

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.4 + phase: 0
(246 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
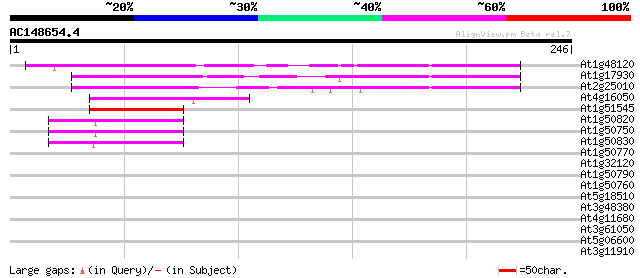
Score E
Sequences producing significant alignments: (bits) Value
At1g48120 serine/threonine phosphatase PP7, putative 83 2e-16
At1g17930 unknown protein 77 6e-15
At2g25010 unknown protein 76 2e-14
At4g16050 hypothetical protein 44 6e-05
At1g51545 unknown protein 43 1e-04
At1g50820 hypothetical protein 42 3e-04
At1g50750 hypothetical protein 41 5e-04
At1g50830 hypothetical protein 40 9e-04
At1g50770 hypothetical protein 40 0.001
At1g32120 hypothetical protein 40 0.001
At1g50790 hypothetical protein 37 0.007
At1g50760 hypothetical protein 37 0.010
At5g18510 putative protein 35 0.028
At3g48380 unknown protein 28 3.4
At4g11680 unknown protein 28 4.5
At3g61050 CaLB protein 28 4.5
At5g06600 ubiquitin carboxyl-terminal hydrolase 27 7.7
At3g11910 ubiquitin carboxyl-terminal hydrolase like protein 27 7.7
>At1g48120 serine/threonine phosphatase PP7, putative
Length = 1338
Score = 82.8 bits (203), Expect = 2e-16
Identities = 67/222 (30%), Positives = 104/222 (46%), Gaps = 25/222 (11%)
Query: 8 GNENWFWDPLR-----ESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVT 62
G +W DPL E GL+ + + + + +AL+ L E W PET TFH+P GE+TVT
Sbjct: 50 GMRDWPLDPLVCQKLIEFGLYGVYKVAFIQLDYALITALVERWRPETHTFHLPAGEITVT 109
Query: 63 LDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSL 122
L DV L L ++G ++ K+ A L LG + +G ++S L
Sbjct: 110 LQDVNILLGLRVDGPAVTGS---TKYNWADLCEDLLGHRPGPKDL-----HGSHVSLAWL 161
Query: 123 MDFYTTYLGRANLLAGTEDPEELVRVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTM 182
R N DP+E V ++ + R ++L L+ +GD+S + L +L +
Sbjct: 162 ---------RENFRNLPADPDE-VTLKCH-TRAFVLALMSGFLYGDKSKHDVALTFLPLL 210
Query: 183 EEGYAGMRNYSWGGMTLAYLYGELAEACRPGDRALGGSVTLL 224
+ + + SWG TLA LY EL A + + G + LL
Sbjct: 211 RD-FDEVAKLSWGSATLALLYRELCRASKRTVSTICGPLVLL 251
>At1g17930 unknown protein
Length = 478
Score = 77.4 bits (189), Expect = 6e-15
Identities = 54/198 (27%), Positives = 95/198 (47%), Gaps = 21/198 (10%)
Query: 28 LGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLK 87
+G ++ ++L+ L E W ET+TFH P GEMT+TLD+V+ + L ++G+ + K+
Sbjct: 57 VGSISLNNSLISALVERWRRETNTFHFPCGEMTITLDEVSLILGLAVDGKPVVGVKEK-D 115
Query: 88 HKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLGRANLLAGTEDPE-ELV 146
+ + +R LG + G+ G ++ L + + E P+ +
Sbjct: 116 EDPSQVCLRLLGKLPK------GELSGNRVTAKWLKESF------------AECPKGATM 157
Query: 147 RVRPYCVRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLYGEL 206
+ Y R YL+Y+V F +I + YL E+ + Y+WG LA+LY ++
Sbjct: 158 KEIEYHTRAYLIYIVGSTIFATTDPSKISVDYLILFED-FEKAGEYAWGAAALAFLYRQI 216
Query: 207 AEACRPGDRALGGSVTLL 224
A + +GG +TLL
Sbjct: 217 GNASQRSQSIIGGCLTLL 234
>At2g25010 unknown protein
Length = 509
Score = 75.9 bits (185), Expect = 2e-14
Identities = 62/201 (30%), Positives = 90/201 (43%), Gaps = 24/201 (11%)
Query: 28 LGYATVPHALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLK 87
+G ++ ++L+ L E W ET+TFH+PLGEMT+TLD+VA + L I+G + K
Sbjct: 66 IGPMSLNNSLISALVERWRRETNTFHLPLGEMTITLDEVALVLGLEIDGDPIVGSK---- 121
Query: 88 HKGATLLMRHLGVSQQEAEKICGQEYGGYISYPSLMDFYTTYLG-RANLLAGT--EDPEE 144
V + A +CG+ G PS + + N L T E PE+
Sbjct: 122 ------------VGDEVAMDMCGRLLG---KLPSAANKEVNCSRVKLNWLKRTFSECPED 166
Query: 145 LVRVRPYC-VRCYLLYLVECLWFGDRSNKRIELVYLTTMEEGYAGMRNYSWGGMTLAYLY 203
C R YLLYL+ F ++ + YL E+ + Y+WG LA LY
Sbjct: 167 ASFDVVKCHTRAYLLYLIGSTIFATTDGDKVSVKYLPLFED-FDQAGRYAWGAAALACLY 225
Query: 204 GELAEACRPGDRALGGSVTLL 224
L A + G +TLL
Sbjct: 226 RALGNASLKSQSNICGCLTLL 246
>At4g16050 hypothetical protein
Length = 900
Score = 44.3 bits (103), Expect = 6e-05
Identities = 26/72 (36%), Positives = 39/72 (54%), Gaps = 2/72 (2%)
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEGRML--SHEKKMLKHKGATL 93
+L+++L + W PET+TF P GE T+TL+DV L I G + S + +K L
Sbjct: 362 SLILSLAQNWCPETNTFVFPWGEATITLEDVNVLLGFSISGSSVFASLQSSEMKEAVEKL 421
Query: 94 LMRHLGVSQQEA 105
R G +QE+
Sbjct: 422 QKRCQGSMKQES 433
>At1g51545 unknown protein
Length = 629
Score = 43.1 bits (100), Expect = 1e-04
Identities = 19/41 (46%), Positives = 26/41 (63%)
Query: 36 ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
+LL+ L E W PET +F P GE T+TL+DV L ++G
Sbjct: 30 SLLLALVEKWCPETKSFLFPWGEATITLEDVLVLLGFSVQG 70
>At1g50820 hypothetical protein
Length = 528
Score = 42.0 bits (97), Expect = 3e-04
Identities = 22/61 (36%), Positives = 33/61 (54%), Gaps = 2/61 (3%)
Query: 18 RESGLHDLVYLGYATVPHA--LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIE 75
R++G+ + V +P L++ L E W P+T TF P GE T+TL+DV L +
Sbjct: 76 RKAGIFEAVIASTYKIPKDTDLVLGLAEKWCPDTKTFIFPWGEATITLEDVMVLLGFSVL 135
Query: 76 G 76
G
Sbjct: 136 G 136
>At1g50750 hypothetical protein
Length = 816
Score = 41.2 bits (95), Expect = 5e-04
Identities = 22/61 (36%), Positives = 32/61 (52%), Gaps = 2/61 (3%)
Query: 18 RESGLHDLVYLGYATVPHA--LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIE 75
RE+G+ + V +P L++ + E W P T TF P GE VTL+DV L+ +
Sbjct: 55 REAGIFEAVMASIYRIPKNPDLILGIAEKWCPYTKTFVFPWGETAVTLEDVMVLSGFSVL 114
Query: 76 G 76
G
Sbjct: 115 G 115
>At1g50830 hypothetical protein
Length = 768
Score = 40.4 bits (93), Expect = 9e-04
Identities = 19/61 (31%), Positives = 36/61 (58%), Gaps = 2/61 (3%)
Query: 18 RESGLHDLVYLGYATVPH--ALLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIE 75
R++G+ + + + ++ +L++++ E W PET +F P GE T+TL+DV L +
Sbjct: 98 RKAGIFEAIKVSTYSITKNPSLILSVSEKWCPETKSFVFPWGEATITLEDVMVLLGFSVL 157
Query: 76 G 76
G
Sbjct: 158 G 158
>At1g50770 hypothetical protein
Length = 632
Score = 39.7 bits (91), Expect = 0.001
Identities = 22/61 (36%), Positives = 34/61 (55%), Gaps = 2/61 (3%)
Query: 18 RESGLHDLVYLG-YATVPHA-LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIE 75
R++G+ + V Y P+ L++ + E W P+T TF P GE T+TL+DV L +
Sbjct: 79 RKAGIFEAVTASTYKINPNTELVLGIAEKWCPDTKTFVFPWGETTITLEDVMLLLGFSVL 138
Query: 76 G 76
G
Sbjct: 139 G 139
>At1g32120 hypothetical protein
Length = 1206
Score = 39.7 bits (91), Expect = 0.001
Identities = 23/61 (37%), Positives = 35/61 (56%), Gaps = 2/61 (3%)
Query: 18 RESGLHDLVYLG-YATVPHA-LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIE 75
++SG++D + Y H L++ L E W ET+TF P GE T+TL+D+ L L +
Sbjct: 100 KKSGVYDAILASRYQIKRHDDLIVALVEKWCIETNTFVFPWGEATLTLEDMIVLGGLSVT 159
Query: 76 G 76
G
Sbjct: 160 G 160
>At1g50790 hypothetical protein
Length = 812
Score = 37.4 bits (85), Expect = 0.007
Identities = 17/40 (42%), Positives = 24/40 (59%)
Query: 37 LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
L+M + E W P+T+TF GE T+TL+DV L + G
Sbjct: 101 LVMGIAEKWCPDTNTFVFSWGEATITLEDVMVLLGFSVLG 140
>At1g50760 hypothetical protein
Length = 649
Score = 37.0 bits (84), Expect = 0.010
Identities = 16/40 (40%), Positives = 24/40 (60%)
Query: 37 LLMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
L++ + E W P+T TF GE T+TL+DV L+ + G
Sbjct: 90 LVLGVAEKWSPDTKTFVFSWGEATITLEDVMVLSGFSVLG 129
>At5g18510 putative protein
Length = 702
Score = 35.4 bits (80), Expect = 0.028
Identities = 15/39 (38%), Positives = 23/39 (58%)
Query: 38 LMTLCEMWHPETSTFHMPLGEMTVTLDDVACLTHLPIEG 76
++++ E W ET +F P GE T+TL+DV L + G
Sbjct: 98 ILSIAEKWCSETKSFIFPWGEATITLEDVMVLLGFSVLG 136
>At3g48380 unknown protein
Length = 645
Score = 28.5 bits (62), Expect = 3.4
Identities = 27/96 (28%), Positives = 40/96 (41%), Gaps = 17/96 (17%)
Query: 46 HPETSTFHMPLGEMTVTLDDVACLTH----LPIEGRML-----------SHEKKMLKHKG 90
HP TS + + GE + DV L H LP++ +L K + +G
Sbjct: 368 HPITSMYELNYGETEMKQVDVRKLLHLRLGLPLDRPLLRIANALDLSVNDDSKSNMNRRG 427
Query: 91 ATLLMR-HLGV-SQQEAEKICGQEYGGYISYPSLMD 124
+TLL H+G+ S +E + G Y Y L D
Sbjct: 428 STLLKDVHIGIPSSGVSEGVASIIQGSYEYYHYLQD 463
>At4g11680 unknown protein
Length = 390
Score = 28.1 bits (61), Expect = 4.5
Identities = 29/136 (21%), Positives = 59/136 (43%), Gaps = 10/136 (7%)
Query: 48 ETSTFHMPLGEMTVTLDDVACLTHLPIEGRMLSHEKKMLKHKGATLLMRH--LGVSQQEA 105
+T+ F P T+++D+ + H + L + L+H G+ +MR + V + A
Sbjct: 34 DTTPFLPPTVTRTISVDEESNPIHRSARRQGLREAARFLRHAGSRRMMREPSMLVRETAA 93
Query: 106 EKICGQEYGGYISYP-----SLMDFYTTYLGRANLLAGTEDPEEL---VRVRPYCVRCYL 157
E++ ++ S P L + +G A L+ ++ + V V Y ++C+L
Sbjct: 94 EQLEERQSDWAYSKPVVFLDILWNLAFVAIGVAVLILSRDEKPNMPLRVWVVGYGIQCWL 153
Query: 158 LYLVECLWFGDRSNKR 173
C+ + R +R
Sbjct: 154 HMACVCVEYRRRRRRR 169
>At3g61050 CaLB protein
Length = 510
Score = 28.1 bits (61), Expect = 4.5
Identities = 15/46 (32%), Positives = 27/46 (58%), Gaps = 3/46 (6%)
Query: 77 RMLSHEKKMLKHKGATLLMRHLG-VSQQEAEKICGQEYGGYISYPS 121
RM++H K + M+ LG +S+ + +KICG + +IS+P+
Sbjct: 23 RMMTHRSS--KRVAKAVDMKLLGSLSRDDLKKICGDNFPQWISFPA 66
>At5g06600 ubiquitin carboxyl-terminal hydrolase
Length = 1126
Score = 27.3 bits (59), Expect = 7.7
Identities = 14/47 (29%), Positives = 23/47 (48%)
Query: 12 WFWDPLRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGE 58
W +D +E+G L G ++LL TL + + + +HMP E
Sbjct: 197 WSYDSKKETGFVGLKNQGATCYMNSLLQTLYHIPYFRKAVYHMPTTE 243
>At3g11910 ubiquitin carboxyl-terminal hydrolase like protein
Length = 1089
Score = 27.3 bits (59), Expect = 7.7
Identities = 14/47 (29%), Positives = 23/47 (48%)
Query: 12 WFWDPLRESGLHDLVYLGYATVPHALLMTLCEMWHPETSTFHMPLGE 58
W +D +E+G L G ++LL TL + + + +HMP E
Sbjct: 187 WSYDSKKETGFVGLKNQGATCYMNSLLQTLYHIPYFRKAVYHMPTTE 233
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,760,458
Number of Sequences: 26719
Number of extensions: 238862
Number of successful extensions: 612
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 590
Number of HSP's gapped (non-prelim): 20
length of query: 246
length of database: 11,318,596
effective HSP length: 97
effective length of query: 149
effective length of database: 8,726,853
effective search space: 1300301097
effective search space used: 1300301097
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC148654.4


