

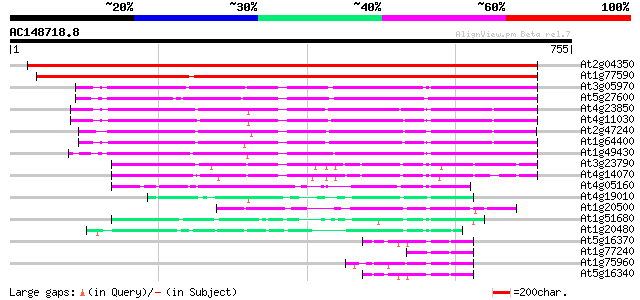
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.8 - phase: 0
(755 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g04350 putative acyl-CoA synthetase 1013 0.0
At1g77590 long chain acyl-CoA synthetase 9 (LACS9) 920 0.0
At3g05970 AMP-binding protein 360 1e-99
At5g27600 long chain acyl-CoA synthetase 7 (LACS7) 360 2e-99
At4g23850 acyl-CoA synthetase - like protein 322 6e-88
At4g11030 putative acyl-CoA synthetase 311 1e-84
At2g47240 putative acyl-CoA synthetase 307 1e-83
At1g64400 putative acyl-CoA synthetase 306 4e-83
At1g49430 acyl CoA synthetase-like protein (At1g49430) 299 3e-81
At3g23790 AMP-binding protein, putative 144 1e-34
At4g14070 A6 anther-specific protein (Z97335.16) 143 4e-34
At4g05160 4-coumarate--CoA ligase - like protein 102 1e-21
At4g19010 4-coumarate-CoA ligase - like 99 9e-21
At1g20500 hypothetical protein 93 5e-19
At1g51680 4-coumarate--coenzyme A ligase (At4CL1) 90 5e-18
At1g20480 unknown protein 81 2e-15
At5g16370 AMP-binding protein 68 2e-11
At1g77240 unknown protein 68 2e-11
At1g75960 adenosine monophosphate binding protein 8 AMPBP8 68 2e-11
At5g16340 AMP-binding protein 66 8e-11
>At2g04350 putative acyl-CoA synthetase
Length = 720
Score = 1013 bits (2618), Expect = 0.0
Identities = 487/687 (70%), Positives = 580/687 (83%)
Query: 24 DHGELGTVFAIVIGIIVPVLFSVFFLGKKKGKVRGVPVEVSGESGYAVRNARYSELVEVP 83
D G G + ++ ++VPVL SV G KKGK RGVP++V GE GY +R+AR ELV+VP
Sbjct: 17 DFGVYGIIGGGIVALLVPVLLSVVLNGTKKGKKRGVPIKVGGEEGYTMRHARAPELVDVP 76
Query: 84 WKGAPTMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVF 143
W+GA TM LFE SCKK++ +R LGTR+ I KEF+T+SDGRKFEK+HLGEY+W++YGEVF
Sbjct: 77 WEGAATMPALFEQSCKKYSKDRLLGTREFIDKEFITASDGRKFEKLHLGEYKWQSYGEVF 136
Query: 144 ARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHS 203
RV NFASGL+ +GH++D VAIFSDTRAEWFIA QGCFRQ+ITVVTIYASLGE+ALI+S
Sbjct: 137 ERVCNFASGLVNVGHNVDDRVAIFSDTRAEWFIAFQGCFRQSITVVTIYASLGEEALIYS 196
Query: 204 LNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCKIASFDE 263
LNET+VSTLICD K L KL AI+S L +++NIIY E+D + + + ++S E
Sbjct: 197 LNETRVSTLICDSKQLKKLSAIQSSLKTVKNIIYIEEDGVDVASSDVNSMGDITVSSISE 256
Query: 264 VEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSK 323
VEKLG+++ V+P LPSKN VAV+M+TSGSTGLPKGVMITHGN+VAT A VM V+P L
Sbjct: 257 VEKLGQKNAVQPILPSKNGVAVIMFTSGSTGLPKGVMITHGNLVATAAGVMKVVPKLDKN 316
Query: 324 DVYLAYLPLAHVFEMAAESVMLAAGVAIGYGSPMTLTDTSNKVKKGTKGDVTVLKPTLLT 383
D Y+AYLPLAHVFE+ AE V+ +G AIGYGS MTLTDTSNKVKKGTKGDV+ LKPT++T
Sbjct: 317 DTYIAYLPLAHVFELEAEIVVFTSGSAIGYGSAMTLTDTSNKVKKGTKGDVSALKPTIMT 376
Query: 384 AVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFK 443
AVPAI+DR+R+GV+KKVEEKGG+AK LF AYKRRLAAV GSW GAWG+EK++WD ++FK
Sbjct: 377 AVPAILDRVREGVLKKVEEKGGMAKTLFDFAYKRRLAAVDGSWFGAWGLEKMLWDALVFK 436
Query: 444 KIRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVG 503
KIR VLGG++RFML GGAPLS DSQ+FINIC+G+PIGQGYGLTET AGA FSE DD +VG
Sbjct: 437 KIRAVLGGHIRFMLVGGAPLSPDSQRFINICMGSPIGQGYGLTETCAGATFSEWDDPAVG 496
Query: 504 RVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEK 563
RVGPPLPC Y+KLVSWEEGGY SDKPMPRGE+VVGG SVTAGYF NQ+KTDEV+KVDEK
Sbjct: 497 RVGPPLPCGYVKLVSWEEGGYRISDKPMPRGEIVVGGNSVTAGYFNNQEKTDEVYKVDEK 556
Query: 564 GVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHAD 623
G RWFYTGDIGRFHPDGCLE+IDRKKDIVKLQHGEY+SLGKVEAAL S +YVD+IMVHAD
Sbjct: 557 GTRWFYTGDIGRFHPDGCLEVIDRKKDIVKLQHGEYVSLGKVEAALGSSNYVDNIMVHAD 616
Query: 624 PFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKT 683
P +SYCVALVV S +LEKWA+E G+++ +F +LC K EAV EV QS++KA KAAKL+K
Sbjct: 617 PINSYCVALVVPSRGALEKWAEEAGVKHSEFAELCEKGEAVKEVQQSLTKAGKAAKLEKF 676
Query: 684 EVPAKIKLLADPWTPESGLVTAALKLK 710
E+PAKIKLL++PWTPESGLVTAALK+K
Sbjct: 677 ELPAKIKLLSEPWTPESGLVTAALKIK 703
>At1g77590 long chain acyl-CoA synthetase 9 (LACS9)
Length = 691
Score = 920 bits (2379), Expect = 0.0
Identities = 437/674 (64%), Positives = 545/674 (80%), Gaps = 6/674 (0%)
Query: 37 GIIVPVLFSVFFLGKKKGKVRGVPVEVSGESGYAVRNARYSELVEVPWKGAPTMAHLFEP 96
G+IVP+ + KK K RGV V+V GE GYA+RN R++E V W+ T+ LFE
Sbjct: 7 GVIVPLALTFLVQKSKKEKKRGVVVDVGGEPGYAIRNHRFTEPVSSHWEHISTLPELFEI 66
Query: 97 SCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSNFASGLLKL 156
SC H+ FLGTRKLI +E TS DG+ FEK+HLG+YEW T+G+ V +FASGL+++
Sbjct: 67 SCNAHSDRVFLGTRKLISREIETSEDGKTFEKLHLGDYEWLTFGKTLEAVCDFASGLVQI 126
Query: 157 GHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQVSTLICDV 216
GH + VAIF+DTR EWFI+LQGCFR+N+TVVTIY+SLGE+AL HSLNET+V+T+IC
Sbjct: 127 GHKTEERVAIFADTREEWFISLQGCFRRNVTVVTIYSSLGEEALCHSLNETEVTTVICGS 186
Query: 217 KLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCKIASFDEVEKLGKESPVEPS 276
K L KL I +L +++ +I +D+ F ++SN SF +V+KLG+E+PV+P+
Sbjct: 187 KELKKLMDISQQLETVKRVICMDDE------FPSDVNSNWMATSFTDVQKLGRENPVDPN 240
Query: 277 LPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAHVF 336
P VAV+MYTSGSTGLPKGVM+THGN++AT +AVMT++P+LG +D+Y+AYLPLAH+
Sbjct: 241 FPLSADVAVIMYTSGSTGLPKGVMMTHGNVLATVSAVMTIVPDLGKRDIYMAYLPLAHIL 300
Query: 337 EMAAESVMLAAGVAIGYGSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGV 396
E+AAESVM G AIGYGSP+TLTDTSNK+KKGTKGDVT LKPT++TAVPAI+DR+RDGV
Sbjct: 301 ELAAESVMATIGSAIGYGSPLTLTDTSNKIKKGTKGDVTALKPTIMTAVPAILDRVRDGV 360
Query: 397 VKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLRFM 456
KKV+ KGGL+K LF AY RRL+A+ GSW GAWG+EKL+WD ++F+KIR VLGG +R++
Sbjct: 361 RKKVDAKGGLSKKLFDFAYARRLSAINGSWFGAWGLEKLLWDVLVFRKIRAVLGGQIRYL 420
Query: 457 LCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKL 516
L GGAPLSGD+Q+FINICVGAPIGQGYGLTET AG FSE +D SVGRVG PLPC ++KL
Sbjct: 421 LSGGAPLSGDTQRFINICVGAPIGQGYGLTETCAGGTFSEFEDTSVGRVGAPLPCSFVKL 480
Query: 517 VSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRF 576
V W EGGYL+SDKPMPRGE+V+GG ++T GYFKN++KT EV+KVDEKG+RWFYTGDIGRF
Sbjct: 481 VDWAEGGYLTSDKPMPRGEIVIGGSNITLGYFKNEEKTKEVYKVDEKGMRWFYTGDIGRF 540
Query: 577 HPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADPFHSYCVALVVAS 636
HPDGCLEIIDRKKDIVKLQHGEY+SLGKVEAAL+ YV++IMVHAD F+SYCVALVVAS
Sbjct: 541 HPDGCLEIIDRKKDIVKLQHGEYVSLGKVEAALSISPYVENIMVHADSFYSYCVALVVAS 600
Query: 637 HQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEVPAKIKLLADPW 696
++E WA + GI++ +F +LC K +AV EV S+ KAAK ++L+K E+PAKIKLLA PW
Sbjct: 601 QHTVEGWASKQGIDFANFEELCTKEQAVKEVYASLVKAAKQSRLEKFEIPAKIKLLASPW 660
Query: 697 TPESGLVTAALKLK 710
TPESGLVTAALKLK
Sbjct: 661 TPESGLVTAALKLK 674
>At3g05970 AMP-binding protein
Length = 701
Score = 360 bits (925), Expect = 1e-99
Identities = 216/624 (34%), Positives = 328/624 (51%), Gaps = 39/624 (6%)
Query: 89 TMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSN 148
T+ FE + ++LGTR + DG +G+Y+W TYGE +
Sbjct: 85 TLHDNFEHAVHDFRDYKYLGTRVRV--------DGT------VGDYKWMTYGEAGTARTA 130
Query: 149 FASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQ 208
SGL+ G + S V I+ R EW I C + V +Y +LG DA+ +N
Sbjct: 131 LGSGLVHHGIPMGSSVGIYFINRPEWLIVDHACSSYSYVSVPLYDTLGPDAVKFIVNHAT 190
Query: 209 VSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCKIASFDEVEKLG 268
V + C + LN L + S++ S++ ++ E S SS K+ S+ + G
Sbjct: 191 VQAIFCVAETLNSLLSCLSEMPSVRLVVVV--GGLIESLPSLPSSSGVKVVSYSVLLNQG 248
Query: 269 KESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLA 328
+ +P P + VA + YTSG+TG PKGV++TH N++A A + S DVY++
Sbjct: 249 RSNPQRFFPPKPDDVATICYTSGTTGTPKGVVLTHANLIANVAGSSFSV-KFFSSDVYIS 307
Query: 329 YLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVP 386
YLPLAH++E A + + + GVA+G+ G M L D D+ L+PT+ ++VP
Sbjct: 308 YLPLAHIYERANQILTVYFGVAVGFYQGDNMKLLD-----------DLAALRPTVFSSVP 356
Query: 387 AIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKKIR 446
+ +RI G++ V+ GGL + LF AY + A+ + +WD ++F KI+
Sbjct: 357 RLYNRIYAGIINAVKTSGGLKERLFNAAYNAKKQALLNGKSAS-----PIWDRLVFNKIK 411
Query: 447 TVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVG 506
LGG +RFM G +PLS + +F+ +C G + +GYG+TET + + D G VG
Sbjct: 412 DRLGGRVRFMTSGASPLSPEVMEFLKVCFGGRVTEGYGMTETSCVISGMDEGDNLTGHVG 471
Query: 507 PPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVR 566
P P C +KLV E Y S+D+P PRGE+ V G + GY+K++ +T EV +DE G
Sbjct: 472 SPNPACEVKLVDVPEMNYTSADQPHPRGEICVRGPIIFTGYYKDEIQTKEV--IDEDG-- 527
Query: 567 WFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADPFH 626
W +TGDIG + P G L+IIDRKK+I KL GEYI+ K+E A +V ++ D F+
Sbjct: 528 WLHTGDIGLWLPGGRLKIIDRKKNIFKLAQGEYIAPEKIENVYAKCKFVGQCFIYGDSFN 587
Query: 627 SYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEVP 686
S VA+V L+ WA GI+ D +LCN P VL + + A+L+ E
Sbjct: 588 SSLVAVVSVDPDVLKSWAASEGIKGGDLRELCNNPRVKAAVLSDMDTVGREAQLRGFEFA 647
Query: 687 AKIKLLADPWTPESGLVTAALKLK 710
+ L+ +P+T E+GL+T K+K
Sbjct: 648 KAVTLVLEPFTLENGLLTPTFKIK 671
>At5g27600 long chain acyl-CoA synthetase 7 (LACS7)
Length = 698
Score = 360 bits (924), Expect = 2e-99
Identities = 218/625 (34%), Positives = 324/625 (50%), Gaps = 43/625 (6%)
Query: 89 TMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSN 148
T+ F + + + N++LGTR SDG +GEY W TYGE +
Sbjct: 85 TLHDNFVHAVETYAENKYLGTR--------VRSDGT------IGEYSWMTYGEAASERQA 130
Query: 149 FASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQ 208
SGLL G + V ++ R EW + C + V +Y +LG DA+ +N
Sbjct: 131 IGSGLLFHGVNQGDCVGLYFINRPEWLVVDHACAAYSFVSVPLYDTLGPDAVKFVVNHAN 190
Query: 209 VSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCKIASFDEVEKLG 268
+ + C + LN + +KL S N I+ +EH S + I S+ ++ G
Sbjct: 191 LQAIFCVPQTLN---IVIAKLPS-GNPIHSSHCGADEHLPSLPRGTGVTIVSYQKLLSQG 246
Query: 269 KESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLA 328
+ S S P +A + YTSG+TG PKGV++THGN++A A +V DVY++
Sbjct: 247 RSSLHPFSPPKPEDIATICYTSGTTGTPKGVVLTHGNLIANVAG-SSVEAEFFPSDVYIS 305
Query: 329 YLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVP 386
YLPLAH++E A + + + GVA+G+ G L D D VL+PT+ +VP
Sbjct: 306 YLPLAHIYERANQIMGVYGGVAVGFYQGDVFKLMD-----------DFAVLRPTIFCSVP 354
Query: 387 AIIDRIRDGVVKKVEEKGGLAKNLFQIAYK-RRLAAVKGSWLGAWGVEKLVWDTIIFKKI 445
+ +RI DG+ V+ G + K LF+IAY ++ A + G A+ WD ++F KI
Sbjct: 355 RLYNRIYDGITSAVKSSGVVKKRLFEIAYNSKKQAIINGRTPSAF------WDKLVFNKI 408
Query: 446 RTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRV 505
+ LGG +RFM G +PLS D F+ IC G + +GYG+TET + + D G V
Sbjct: 409 KEKLGGRVRFMGSGASPLSPDVMDFLRICFGCSVREGYGMTETSCVISAMDDGDNLSGHV 468
Query: 506 GPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGV 565
G P P C +KLV E Y S D+P PRGE+ V G + GY+K++++T E+ G
Sbjct: 469 GSPNPACEVKLVDVPEMNYTSDDQPYPRGEICVRGPIIFKGYYKDEEQTREIL----DGD 524
Query: 566 RWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADPF 625
W +TGDIG + P G L+IIDRKK+I KL GEYI+ K+E +V +H D F
Sbjct: 525 GWLHTGDIGLWLPGGRLKIIDRKKNIFKLAQGEYIAPEKIENVYTKCRFVSQCFIHGDSF 584
Query: 626 HSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEV 685
+S VA+V + ++ WA GI+Y+ LCN P VL + + A+L+ E
Sbjct: 585 NSSLVAIVSVDPEVMKDWAASEGIKYEHLGQLCNDPRVRKTVLAEMDDLGREAQLRGFEF 644
Query: 686 PAKIKLLADPWTPESGLVTAALKLK 710
+ L+ +P+T E+GL+T K+K
Sbjct: 645 AKAVTLVPEPFTLENGLLTPTFKIK 669
>At4g23850 acyl-CoA synthetase - like protein
Length = 666
Score = 322 bits (824), Expect = 6e-88
Identities = 207/637 (32%), Positives = 338/637 (52%), Gaps = 46/637 (7%)
Query: 83 PWKGAPTMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEV 142
P +G + +F S +K+ +N LG R+++ DG+ G+Y W+TY EV
Sbjct: 40 PIEGMDSCWDVFRMSVEKYPNNPMLGRREIV--------DGKP------GKYVWQTYQEV 85
Query: 143 FARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIH 202
+ V + L +G ++ I+ EW I+++ C + V +Y +LG DA+
Sbjct: 86 YDIVMKLGNSLRSVGVKDEAKCGIYGANSPEWIISMEACNAHGLYCVPLYDTLGADAVEF 145
Query: 203 SLNETQVSTLICDVKLLNKLDAIRSKLTS-LQNIIYFEDDSKEEHTFSEGLSSNCKIASF 261
++ ++VS + + K +++L T ++ ++ F S+E+ +E + I ++
Sbjct: 146 IISHSEVSIVFVEEKKISELFKTCPNSTEYMKTVVSFGGVSREQKEEAE--TFGLVIYAW 203
Query: 262 DEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPN-- 319
DE KLG+ + + K+ + +MYTSG+TG PKGVMI++ +IV A V+ ++ +
Sbjct: 204 DEFLKLGEGKQYDLPIKKKSDICTIMYTSGTTGDPKGVMISNESIVTLIAGVIRLLKSAN 263
Query: 320 --LGSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVT 375
L KDVYL+YLPLAH+F+ E + G AIG+ G L + D+
Sbjct: 264 EALTVKDVYLSYLPLAHIFDRVIEECFIQHGAAIGFWRGDVKLLIE-----------DLA 312
Query: 376 VLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKL 435
LKPT+ AVP ++DR+ G+ KK+ + G L K +F A+ + +K G VE
Sbjct: 313 ELKPTIFCAVPRVLDRVYSGLQKKLSDGGFLKKFIFDSAFSYKFGYMKK---GQSHVEAS 369
Query: 436 -VWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAF 494
++D ++F K++ LGGN+R +L G APL+ + F+ + + QGYGLTE+ AG
Sbjct: 370 PLFDKLVFSKVKQGLGGNVRIILSGAAPLASHVESFLRVVACCHVLQGYGLTESCAGTFV 429
Query: 495 SEADDYSV-GRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDK 553
S D+ + G VGPP+P I+L S E Y + RGE+ + G ++ +GY+K +D
Sbjct: 430 SLPDELGMLGTVGPPVPNVDIRLESVPEMEY-DALASTARGEICIRGKTLFSGYYKREDL 488
Query: 554 TDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGD 613
T EV +D W +TGD+G + PDG ++IIDRKK+I KL GEY+++ +E
Sbjct: 489 TKEVL-IDG----WLHTGDVGEWQPDGSMKIIDRKKNIFKLSQGEYVAVENIENIYGEVQ 543
Query: 614 YVDSIMVHADPFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISK 673
VDS+ V+ + F S+ +A+ + LE+WA E G+ D+ LC +A +L + K
Sbjct: 544 AVDSVWVYGNSFESFLIAIANPNQHILERWAAENGVS-GDYDALCQNEKAKEFILGELVK 602
Query: 674 AAKAAKLQKTEVPAKIKLLADPWTPESGLVTAALKLK 710
AK K++ E+ I L P+ E L+T K K
Sbjct: 603 MAKEKKMKGFEIIKAIHLDPVPFDMERDLLTPTFKKK 639
>At4g11030 putative acyl-CoA synthetase
Length = 666
Score = 311 bits (796), Expect = 1e-84
Identities = 203/636 (31%), Positives = 328/636 (50%), Gaps = 44/636 (6%)
Query: 83 PWKGAPTMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEV 142
P G + +F + +K+ +NR LG R++ S+G+ G+Y W+TY EV
Sbjct: 40 PIDGIQSCWDIFRTAVEKYPNNRMLGRREI--------SNGKA------GKYVWKTYKEV 85
Query: 143 FARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIH 202
+ V + L G I+ EW I+++ C + V +Y +LG A+
Sbjct: 86 YDIVIKLGNSLRSCGIKEGEKCGIYGINCCEWIISMEACNAHGLYCVPLYDTLGAGAVEF 145
Query: 203 SLNETQVSTLICDVKLLNKLDAIRSKLTS-LQNIIYFEDDSKEEHTFSEGLSSNCKIASF 261
++ +VS + K + +L T ++ ++ F E+ +E L I S+
Sbjct: 146 IISHAEVSIAFVEEKKIPELFKTCPNSTKYMKTVVSFGGVKPEQKEEAEKLG--LVIHSW 203
Query: 262 DEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPN-- 319
DE KLG+ E + + + +MYTSG+TG PKGVMI++ +IV T VM + N
Sbjct: 204 DEFLKLGEGKQYELPIKKPSDICTIMYTSGTTGDPKGVMISNESIVTITTGVMHFLGNVN 263
Query: 320 --LGSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVT 375
L KDVY++YLPLAHVF+ A E ++ G +IG+ G L + D+
Sbjct: 264 ASLSEKDVYISYLPLAHVFDRAIEECIIQVGGSIGFWRGDVKLLIE-----------DLG 312
Query: 376 VLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKL 435
LKP++ AVP ++DR+ G+ +K+ G K +F +A+ + +K
Sbjct: 313 ELKPSIFCAVPRVLDRVYTGLQQKLSGGGFFKKKVFDVAFSYKFGNMKKGQSHV--AASP 370
Query: 436 VWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFS 495
D ++F K++ LGGN+R +L G APL+ + F+ + + QGYGLTE+ AG +
Sbjct: 371 FCDKLVFNKVKQGLGGNVRIILSGAAPLASHIESFLRVVACCNVLQGYGLTESCAGTFAT 430
Query: 496 EADDYSV-GRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKT 554
D+ + G VGPP+P I+L S E Y + PRGE+ + G ++ +GY+K +D T
Sbjct: 431 FPDELDMLGTVGPPVPNVDIRLESVPEMNYDALGST-PRGEICIRGKTLFSGYYKREDLT 489
Query: 555 DEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDY 614
EVF +D W +TGD+G + P+G ++IIDRKK+I KL GEY+++ +E + +
Sbjct: 490 KEVF-IDG----WLHTGDVGEWQPNGSMKIIDRKKNIFKLAQGEYVAVENLENVYSQVEV 544
Query: 615 VDSIMVHADPFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKA 674
++SI V+ + F S+ VA+ + Q+LE+WA E G+ DF +C +A +L + K
Sbjct: 545 IESIWVYGNSFESFLVAIANPAQQTLERWAVENGVN-GDFNSICQNAKAKAFILGELVKT 603
Query: 675 AKAAKLQKTEVPAKIKLLADPWTPESGLVTAALKLK 710
AK KL+ E+ + L + E L+T K K
Sbjct: 604 AKENKLKGFEIIKDVHLEPVAFDMERDLLTPTYKKK 639
>At2g47240 putative acyl-CoA synthetase
Length = 660
Score = 307 bits (787), Expect = 1e-83
Identities = 198/625 (31%), Positives = 312/625 (49%), Gaps = 46/625 (7%)
Query: 93 LFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSNFASG 152
+F S +K N LG R+++ ++ +G Y W+TY EV+ V S
Sbjct: 47 IFSKSVEKFPDNNMLGWRRIVDEK--------------VGPYMWKTYKEVYEEVLQIGSA 92
Query: 153 LLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQVS-T 211
L G + S V I+ +W IA++ C + V +Y +LG A+ + + ++
Sbjct: 93 LRAAGAEPGSRVGIYGVNCPQWIIAMEACAAHTLICVPLYDTLGSGAVDYIVEHAEIDFV 152
Query: 212 LICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKE-EHTFSEGLSSNCKIASFDEVEKLGKE 270
+ D K+ L+ L+ I+ F + S E H SE K S+ + +G+E
Sbjct: 153 FVQDTKIKGLLEPDCKCAKRLKAIVSFTNVSDELSHKASE---IGVKTYSWIDFLHMGRE 209
Query: 271 SPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSK----DVY 326
P + + P + +MYTSG++G PKGV++TH + + + K DVY
Sbjct: 210 KPEDTNPPKAFNICTIMYTSGTSGDPKGVVLTHQAVATFVVGMDLYMDQFEDKMTHDDVY 269
Query: 327 LAYLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVTVLKPTLLTA 384
L++LPLAH+ + E G ++GY G+ L D D+ LKPT L
Sbjct: 270 LSFLPLAHILDRMNEEYFFRKGASVGYYHGNLNVLRD-----------DIQELKPTYLAG 318
Query: 385 VPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKK 444
VP + +RI +G+ K ++E + +F YK +LA + + + + D I F+K
Sbjct: 319 VPRVFERIHEGIQKALQELNPRRRFIFNALYKHKLAWLNRGY--SHSKASPMADFIAFRK 376
Query: 445 IRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSV-G 503
IR LGG +R ++ GGAPLS + ++F+ + + QGYGLTET G A D+ + G
Sbjct: 377 IRDKLGGRIRLLVSGGAPLSPEIEEFLRVTCCCFVVQGYGLTETLGGTALGFPDEMCMLG 436
Query: 504 RVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEK 563
VG P I+L E GY + P GE+ + G + +GY+KN + T+EV K
Sbjct: 437 TVGIPAVYNEIRLEEVSEMGYDPLGEN-PAGEICIRGQCMFSGYYKNPELTEEVMKDG-- 493
Query: 564 GVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHAD 623
WF+TGDIG P+G L+IIDRKK+++KL GEY++L +E V I V+ D
Sbjct: 494 ---WFHTGDIGEILPNGVLKIIDRKKNLIKLSQGEYVALEHLENIFGQNSVVQDIWVYGD 550
Query: 624 PFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKT 683
F S VA+VV + +++ +WA++ G K F +LC+ PE ++ + A+ KL+K
Sbjct: 551 SFKSMLVAVVVPNPETVNRWAKDLGFT-KPFEELCSFPELKEHIISELKSTAEKNKLRKF 609
Query: 684 EVPAKIKLLADPWTPESGLVTAALK 708
E + + P+ E LVTA LK
Sbjct: 610 EYIKAVTVETKPFDVERDLVTATLK 634
>At1g64400 putative acyl-CoA synthetase
Length = 665
Score = 306 bits (783), Expect = 4e-83
Identities = 205/629 (32%), Positives = 329/629 (51%), Gaps = 50/629 (7%)
Query: 93 LFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETYGEVFARVSNFASG 152
+F S +K +N LG R+++ DG+ G+Y W+TY EV V +
Sbjct: 50 IFRLSVEKSPNNPMLGRREIV--------DGKA------GKYVWQTYKEVHNVVIKLGNS 95
Query: 153 LLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQVSTL 212
+ +G I+ EW I+++ C + V +Y +LG A+ + +VS
Sbjct: 96 IRTIGVGKGDKCGIYGANSPEWIISMEACNAHGLYCVPLYDTLGAGAIEFIICHAEVSLA 155
Query: 213 ICDVKLLNKLDAIRSKLTS-LQNIIYFEDDSKEEHTFSEGLSSNCKIASFDEVEKLGKES 271
+ +++L K T L+ I+ F + + + +E I S+D+ KLG+
Sbjct: 156 FAEENKISELLKTAPKSTKYLKYIVSFGEVTNNQRVEAE--RHRLTIYSWDQFLKLGEGK 213
Query: 272 PVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVM----TVIPNLGSKDVYL 327
E ++ V +MYTSG+TG PKGV++T+ +I+ V T+ L SKDVYL
Sbjct: 214 HYELPEKRRSDVCTIMYTSGTTGDPKGVLLTNESIIHLLEGVKKLLKTIDEELTSKDVYL 273
Query: 328 AYLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAV 385
+YLPLAH+F+ E + + +IG+ G L + D+ LKPT+ AV
Sbjct: 274 SYLPLAHIFDRVIEELCIYEAASIGFWRGDVKILIE-----------DIAALKPTVFCAV 322
Query: 386 PAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAV-KGSWLGAWGVEKLVWDTIIFKK 444
P +++RI G+ +K+ + G + K LF A+K + + KG + D I+FKK
Sbjct: 323 PRVLERIYTGLQQKLSDGGFVKKKLFNFAFKYKHKNMEKGQ---PHEQASPIADKIVFKK 379
Query: 445 IRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSV-G 503
++ LGGN+R +L G APL+ + F+ + A + QGYGLTE+ G S ++ S+ G
Sbjct: 380 VKEGLGGNVRLILSGAAPLAAHIESFLRVVACAHVLQGYGLTESCGGTFVSIPNELSMLG 439
Query: 504 RVGPPLPCCYIKLVSWEEGGY--LSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVD 561
VGPP+P I+L S E GY L+S+ PRGE+ + G ++ +GY+K +D T EVF +D
Sbjct: 440 TVGPPVPNVDIRLESVPEMGYDALASN---PRGEICIRGKTLFSGYYKREDLTQEVF-ID 495
Query: 562 EKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVH 621
W +TGD+G + PDG ++IIDRKK+I KL GEY+++ +E + ++SI V+
Sbjct: 496 G----WLHTGDVGEWQPDGAMKIIDRKKNIFKLSQGEYVAVENLENIYSHVAAIESIWVY 551
Query: 622 ADPFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQ 681
+ + SY VA+V S +E WA+E + DF +C + VL ++ AK KL+
Sbjct: 552 GNSYESYLVAVVCPSKIQIEHWAKEHKVS-GDFESICRNQKTKEFVLGEFNRVAKDKKLK 610
Query: 682 KTEVPAKIKLLADPWTPESGLVTAALKLK 710
E+ + L P+ E L+T + K+K
Sbjct: 611 GFELIKGVHLDTVPFDMERDLITPSYKMK 639
>At1g49430 acyl CoA synthetase-like protein (At1g49430)
Length = 665
Score = 299 bits (766), Expect = 3e-81
Identities = 198/640 (30%), Positives = 317/640 (48%), Gaps = 51/640 (7%)
Query: 80 VEVPWKGAPTMAHLFEPSCKKHTHNRFLGTRKLIGKEFVTSSDGRKFEKVHLGEYEWETY 139
++ PW+ F + KK+ + + LG R ++D + +G Y W TY
Sbjct: 44 IDSPWQ-------FFSEAVKKYPNEQMLGQR--------VTTDSK------VGPYTWITY 82
Query: 140 GEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDA 199
E S + G D I+ EW IA++ C Q IT V +Y SLG +A
Sbjct: 83 KEAHDAAIRIGSAIRSRGVDPGHCCGIYGANCPEWIIAMEACMSQGITYVPLYDSLGVNA 142
Query: 200 LIHSLNETQVSTLICDVKLLNKLDAIRSKLTS-LQNIIYFEDDSKEEHTFSEGLSSNC-K 257
+ +N +VS + K ++ + + + +S L+ I+ F + S T E + C
Sbjct: 143 VEFIINHAEVSLVFVQEKTVSSILSCQKGCSSNLKTIVSFGEVSS---TQKEEAKNQCVS 199
Query: 258 IASFDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVI 317
+ S++E +G K + +MYTSG+TG PKGV++ + I ++ ++
Sbjct: 200 LFSWNEFSLMGNLDEANLPRKRKTDICTIMYTSGTTGEPKGVILNNAAISVQVLSIDKML 259
Query: 318 P----NLGSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGY--GSPMTLTDTSNKVKKGTK 371
+ + DV+ +YLPLAH ++ E L+ G ++GY G L D
Sbjct: 260 EVTDRSCDTSDVFFSYLPLAHCYDQVMEIYFLSRGSSVGYWRGDIRYLMD---------- 309
Query: 372 GDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWG 431
DV LKPT+ VP + D++ G+++K+ G + K LF AY +L ++ +
Sbjct: 310 -DVQALKPTVFCGVPRVYDKLYAGIMQKISASGLIRKKLFDFAYNYKLGNMRKGFSQEEA 368
Query: 432 VEKLVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAG 491
+L D ++F KI+ LGG +L G APL ++F+ I + + QGYGLTE+ G
Sbjct: 369 SPRL--DRLMFDKIKEALGGRAHMLLSGAAPLPRHVEEFLRIIPASNLSQGYGLTESCGG 426
Query: 492 AAFSEADDYS-VGRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKN 550
+ + A +S VG VG P+P +LVS E GY + +PRGE+ + G S+ +GY K
Sbjct: 427 SFTTLAGVFSMVGTVGVPMPTVEARLVSVPEMGYDAFSADVPRGEICLRGNSMFSGYHKR 486
Query: 551 QDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALA 610
QD TD+V +D WF+TGDIG + DG ++IIDRKK+I KL GEY+++ +E +
Sbjct: 487 QDLTDQVL-IDG----WFHTGDIGEWQEDGSMKIIDRKKNIFKLSQGEYVAVENLENTYS 541
Query: 611 SGDYVDSIMVHADPFHSYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQS 670
+ I V+ + F S+ V +VV +++E WA+ DF LC +A L
Sbjct: 542 RCPLIAQIWVYGNSFESFLVGVVVPDRKAIEDWAKLNYQSPNDFESLCQNLKAQKYFLDE 601
Query: 671 ISKAAKAAKLQKTEVPAKIKLLADPWTPESGLVTAALKLK 710
++ AK +L+ E+ I L +P+ E L+T KLK
Sbjct: 602 LNSTAKQYQLKGFEMLKAIHLEPNPFDIERDLITPTFKLK 641
>At3g23790 AMP-binding protein, putative
Length = 722
Score = 144 bits (364), Expect = 1e-34
Identities = 148/624 (23%), Positives = 258/624 (40%), Gaps = 81/624 (12%)
Query: 138 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQN-ITVVTIYASLG 196
TY ++ + +F GL +G D +A+F+D W +A QG + VV S
Sbjct: 112 TYRQLEQEILDFVEGLRVVGVKADEKIALFADNSCRWLVADQGIMATGAVNVVRGSRSSV 171
Query: 197 EDALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNC 256
E+ L + V+ ++ + + N++ S + + +I + T +
Sbjct: 172 EELLQIYCHSESVALVVDNPEFFNRIAESFSYKAAPKFVILLWGEKSSLVTAGR----HT 227
Query: 257 KIASFDEVEKLGKE-----------SPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGN 305
+ S++E++K G+E E + +A +MYTSG+TG PKGVM+TH N
Sbjct: 228 PVYSYNEIKKFGQERRAKFARSNDSGKYEYEYIDPDDIATIMYTSGTTGNPKGVMLTHQN 287
Query: 306 IVATTAAVMTVIPNLGSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGYGSPMTLTDTSNK 365
++ + +P + + +L+ LP H +E A E + GV Y S L D
Sbjct: 288 LLHQIRNLSDFVP-AEAGERFLSMLPSWHAYERACEYFIFTCGVEQKYTSIRFLKD---- 342
Query: 366 VKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNL------FQIAYKRRL 419
D+ +P L +VP + + + G+ K++ K L +AY
Sbjct: 343 -------DLKRYQPHYLISVPLVYETLYSGIQKQISASSPARKFLALTLIKVSLAYTEMK 395
Query: 420 AAVKG-----------------SWLGAWGVEKLVW------DTIIFKKIRTVLGGNLRFM 456
+G WL A V +W + ++ +KIR+ + G +
Sbjct: 396 RVYEGLCLTKNQKPPMYIVSLVDWLWARVVAFFLWPLHMLAEKLVHRKIRSSI-GITKAG 454
Query: 457 LCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKL 516
+ GG L +F +G + GYGLTET + +G VG P+ K+
Sbjct: 455 VSGGGSLPMHVDKFFE-AIGVNVQNGYGLTETSPVVSARRLRCNVLGSVGHPIKDTEFKI 513
Query: 517 VSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRF 576
V E G L P +G V V G V GY+KN T +V +D+ G WF TGD+G
Sbjct: 514 VDHETGTVL---PPGSKGIVKVRGPPVMKGYYKNPLATKQV--IDDDG--WFNTGDMGWI 566
Query: 577 HPD----------GCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADPFH 626
P G + + R KD + L GE + ++E A + + I+V
Sbjct: 567 TPQHSTGRSRSCGGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSNLIQQIVVIGQD-Q 625
Query: 627 SYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEVP 686
A+V+ + ++ E A++ + +K + V + + K Q
Sbjct: 626 RRLGAIVIPNKEAAEGAAKQKISPVDSEVNELSKETITSMVYEELRKWTSQCSFQ----V 681
Query: 687 AKIKLLADPWTPESGLVTAALKLK 710
+ ++ +P+T ++GL+T +K++
Sbjct: 682 GPVLIVDEPFTIDNGLMTPTMKIR 705
>At4g14070 A6 anther-specific protein (Z97335.16)
Length = 727
Score = 143 bits (360), Expect = 4e-34
Identities = 151/624 (24%), Positives = 259/624 (41%), Gaps = 92/624 (14%)
Query: 138 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQN-ITVVTIYASLG 196
TY ++ + +FA GL LG D +A+F+D W ++ QG + VV S
Sbjct: 128 TYKQLEQEILDFAEGLRVLGVKADEKIALFADNSCRWLVSDQGIMATGAVNVVRGSRSSV 187
Query: 197 EDALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNC 256
E+ L + V+ ++ + + N++ + SL+ +I + T +G+
Sbjct: 188 EELLQIYRHSESVAIVVDNPEFFNRIAESFTSKASLRFLILLWGEKSSLVT--QGMQ--I 243
Query: 257 KIASFDEVEKLGKESPVEPSLPS-----------KNAVAVVMYTSGSTGLPKGVMITHGN 305
+ S+ E+ G+ES + S + + A +MYTSG+TG PKGVM+TH N
Sbjct: 244 PVYSYAEIINQGQESRAKLSASNDTRSYRNQFIDSDDTAAIMYTSGTTGNPKGVMLTHRN 303
Query: 306 IVATTAAVMTVIPNLGSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGYGSPMTLTDTSNK 365
++ + +P + D +L+ LP H +E A+E + GV Y S L D
Sbjct: 304 LLHQIKHLSKYVP-AQAGDKFLSMLPSWHAYERASEYFIFTCGVEQMYTSIRYLKD---- 358
Query: 366 VKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGG----LAKNLFQI--AYKRRL 419
D+ +P + +VP + + + G+ K++ LA L ++ AY
Sbjct: 359 -------DLKRYQPNYIVSVPLVYETLYSGIQKQISASSAGRKFLALTLIKVSMAYMEMK 411
Query: 420 AAVKG-----------------SWLGAWGVEKLVW------DTIIFKKIRTVLGGNLRFM 456
+G WL A + L+W +I+KKI + +G + +
Sbjct: 412 RIYEGMCLTKEQKPPMYIVAFVDWLWARVIAALLWPLHMLAKKLIYKKIHSSIGIS-KAG 470
Query: 457 LCGGAPLSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKL 516
+ GG L +F +G + GYGLTET +G G P+ K+
Sbjct: 471 ISGGGSLPIHVDKFFE-AIGVILQNGYGLTETSPVVCARTLSCNVLGSAGHPMHGTEFKI 529
Query: 517 VSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRF 576
V E L P +G + V G V GY+KN T +V ++E G WF TGD G
Sbjct: 530 VDPETNNVL---PPGSKGIIKVRGPQVMKGYYKNPSTTKQV--LNESG--WFNTGDTGWI 582
Query: 577 ----------HPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADPFH 626
H G + + R KD + L GE + ++E A ++ I+V
Sbjct: 583 APHHSKGRSRHCGGVIVLEGRAKDTIVLSTGENVEPLEIEEAAMRSRVIEQIVVIGQD-R 641
Query: 627 SYCVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQKTEVP 686
A+++ + + ++ ET +K + V Q + K Q
Sbjct: 642 RRLGAIIIPNKEEAQRVDPET-----------SKETLKSLVYQELRKWTSECSFQ----V 686
Query: 687 AKIKLLADPWTPESGLVTAALKLK 710
+ ++ DP+T ++GL+T +K++
Sbjct: 687 GPVLIVDDPFTIDNGLMTPTMKIR 710
>At4g05160 4-coumarate--CoA ligase - like protein
Length = 544
Score = 102 bits (253), Expect = 1e-21
Identities = 118/490 (24%), Positives = 211/490 (42%), Gaps = 80/490 (16%)
Query: 138 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGE 197
T+ ++ + V+ A G +LG + V IF+ ++ + CF + ++ +
Sbjct: 56 TFSQLKSAVARLAHGFHRLGIRKNDVVLIFAPNSYQFPL----CFLAVTAIGGVFTTANP 111
Query: 198 DALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCK 257
++++NE + K++ ++ + K+ + SK+ G SN K
Sbjct: 112 ---LYTVNEVSKQIKDSNPKIIISVNQLFDKIKGFDLPVVLLG-SKDTVEIPPG--SNSK 165
Query: 258 IASFDEVEKLGKESPVEPSLPSKNA-VAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTV 316
I SFD V +L + P + K + A ++Y+SG+TG KGV +THGN +A + V
Sbjct: 166 ILSFDNVMELSEPVSEYPFVEIKQSDTAALLYSSGTTGTSKGVELTHGNFIAASLMVTMD 225
Query: 317 IPNLGS-KDVYLAYLPLAHVFEMAAESV-MLAAGVAIGYGSPMTLTDTSNKVKKGTKGDV 374
+G V+L +LP+ HVF +A + L G A+ + L ++K +
Sbjct: 226 QDLMGEYHGVFLCFLPMFHVFGLAVITYSQLQRGNALVSMARFELELVLKNIEKFRVTHL 285
Query: 375 TVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEK 434
V+ P L + +VKK + L+++K ++G+
Sbjct: 286 WVVPPVFLAL-------SKQSIVKKFD-----------------LSSLK--YIGS----- 314
Query: 435 LVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINICV-GAPIGQGYGLTETFAGAA 493
G APL D + + + QGYG+TET +
Sbjct: 315 ------------------------GAAPLGKDLMEECGRNIPNVLLMQGYGMTETCGIVS 350
Query: 494 FSE--ADDYSVGRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQ 551
+ + G G P ++VS E G S P +GE+ V G ++ GY N
Sbjct: 351 VEDPRLGKRNSGSAGMLAPGVEAQIVSVETG---KSQPPNQQGEIWVRGPNMMKGYLNNP 407
Query: 552 DKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALAS 611
T E +D+K W +TGD+G F+ DG L ++DR K+++K + G ++ ++E L S
Sbjct: 408 QATKET--IDKKS--WVHTGDLGYFNEDGNLYVVDRIKELIKYK-GFQVAPAELEGLLVS 462
Query: 612 -GDYVDSIMV 620
D +D++++
Sbjct: 463 HPDILDAVVI 472
>At4g19010 4-coumarate-CoA ligase - like
Length = 566
Score = 99.0 bits (245), Expect = 9e-21
Identities = 115/447 (25%), Positives = 178/447 (39%), Gaps = 86/447 (19%)
Query: 186 ITVVTIYASLGEDALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEE 245
+T + +SLGE + ++E V + + KL ++ + S+ F+ E
Sbjct: 125 VTTMNPSSSLGE--IKKQVSECSVGLAFTSTENVEKLSSLGVSVISVSESYDFDSIRIEN 182
Query: 246 HTFSEGLSSNCKIASFDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGN 305
F + SF V K L ++ VA +MY+SG+TG KGV++TH N
Sbjct: 183 PKFYSIMKE-----SFGFVPK---------PLIKQDDVAAIMYSSGTTGASKGVLLTHRN 228
Query: 306 IVATTAAVMTVIPNL----GSKDVYLAYLPLAHVFEMAAESV-MLAAGVAIGYGSPMTLT 360
++A+ + + GS +VYLA LPL H++ ++ + +L+ G I +
Sbjct: 229 LIASMELFVRFEASQYEYPGSSNVYLAALPLCHIYGLSLFVMGLLSLGSTIVVMKRFDAS 288
Query: 361 DTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLA 420
D N +++ K T VP ++ +A ++
Sbjct: 289 DVVNVIER--------FKITHFPVVPPML-----------------------MALTKKAK 317
Query: 421 AVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDS-QQFINICVGAPI 479
V G +FK ++ V G APLS + F+ +
Sbjct: 318 GVCGE---------------VFKSLKQVSSG--------AAPLSRKFIEDFLQTLPHVDL 354
Query: 480 GQGYGLTETFAGAA--FSEADDYSVGRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVV 537
QGYG+TE+ A F+ VG P K+V W G +L P RGE+
Sbjct: 355 IQGYGMTESTAVGTRGFNSEKLSRYSSVGLLAPNMQAKVVDWSSGSFLP---PGNRGELW 411
Query: 538 VGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHG 597
+ G V GY N T D W TGDI F DG L I+DR K+I+K + G
Sbjct: 412 IQGPGVMKGYLNNPKATQMSIVEDS----WLRTGDIAYFDEDGYLFIVDRIKEIIKYK-G 466
Query: 598 EYISLGKVEAALASGDYVDSIMVHADP 624
I+ +EA L S + V A P
Sbjct: 467 FQIAPADLEAVLVSHPLIIDAAVTAAP 493
>At1g20500 hypothetical protein
Length = 550
Score = 93.2 bits (230), Expect = 5e-19
Identities = 101/414 (24%), Positives = 179/414 (42%), Gaps = 78/414 (18%)
Query: 279 SKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAHVFEM 338
+++ A+++Y+SG+TG KGV+ +H N+ A A ++ NL D+++ +P+ H + +
Sbjct: 195 NQDDTAMMLYSSGTTGPSKGVISSHRNLTAHVARFIS--DNLKRDDIFICTVPMFHTYGL 252
Query: 339 AAESV-MLAAGVAIGYGSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRDGVV 397
++ +A G + L D + V+K L A P ++ I D +
Sbjct: 253 LTFAMGTVALGSTVVILRRFQLHDMMDAVEKHRA-------TALALAPPVLVAMINDADL 305
Query: 398 KKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLRFML 457
K + +D K +R
Sbjct: 306 IKAK-----------------------------------YDLSSLKTVR----------- 319
Query: 458 CGGAPLSGD-SQQFINICVGAPIGQGYGLTETFAGAAFSEADDYS--VGRVGPPLPCCYI 514
CGGAPLS + ++ F+ I QGY LTE+ G AF+ + + S G G
Sbjct: 320 CGGAPLSKEVTEGFLEKYPTVDILQGYALTESNGGGAFTNSAEESRRYGTAGTLTSDVEA 379
Query: 515 KLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIG 574
++V G ++ ++ GE+ + G S++ GYFKNQ+ T+E ++ W TGD+
Sbjct: 380 RIVDPNTGRFMGINQT---GELWLKGPSISKGYFKNQEATNETINLE----GWLKTGDLC 432
Query: 575 RFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALAS-GDYVDSIMVHADPF-----HSY 628
DG L ++DR K+++K + G + ++EA L + D +D+ ++ PF Y
Sbjct: 433 YIDEDGFLFVVDRLKELIKYK-GYQVPPAELEALLITHPDILDAAVI---PFPDKEAGQY 488
Query: 629 CVALVVASHQSLEKWAQETGIEYKDFPDLCNKPEAVTEVLQSISKAAKAAKLQK 682
+A VV H+S +++ I++ K + SI K A L+K
Sbjct: 489 PMAYVVRKHES--NLSEKQVIDFISKQVAPYKKIRKVSFINSIPKTASGKTLRK 540
>At1g51680 4-coumarate--coenzyme A ligase (At4CL1)
Length = 561
Score = 89.7 bits (221), Expect = 5e-18
Identities = 126/515 (24%), Positives = 202/515 (38%), Gaps = 84/515 (16%)
Query: 138 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGE 197
TY +V A+ KLG + + V + E+ ++ + T
Sbjct: 67 TYSDVHVISRQIAANFHKLGVNQNDVVMLLLPNCPEFVLSFLAASFRGATATAANPFFTP 126
Query: 198 DALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCK 257
+ + +I + + ++K+ +++ ++ D E EG +
Sbjct: 127 AEIAKQAKASNTKLIITEARYVDKIKPLQND----DGVVIVCIDDNESVPIPEGCLRFTE 182
Query: 258 IA-SFDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTA-AVMT 315
+ S E ++ + P + V + Y+SG+TGLPKGVM+TH +V + A V
Sbjct: 183 LTQSTTEASEVIDSVEISP-----DDVVALPYSSGTTGLPKGVMLTHKGLVTSVAQQVDG 237
Query: 316 VIPNL--GSKDVYLAYLPLAHVFEMAAESVMLAAGVAIGYGSPMTLTDTSNKVKKGTKGD 373
PNL S DV L LP+ H++ A S+ML G+ +G + N + +
Sbjct: 238 ENPNLYFHSDDVILCVLPMFHIY--ALNSIML-CGLRVGAAILIMPKFEINLLLE----L 290
Query: 374 VTVLKPTLLTAVPAIIDRIRDGVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVE 433
+ K T+ VP I+ LA K S E
Sbjct: 291 IQRCKVTVAPMVPPIV-----------------------------LAIAKSS-------E 314
Query: 434 KLVWDTIIFKKIRTVLGGNLRFMLCGGAPLSGDSQQFINI-CVGAPIGQGYGLTETFAGA 492
+D IR V G APL + + +N A +GQGYG+TE
Sbjct: 315 TEKYD---LSSIRVVKSG--------AAPLGKELEDAVNAKFPNAKLGQGYGMTEAGPVL 363
Query: 493 AFS-----EADDYSVGRVGPPLPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGY 547
A S E G G + +K+V + G LS ++P GE+ + G + GY
Sbjct: 364 AMSLGFAKEPFPVKSGACGTVVRNAEMKIVDPDTGDSLSRNQP---GEICIRGHQIMKGY 420
Query: 548 FKNQDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEA 607
N T E +D+ G W +TGDIG D L I+DR K+++K + G ++ ++EA
Sbjct: 421 LNNPAATAET--IDKDG--WLHTGDIGLIDDDDELFIVDRLKELIKYK-GFQVAPAELEA 475
Query: 608 ALASGDYVDSIMVHA---DPFHSYCVALVVASHQS 639
L + + V A + VA VV S S
Sbjct: 476 LLIGHPDITDVAVVAMKEEAAGEVPVAFVVKSKDS 510
>At1g20480 unknown protein
Length = 565
Score = 81.3 bits (199), Expect = 2e-15
Identities = 117/518 (22%), Positives = 209/518 (39%), Gaps = 94/518 (18%)
Query: 104 NRFLGTRKLIGKE-------FVTSSDGRKFEKVHLGEYEWETYGEVFARVSNFASGLLKL 156
N+FL I + FV + GR+ ++ E++ V A L L
Sbjct: 46 NQFLDVTSFIASQPHRGKTVFVDAVTGRRL-----------SFPELWLGVERVAGCLYAL 94
Query: 157 GHDIDSHVAIFSDTRAEWFIALQGCFRQNITVVTIYASLGEDALIHSLNETQVSTLICDV 216
G + V I S + I + T D + + +++
Sbjct: 95 GVRKGNVVIILSPNSILFPIVSLSVMSLGAIITTANPINTSDEISKQIGDSRPVLAFTTC 154
Query: 217 KLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGLSSNCKIASFDE--VEKLGKESPVE 274
KL++KL A ++ + DD H S+ K+ E +E ES V+
Sbjct: 155 KLVSKLAAA----SNFNLPVVLMDDY---HVPSQSYGDRVKLVGRLETMIETEPSESRVK 207
Query: 275 PSLPSKNAVAVVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAH 334
+ +++ A ++Y+SG+TG KGVM++H N++A A G + + +P+ H
Sbjct: 208 QRV-NQDDTAALLYSSGTTGTSKGVMLSHRNLIALVQAYRA---RFGLEQRTICTIPMCH 263
Query: 335 VFEMAAESVMLAAGVAIGYGSPMTLTDTSNKVKKGTKGDVTVLKPTLLTAVPAIIDRIRD 394
+F + L +A+G+ + K+ V + + L+ VP I+ + +
Sbjct: 264 IFGFGGFATGL---IALGWTIVVLPKFDMAKLLSA----VETHRSSYLSLVPPIVVAMVN 316
Query: 395 GVVKKVEEKGGLAKNLFQIAYKRRLAAVKGSWLGAWGVEKLVWDTIIFKKIRTVLGGNLR 454
G ++ K L+ +L
Sbjct: 317 GA-NEINSKYDLS--------------------------------------------SLH 331
Query: 455 FMLCGGAPLSGD-SQQFINICVGAPIGQGYGLTETFAGAA--FSEADDYSVGRVGPPLPC 511
++ GGAPLS + +++F+ I QGYGLTE+ A AA F++ + G G P
Sbjct: 332 TVVAGGAPLSREVTEKFVENYPKVKILQGYGLTESTAIAASMFNKEETKRYGASGLLAPN 391
Query: 512 CYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTG 571
K+V + G L ++ GE+ + +V GYFKN++ T +D +G W TG
Sbjct: 392 VEGKIVDPDTGRVLGVNQT---GELWIRSPTVMKGYFKNKEATAST--IDSEG--WLKTG 444
Query: 572 DIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAAL 609
D+ DG + ++DR K+++K +G ++ ++EA L
Sbjct: 445 DLCYIDGDGFVFVVDRLKELIKC-NGYQVAPAELEALL 481
>At5g16370 AMP-binding protein
Length = 552
Score = 68.2 bits (165), Expect = 2e-11
Identities = 58/166 (34%), Positives = 76/166 (44%), Gaps = 27/166 (16%)
Query: 475 VGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKLVSWEEG----GYLSSDKP 530
+G I GYGLTET AG S A R LP + +G G+ D
Sbjct: 320 IGFVISHGYGLTET-AGLNVSCAWKPQWNR----LPASDRARLKARQGVRTVGFTEIDVV 374
Query: 531 MPR------------GEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHP 578
P GE+V+ G S+ GY K+ T++ K WFYTGD+G H
Sbjct: 375 DPESGRSVERNGETVGEIVMRGSSIMLGYLKDPVGTEKALKNG-----WFYTGDVGVIHS 429
Query: 579 DGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADP 624
DG LEI DR KDI+ + GE +S +VE L + V+ + V A P
Sbjct: 430 DGYLEIKDRSKDII-ITGGENVSSVEVETVLYTNPAVNEVAVVARP 474
Score = 30.8 bits (68), Expect = 2.9
Identities = 19/52 (36%), Positives = 28/52 (53%), Gaps = 5/52 (9%)
Query: 285 VVMYTSGSTGLPKGVMITHGNI--VATTAAVMTVIPNLGSKDVYLAYLPLAH 334
V+ YTSG+T PKGV+ H I ++ + + +P VYL LP+ H
Sbjct: 189 VLNYTSGTTSAPKGVVHCHRGIFVMSIDSLIDWTVP---KNPVYLWTLPIFH 237
>At1g77240 unknown protein
Length = 545
Score = 68.2 bits (165), Expect = 2e-11
Identities = 40/91 (43%), Positives = 52/91 (56%), Gaps = 5/91 (5%)
Query: 534 GEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVK 593
GE+V G SV GY+K+ + T + D WFYTGDIG HPDG LE+ DR KD+V
Sbjct: 390 GEIVFRGGSVMLGYYKDPEGTAASMREDG----WFYTGDIGVMHPDGYLEVKDRSKDVV- 444
Query: 594 LQHGEYISLGKVEAALASGDYVDSIMVHADP 624
+ GE IS ++EA L + + V A P
Sbjct: 445 ICGGENISSTELEAVLYTNPAIKEAAVVAKP 475
Score = 34.3 bits (77), Expect = 0.26
Identities = 19/50 (38%), Positives = 32/50 (64%), Gaps = 1/50 (2%)
Query: 285 VVMYTSGSTGLPKGVMITHGNIVATTAAVMTVIPNLGSKDVYLAYLPLAH 334
V+ YTSG+T PKGV+ +H ++ +T + + +L ++ VYL LP+ H
Sbjct: 189 VLNYTSGTTSSPKGVVHSHRSVFMSTINSL-LDWSLPNRPVYLWTLPMFH 237
>At1g75960 adenosine monophosphate binding protein 8 AMPBP8
Length = 544
Score = 68.2 bits (165), Expect = 2e-11
Identities = 63/190 (33%), Positives = 85/190 (44%), Gaps = 33/190 (17%)
Query: 453 LRFMLCGGAP-----LSGDSQQFINICVGAPIGQGYGLTETFAGAAFSEADDYSVGRVGP 507
+ F+ G +P L +S FI + GYGLTET AG S A + R+
Sbjct: 300 VNFLTAGSSPPATVLLRAESLGFI-------VSHGYGLTET-AGVIVSCAWKPNWNRLPA 351
Query: 508 P-------------LPCCYIKLVSWEEGGYLSSDKPMPRGEVVVGGFSVTAGYFKNQDKT 554
+ I +V E G + D GE+V+ G S+ GY KN T
Sbjct: 352 SDQAQLKSRQGVRTVGFSEIDVVDPESGRSVERDGETV-GEIVLRGSSIMLGYLKNPIGT 410
Query: 555 DEVFKVDEKGVRWFYTGDIGRFHPDGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDY 614
FK WF+TGD+G H DG LEI DR KD++ + GE +S +VEA L +
Sbjct: 411 QNSFKNG-----WFFTGDLGVIHGDGYLEIKDRSKDVI-ISGGENVSSVEVEAVLYTNPA 464
Query: 615 VDSIMVHADP 624
V+ V A P
Sbjct: 465 VNEAAVVARP 474
Score = 33.9 bits (76), Expect = 0.35
Identities = 22/52 (42%), Positives = 27/52 (51%), Gaps = 5/52 (9%)
Query: 285 VVMYTSGSTGLPKGVMITHGNIVATTAAVMT--VIPNLGSKDVYLAYLPLAH 334
VV YTSG+T PKGV+ H I T +T +P VYL LP+ H
Sbjct: 188 VVNYTSGTTSSPKGVVHCHRGIFVMTLDSLTDWAVP---KTPVYLWTLPIFH 236
>At5g16340 AMP-binding protein
Length = 550
Score = 65.9 bits (159), Expect = 8e-11
Identities = 58/166 (34%), Positives = 75/166 (44%), Gaps = 27/166 (16%)
Query: 475 VGAPIGQGYGLTETFAGAAFSEADDYSVGRVGPPLPCCYIKLVSWEEG----GYLSSDKP 530
+G I GYGLTET AG S A LP + +G G+ D
Sbjct: 320 IGFVISHGYGLTET-AGVIVSCAWKPKWNH----LPASDRARLKARQGVRTVGFTEIDVV 374
Query: 531 MPR------------GEVVVGGFSVTAGYFKNQDKTDEVFKVDEKGVRWFYTGDIGRFHP 578
P GE+V+ G SV GY K+ T++ K WFYTGD+G H
Sbjct: 375 DPESGLSVERNGETVGEIVMRGSSVMLGYLKDPVGTEKALKNG-----WFYTGDVGVIHS 429
Query: 579 DGCLEIIDRKKDIVKLQHGEYISLGKVEAALASGDYVDSIMVHADP 624
DG LEI DR KDI+ + GE +S +VE L + V+ + V A P
Sbjct: 430 DGYLEIKDRSKDII-ITGGENVSSVEVETVLYTIPAVNEVAVVARP 474
Score = 32.0 bits (71), Expect = 1.3
Identities = 45/204 (22%), Positives = 79/204 (38%), Gaps = 15/204 (7%)
Query: 138 TYGEVFARVSNFASGLLKLGHDIDSHVAIFSDT-----RAEWFIALQGCFRQNITVVTIY 192
T+ E R AS L +G V++ S ++ + + G NI
Sbjct: 42 TWRETNLRCLRVASSLSSIGIGRSDVVSVLSPNTPAMYELQFAVPMSGAILNNINT---- 97
Query: 193 ASLGEDALIHSLNETQVSTLICDVKLLNKLDAIRSKLTSLQNIIYFEDDSKEEHTFSEGL 252
L + L + L DV ++ S +T+ I+ D +EE ++
Sbjct: 98 -RLDARTVSVLLRHCESKLLFVDVFSVDLAVEAVSMMTTDPPILVVIADKEEEGGVADVA 156
Query: 253 SSNCKIASFDEVEKLGKESPVEPSLPSKNAVAVVMYTSGSTGLPKGVMITHGNI--VATT 310
+ ++D++ + G S+ V+ YTSG+T PKGV+ H I ++
Sbjct: 157 DLSKFSYTYDDLIERGDPGFKWIRPESEWDPVVLNYTSGTTSAPKGVVHCHRGIFVMSVD 216
Query: 311 AAVMTVIPNLGSKDVYLAYLPLAH 334
+ + +P VYL LP+ H
Sbjct: 217 SLIDWAVP---KNPVYLWTLPIFH 237
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,361,710
Number of Sequences: 26719
Number of extensions: 714580
Number of successful extensions: 1927
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1734
Number of HSP's gapped (non-prelim): 94
length of query: 755
length of database: 11,318,596
effective HSP length: 107
effective length of query: 648
effective length of database: 8,459,663
effective search space: 5481861624
effective search space used: 5481861624
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148718.8


