

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148654.3 - phase: 0 /pseudo
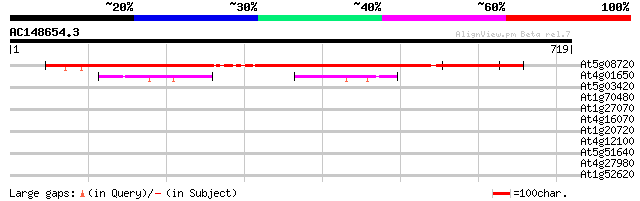
(719 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g08720 unknown protein 793 0.0
At4g01650 unknown protein 81 2e-15
At5g03420 putative protein 42 0.001
At1g70480 OBp32 protein 35 0.19
At1g27070 unknown protein 32 1.6
At4g16070 unknown protein 31 2.8
At1g20720 hypothetical protein 31 2.8
At4g12100 unknown protein (At4g12100) 30 6.2
At5g51640 unknown protein (At5g51640) 29 8.1
At4g27980 putative protein 29 8.1
At1g52620 29 8.1
>At5g08720 unknown protein
Length = 719
Score = 793 bits (2047), Expect = 0.0
Identities = 423/631 (67%), Positives = 482/631 (76%), Gaps = 36/631 (5%)
Query: 46 SKPSFLSLSLFFPRHFHKSIALSST------TQCKPRSH--LGGNLNNGLEED------- 90
++P FLS+ L P ++S++ +C H GG +NGL D
Sbjct: 20 NEPVFLSVLLPSPSRIRVFSSISTSGIGGGVAKCHGTRHSGAGGRGDNGLRRDSGLGFDE 79
Query: 91 -GDREVHCELQVVSWRERRVKAEISINADINSVWNALTDYEHLADFIPNLVWSGRIPCPF 149
G+R+V CE+ V+SWRERR++ EI +++D SVWN LTDYE LADFIPNLVWSGRIPCP
Sbjct: 80 RGERKVRCEVDVISWRERRIRGEIWVDSDSQSVWNVLTDYERLADFIPNLVWSGRIPCPH 139
Query: 150 PGRIWLEQRGFQRAMYWHIEARVVLDLQELLNSEWDRELHFSMVDGDFKKFEGKWSVKSG 209
PGRIWLEQRG QRA+YWHIEARVVLDL E L+S RELHFSMVDGDFKKFEGKWSVKSG
Sbjct: 140 PGRIWLEQRGLQRALYWHIEARVVLDLHECLDSPNGRELHFSMVDGDFKKFEGKWSVKSG 199
Query: 210 TRSSSTNLSYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVERNLLGNQKLPQPEDD 269
RS T LSYEVNVIPRFNFPAIFLERI+RSDLPVNLRA+A + E+ K P +D
Sbjct: 200 IRSVGTVLSYEVNVIPRFNFPAIFLERIIRSDLPVNLRAVARQAEKIYKDCGK-PSIIED 258
Query: 270 LHKTSLVVNGSSVKKINGSLCETDKLAPGQDKEGLDTSISGSLPASSSELNSNWGIFGKV 329
L L + S NG E D LA E S GSL A S+ELN+NWG++GK
Sbjct: 259 L----LGIISSQPAPSNG--IEFDSLAT----ERSVASSVGSL-AHSNELNNNWGVYGKA 307
Query: 330 CSLDKPCVVDEVHLRRFDGLLENGGVHRCVVASITVKAPVRDVWNVMSSYETLPEIVPNL 389
C LDKPC VDEVHLRRFDGLLENGGVHRC VASITVKAPV +VW V++SYE+LPEIVPNL
Sbjct: 308 CKLDKPCTVDEVHLRRFDGLLENGGVHRCAVASITVKAPVCEVWKVLTSYESLPEIVPNL 367
Query: 390 AISKILSRDNNKVRILQEGCKGLLYMVLHARVVLDLCEQLEQEISFEQAEGDFDSFHGKW 449
AISKILSRDNNKVRILQEGCKGLLYMVLHAR VLDL E EQEI FEQ EGDFDS GKW
Sbjct: 368 AISKILSRDNNKVRILQEGCKGLLYMVLHARAVLDLHEIREQEIRFEQVEGDFDSLEGKW 427
Query: 450 TFEQLGNHHTLLKYSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRDYVENQKASQFLE 509
FEQLG+HHTLLKY+V+SKMR+D+FLSEAIMEEVIYEDLPSNLCAIRDY+E ++ + E
Sbjct: 428 IFEQLGSHHTLLKYTVESKMRKDSFLSEAIMEEVIYEDLPSNLCAIRDYIE-KRGEKSSE 486
Query: 510 VCEQNTNSGQQIILSGSGDDNNSS--SADDISDCNVQSSSNQRSRVPGLQRDIEVLKSEL 567
C+ T + S S + + + DD SD + QR R+PGLQRDIEVLKSE+
Sbjct: 487 SCKLETCQVSEETCSSSRAKSVETVYNNDDGSD-----QTKQRRRIPGLQRDIEVLKSEI 541
Query: 568 LKFVAEYGQEGFMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNLSLAYKYRKPKGYWD 627
LKF++E+GQEGFMPMRKQLRLHGRVDIEKAITRMGGFR+IA +MNLSLAYK+RKPKGYWD
Sbjct: 542 LKFISEHGQEGFMPMRKQLRLHGRVDIEKAITRMGGFRRIALMMNLSLAYKHRKPKGYWD 601
Query: 628 NLENLQDEISRFQRCWGMDPSFMPSRKSFER 658
NLENLQ+EI RFQ+ WGMDPSFMPSRKSFER
Sbjct: 602 NLENLQEEIGRFQQSWGMDPSFMPSRKSFER 632
Score = 42.7 bits (99), Expect = 7e-04
Identities = 22/75 (29%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query: 555 GLQRDIEVLKSELLKFVAEYGQE-GFMPMRKQLRLHGRVDIEKAITRMGGFRKIATIMNL 613
G ++E L+ E+ +F +G + FMP RK GR DI +A+ + GG +++ ++ L
Sbjct: 598 GYWDNLENLQEEIGRFQQSWGMDPSFMPSRKSFERAGRYDIARALEKWGGLHEVSRLLAL 657
Query: 614 SLAYKYRKPKGYWDN 628
++ + R+ DN
Sbjct: 658 NVRHPNRQLNSRKDN 672
>At4g01650 unknown protein
Length = 166
Score = 81.3 bits (199), Expect = 2e-15
Identities = 53/155 (34%), Positives = 88/155 (56%), Gaps = 24/155 (15%)
Query: 365 VKAPVRDVWNVMSSYETLPEIVPNLAISKILSRDNNKVRILQEGCKGL-LYMVLHARVVL 423
++A + VW+V++ YE L + +P L +S+++ ++ N+VR+ Q G + L L + +A+ VL
Sbjct: 1 MEASLDSVWSVLTDYEKLSDFIPGLVVSELVEKEGNRVRLFQMGQQNLALGLKFNAKAVL 60
Query: 424 DLCE-QLE-------QEISFEQAEGDFDSFHGKWTFEQL--GNH-----------HTLLK 462
D E +LE +EI F+ EGDF F GKW+ EQL G H T L
Sbjct: 61 DCYEKELEVLPHGRRREIDFKMVEGDFQLFEGKWSIEQLDKGIHGEALDLQFKDFRTTLA 120
Query: 463 YSVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRD 497
Y+VD K + +L ++E + +++ +NL +IRD
Sbjct: 121 YTVDVKPK--MWLPVRLVEGRLCKEIRTNLMSIRD 153
Score = 75.1 bits (183), Expect = 1e-13
Identities = 54/162 (33%), Positives = 82/162 (50%), Gaps = 18/162 (11%)
Query: 115 INADINSVWNALTDYEHLADFIPNLVWSGRIPCPFPGRIWLEQRGFQR-AMYWHIEARVV 173
+ A ++SVW+ LTDYE L+DFIP LV S + R+ L Q G Q A+ A+ V
Sbjct: 1 MEASLDSVWSVLTDYEKLSDFIPGLVVSELVE-KEGNRVRLFQMGQQNLALGLKFNAKAV 59
Query: 174 LDLQ----ELLNSEWDRELHFSMVDGDFKKFEGKWSVKS------------GTRSSSTNL 217
LD E+L RE+ F MV+GDF+ FEGKWS++ + T L
Sbjct: 60 LDCYEKELEVLPHGRRREIDFKMVEGDFQLFEGKWSIEQLDKGIHGEALDLQFKDFRTTL 119
Query: 218 SYEVNVIPRFNFPAIFLERIVRSDLPVNLRALAYRVERNLLG 259
+Y V+V P+ P +E + ++ NL ++ ++ + G
Sbjct: 120 AYTVDVKPKMWLPVRLVEGRLCKEIRTNLMSIRDAAQKVIEG 161
>At5g03420 putative protein
Length = 583
Score = 42.4 bits (98), Expect = 0.001
Identities = 23/57 (40%), Positives = 35/57 (61%), Gaps = 1/57 (1%)
Query: 547 SNQRSRVPGLQRDIEVLKSELLKFVAEYG-QEGFMPMRKQLRLHGRVDIEKAITRMG 602
S +++RV + E L+SE+++FVA G EG +P K+L HGRVD+ + R G
Sbjct: 49 STKQTRVRKRVKSNEELRSEIMQFVALAGLPEGHVPSMKELSAHGRVDLANIVRRRG 105
>At1g70480 OBp32 protein
Length = 320
Score = 34.7 bits (78), Expect = 0.19
Identities = 35/148 (23%), Positives = 66/148 (43%), Gaps = 27/148 (18%)
Query: 81 GNLNNGLEEDGDREVHCELQVVSWRER--------------RVKAEISINADINSVWNAL 126
GN + ++ D ++++ C + SW ++ +KAE+++ ++V+N +
Sbjct: 61 GNSSISMQIDLEKQLDCWRENPSWTDQIPVVKVGIPKGSLCNLKAEVNVGLPPDAVYNIV 120
Query: 127 TDYEHLADFIPNL--VWSGRIPCPFPGR--IWLEQRGFQRAMYWH--IEARVVLDLQELL 180
D ++ F N+ V S ++ R + +EQ R ++W I V++D
Sbjct: 121 IDPDNRRVF-KNIKEVMSRKVLVDDGLRQVVEVEQAALWRFLWWSGTISVHVLVDQDRA- 178
Query: 181 NSEWDRELHFSMVDGDF-KKFEGKWSVK 207
D + F V F K+FEG W VK
Sbjct: 179 ----DHSMKFKQVKSGFMKRFEGSWQVK 202
>At1g27070 unknown protein
Length = 532
Score = 31.6 bits (70), Expect = 1.6
Identities = 14/42 (33%), Positives = 24/42 (56%)
Query: 563 LKSELLKFVAEYGQEGFMPMRKQLRLHGRVDIEKAITRMGGF 604
L++E+L+F+ + G P +K L GR D+ + I GG+
Sbjct: 94 LETEILEFMKNSEKPGMFPSKKDLIRSGRFDLVERIVNQGGW 135
>At4g16070 unknown protein
Length = 654
Score = 30.8 bits (68), Expect = 2.8
Identities = 19/94 (20%), Positives = 43/94 (45%), Gaps = 6/94 (6%)
Query: 464 SVDSKMRRDTFLSEAIMEEVIYEDLPSNLCAIRDYVENQKASQFLE--VCEQNTNSGQQI 521
S+D + + + E + E ++++L L EN++ S+ +E + ++
Sbjct: 488 SIDQVIAETSSIEEDVTEGELWDELDRELTR----QENERDSEAMEEEAAAAKEITEEET 543
Query: 522 ILSGSGDDNNSSSADDISDCNVQSSSNQRSRVPG 555
+++G GD + + +S ++ NQR PG
Sbjct: 544 VITGGGDSSTGQNQSPVSASSMDLIENQRFYPPG 577
>At1g20720 hypothetical protein
Length = 986
Score = 30.8 bits (68), Expect = 2.8
Identities = 24/114 (21%), Positives = 53/114 (46%), Gaps = 7/114 (6%)
Query: 500 ENQKASQFLEVCEQNTNSGQQIILSGSGDDNNSSSADDISDCNVQSSSNQRSRVPGLQRD 559
+++K + FL VC + G DDN + +D ++ N + S+ G RD
Sbjct: 612 DSKKGAAFLAVCRGKVSEGIDF-----ADDNARAVVEDYTNSNPKYHFMYESKAFGYHRD 666
Query: 560 IEVLKSELLKFVAEYGQEGFMPMRKQLRLHGRV-DIEKAITRMGGFRKIATIMN 612
++ +E L+ + Q F+ ++++ V D+E + G+ +++++ N
Sbjct: 667 VKPKIAEDLRSMGHSAQT-FVQVKEEAECCREVIDLECGVQPDLGYCEVSSVTN 719
>At4g12100 unknown protein (At4g12100)
Length = 434
Score = 29.6 bits (65), Expect = 6.2
Identities = 22/64 (34%), Positives = 34/64 (52%), Gaps = 15/64 (23%)
Query: 547 SNQRSRVPGLQRDIEVLKSELLKFVAEYGQEGFM---------PMRKQLRLHGRVDIEKA 597
S+ RSR+ ++VLKS+LL+ + + +EGFM +R+ RL VD E
Sbjct: 305 SSSRSRL------MKVLKSQLLEAHSSFLEEGFMLLMDESLIDDLRRMYRLFSMVDSEDY 358
Query: 598 ITRM 601
I R+
Sbjct: 359 IDRI 362
>At5g51640 unknown protein (At5g51640)
Length = 501
Score = 29.3 bits (64), Expect = 8.1
Identities = 22/87 (25%), Positives = 42/87 (47%), Gaps = 15/87 (17%)
Query: 376 MSSYETLPEIVPNLAISKIL---SRDNNKVRILQEGCKGLLYMVLHARVVLDLCEQLEQE 432
+SS E+ P ++ + K+L S D+N VR+ +E G V +++E
Sbjct: 81 VSSSES-PPVLTQDSDDKVLPKGSHDSNDVRLGEETNSGKSSNV-----------SIDEE 128
Query: 433 ISFEQAEGDFDSFHGKWTFEQLGNHHT 459
+ + E + D +HG W ++ +G +T
Sbjct: 129 ATQDHVETECDLYHGNWFYDPMGPLYT 155
>At4g27980 putative protein
Length = 565
Score = 29.3 bits (64), Expect = 8.1
Identities = 15/42 (35%), Positives = 22/42 (51%)
Query: 486 EDLPSNLCAIRDYVENQKASQFLEVCEQNTNSGQQIILSGSG 527
+D+P N IR+ E+Q + Q + T +Q ILSG G
Sbjct: 346 DDMPRNYAQIREIFESQLSLQVTLLEHVKTTKDEQSILSGCG 387
>At1g52620
Length = 819
Score = 29.3 bits (64), Expect = 8.1
Identities = 21/68 (30%), Positives = 35/68 (50%), Gaps = 8/68 (11%)
Query: 599 TRMGGFRKIATIMNLSLAYKYRKPKGYWDNLENLQDEISRFQRCWGMDPSFMPSRKSFER 658
T +GG+ K+ I N L +K K KG+ LE I+ F + + F+ S +R
Sbjct: 245 TIIGGYCKLGDIENAYLVFKELKLKGFMPTLETFGTMINGFCK----EGDFVAS----DR 296
Query: 659 VVSQGKEK 666
++S+ KE+
Sbjct: 297 LLSEVKER 304
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.137 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,356,835
Number of Sequences: 26719
Number of extensions: 725127
Number of successful extensions: 1994
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 5
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1976
Number of HSP's gapped (non-prelim): 13
length of query: 719
length of database: 11,318,596
effective HSP length: 106
effective length of query: 613
effective length of database: 8,486,382
effective search space: 5202152166
effective search space used: 5202152166
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148654.3


