaps = 11/101 (10%)
Query: 38 PSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQP---------- 87
P + K E++ K P K+ PK E K PK E KP QP
Sbjct: 533 PPKQETPKPEESPKPQP-PKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQEQPPKTE 591
Query: 88 GASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLT 128
S S P+ P S P P +PST ++ T
Sbjct: 592 APKMGSPPLESPVPNDPYDASPIKKRRPQPPSPSTEETKTT 632
Score = 29.6 bits (65), Expect = 1.0
Identities = 22/76 (28%), Positives = 28/76 (35%), Gaps = 1/76 (1%)
Query: 45 KEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSP 104
K E++ K P K+ PK E K PK E KP QP + K S P P
Sbjct: 524 KPEESPKPQP-PKQETPKPEESPKPQPPKQETPKPEESPKPQPPKQETPKPEESPKPQPP 582
Query: 105 VSGSGSSSLLPSPTTP 120
+ P +P
Sbjct: 583 KQEQPPKTEAPKMGSP 598
Score = 29.3 bits (64), Expect = 1.3
Identities = 26/83 (31%), Positives = 32/83 (38%), Gaps = 9/83 (10%)
Query: 38 PSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRN 97
PS K E + +P T++ PK E K PK E KP +QP P S +
Sbjct: 452 PSNPKEPKPESPKQESPKTEQPKPK-PESPKQESPKQEAPKP-----EQPKPKPESPKQE 505
Query: 98 SSFPSSPVSGSGSSSLLPSPTTP 120
SS P S P P P
Sbjct: 506 SSKQEPP---KPEESPKPEPPKP 525
Score = 28.9 bits (63), Expect = 1.7
Identities = 19/60 (31%), Positives = 25/60 (41%)
Query: 61 PKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTP 120
PK K +PK EI PNL+ +P S K + S S S+ P P +P
Sbjct: 404 PKPTPTPKAPEPKKEINPPNLEEPSKPKPEESPKPQQPSPKPETPSHEPSNPKEPKPESP 463
>At5g46780 unknown protein
Length = 237
Score = 32.7 bits (73), Expect = 0.12
Identities = 33/123 (26%), Positives = 47/123 (37%), Gaps = 17/123 (13%)
Query: 16 PLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQT------------AKVAPITKRT---- 59
P + N FR +VQ+LTG ++ QT + AP+T+
Sbjct: 44 PQALVYNINKTDFRSIVQQLTGLGSTSSVNPPQTNHPKPPNSRLVKVRPAPLTQLNHPPP 103
Query: 60 PPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTT 119
PP + + E V+P Q+ P SP S SSPV G+ +P
Sbjct: 104 PPPPPPPVQSVPIASEPVQPVNQFSSNPAESPISAYMRYLIESSPV-GNRVQPQNQNPVQ 162
Query: 120 PST 122
PST
Sbjct: 163 PST 165
>At4g18670 extensin-like protein
Length = 839
Score = 32.7 bits (73), Expect = 0.12
Identities = 27/89 (30%), Positives = 37/89 (41%), Gaps = 4/89 (4%)
Query: 36 TGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGA---SPS 92
T PS G + + PIT +PP P IV PG+ SP+
Sbjct: 415 TTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGS-PPSPSIVPSPPSTTPSPGSPPTSPT 473
Query: 93 SKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
+ + S PSSP + + S SPTTP+
Sbjct: 474 TPTPGGSPPSSPTTPTPGGSPPSSPTTPT 502
Score = 30.0 bits (66), Expect = 0.77
Identities = 15/35 (42%), Positives = 21/35 (59%)
Query: 87 PGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPS 121
P +SP++ + S PSSP + + S SPTTPS
Sbjct: 481 PPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPS 515
Score = 27.3 bits (59), Expect = 5.0
Identities = 22/85 (25%), Positives = 30/85 (34%), Gaps = 23/85 (27%)
Query: 36 TGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHKQPGASPSSKS 95
T PS G + + PIT +PP PG+ PS S
Sbjct: 512 TTPSPGGSPPSPSISPSPPITVPSPPSTPT--------------------SPGSPPSPSS 551
Query: 96 RNSSFPSSPVSGSGSSSLLPSPTTP 120
PSSP+ + S P+P +P
Sbjct: 552 PT---PSSPIPSPPTPSTPPTPISP 573
Score = 26.9 bits (58), Expect = 6.5
Identities = 21/51 (41%), Positives = 23/51 (44%), Gaps = 15/51 (29%)
Query: 87 PGASPSSKSRNSS-------------FPSSPVSGSG--SSSLLPSPTTPST 122
PG SP S S + S P SP S S SS +PSP TPST
Sbjct: 516 PGGSPPSPSISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPSPPTPST 566
Score = 26.9 bits (58), Expect = 6.5
Identities = 21/57 (36%), Positives = 26/57 (44%), Gaps = 11/57 (19%)
Query: 76 IVKPNLQYHKQPGASPSSKSRNSSFP---------SSPVSGSGSSSLLPSP--TTPS 121
+V P+ PG SP S S + S P SP S S++PSP TTPS
Sbjct: 408 VVVPSPPTTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPSTTPS 464
>At4g32420 cyclophilin (AtCYP95)
Length = 837
Score = 31.6 bits (70), Expect = 0.26
Identities = 22/83 (26%), Positives = 36/83 (42%), Gaps = 17/83 (20%)
Query: 53 APITKRTPPKLHERRKCMKPKL------------EIVKPNLQYHKQPGASPS-----SKS 95
+P+ RTPP+ R + + P L P + + P AS S S+S
Sbjct: 728 SPLRGRTPPRYRRRSRSVSPGLCYRNRRYSRSPIRSRSPPYRKRRSPSASHSLSPSRSRS 787
Query: 96 RNSSFPSSPVSGSGSSSLLPSPT 118
R+ S+ SP+ + S+ SP+
Sbjct: 788 RSKSYSKSPIGTGKARSVSRSPS 810
>At3g56880 unknown protein
Length = 245
Score = 31.2 bits (69), Expect = 0.35
Identities = 32/127 (25%), Positives = 52/127 (40%), Gaps = 18/127 (14%)
Query: 9 STTGDCKPLTTFVHTNTGAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHER-- 66
S + K TTF+ + FR++VQ++TG ++ + API K P +L R
Sbjct: 113 SRVSNKKSQTTFITADAANFRQMVQQVTGAKFLGSS----NSIFAPIVKPEPHRLASRLP 168
Query: 67 -------RKCMKPKLEIVKPNLQYHKQP-----GASPSSKSRNSSFPSSPVSGSGSSSLL 114
R P L+ +H++ GA S SS ++ + G SS +
Sbjct: 169 PSCGNLDRSSAVPTLDTSSFLSNHHQENIITDLGAPTGSFHHQSSAGTTTANVGGGSSAV 228
Query: 115 PSPTTPS 121
+ PS
Sbjct: 229 ELDSYPS 235
>At1g32810 hypothetical protein
Length = 654
Score = 30.8 bits (68), Expect = 0.45
Identities = 35/123 (28%), Positives = 48/123 (38%), Gaps = 13/123 (10%)
Query: 72 PKLEIVKPNLQYHKQPGASPSSKSRNSSFPSSPVSG---SGSSSLLPSPTTPSTLFSRLT 128
PK I H + S SK+R SS S S +G SS PS + S F +
Sbjct: 217 PKFSITSKKSMQHNRTSHSSVSKTRESSSSSKTSSATRINGGSSEAPSKHSLSGTFPKNE 276
Query: 129 IMEDEKKQGSVINEI--------NIEEEEKAIKERRFYLHPSPR-SKASGFNEPELLNLF 179
Q S N + N+ +EE A++ L+ SPR + +P L L
Sbjct: 277 KPGQSIFQSSTKNPVQSIISLAPNLSDEELALR-LHHQLNSSPRVPRVPRMRQPGSLPLS 335
Query: 180 PLA 182
P A
Sbjct: 336 PTA 338
>At1g21730 kinesin-related protein (MKRP1)
Length = 890
Score = 30.8 bits (68), Expect = 0.45
Identities = 32/144 (22%), Positives = 56/144 (38%), Gaps = 3/144 (2%)
Query: 26 GAFREVVQRLTGPSEGNATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHK 85
GA V + L P +GN++ +E T +T L + + P
Sbjct: 512 GAVSTVSEHLKEPRDGNSSLDEMTKDRRK--NKTRGMLGWLKLKKSDGVAGTLPTDGNQS 569
Query: 86 QPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQGSVI-NEIN 144
Q SPSS S+ + ++ + + +P T LFS ED G+ I ++++
Sbjct: 570 QASGSPSSSSKYTQTKTTRRENAAAIKSIPEKTVAGDLFSATVGPEDSSPTGTTIADQMD 629
Query: 145 IEEEEKAIKERRFYLHPSPRSKAS 168
+ E+ I L S ++ S
Sbjct: 630 LLHEQTKILVGEVALRTSSLNRLS 653
>At2g27640 unknown protein
Length = 404
Score = 30.4 bits (67), Expect = 0.59
Identities = 23/66 (34%), Positives = 31/66 (46%), Gaps = 16/66 (24%)
Query: 85 KQPGASP--------SSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQ 136
K P SP +SK RNS+ SSP+S G L P T+P T+ ED +
Sbjct: 21 KNPKPSPIQSKKKKNTSKKRNSTSMSSPLSKPG-EHLEPETTSP-------TVEEDSMEL 72
Query: 137 GSVINE 142
G +N+
Sbjct: 73 GDTVNQ 78
>At1g17300 unknown protein
Length = 116
Score = 30.4 bits (67), Expect = 0.59
Identities = 15/33 (45%), Positives = 21/33 (63%)
Query: 83 YHKQPGASPSSKSRNSSFPSSPVSGSGSSSLLP 115
+ + P +P+S SR+ S SS +S S SSS LP
Sbjct: 58 FRRVPTPTPASSSRHLSASSSDISASDSSSSLP 90
>At1g03960 protein phosphatase 2A like protein
Length = 529
Score = 30.4 bits (67), Expect = 0.59
Identities = 19/55 (34%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query: 90 SPSSKSRN--------SSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTIMEDEKKQ 136
+P++KS+N S+ P SP S SGS T+P +L S L +++ K+Q
Sbjct: 63 TPTNKSKNLPSVFLSSSTPPLSPRSSSGSPRFSRQRTSPPSLHSPLRSLKEPKRQ 117
>At4g30710 putative protein
Length = 644
Score = 30.0 bits (66), Expect = 0.77
Identities = 28/104 (26%), Positives = 45/104 (42%), Gaps = 18/104 (17%)
Query: 30 EVVQRLTGPSEGN----ATKEEQTAKVAPITKRTPPKLHERRKCMKPKLEIVKPNLQYHK 85
+ +R PS+ N AT+ +T +V+ + P + R C P + +P +
Sbjct: 6 DTTRRRLLPSDKNNAVVATRRPRTMEVSSRYRSPTPTKNGR--CPSPS--VTRPTVSSSS 61
Query: 86 QPGASPSSKSRNSSFPSSPVSGSGSSSLLPSPTTPSTLFSRLTI 129
Q A+ + S PS+P PSPT+PST L+I
Sbjct: 62 QSVAAKRAVSAERKRPSTP----------PSPTSPSTPIRDLSI 95
>At4g20260 endomembrane-associated protein
Length = 225
Score = 30.0 bits (66), Expect = 0.77
Identities = 14/53 (26%), Positives = 26/53 (48%)
Query: 132 DEKKQGSVINEINIEEEEKAIKERRFYLHPSPRSKASGFNEPELLNLFPLASP 184
+E K+G +EE++ ++E++ P+P + EPE P+A P
Sbjct: 170 EETKKGETPETAVVEEKKPEVEEKKEEATPAPAVVETPVKEPETTTTAPVAEP 222
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.309 0.127 0.360
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,964,032
Number of Sequences: 26719
Number of extensions: 226209
Number of successful extensions: 1147
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 57
Number of HSP's that attempted gapping in prelim test: 995
Number of HSP's gapped (non-prelim): 175
length of query: 190
length of database: 11,318,596
effective HSP length: 94
effective length of query: 96
effective length of database: 8,807,010
effective search space: 845472960
effective search space used: 845472960
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC148755.1


