

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148653.2 + phase: 0
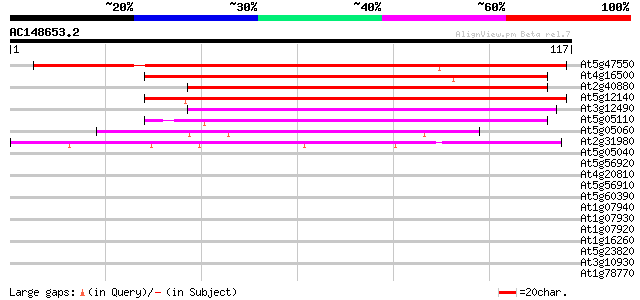
(117 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g47550 unknown protein 125 4e-30
At4g16500 cysteine proteinase inhibitor like protein 82 4e-17
At2g40880 putative cysteine proteinase inhibitor B (cystatin B) 73 3e-14
At5g12140 cystatin (emb|CAA03929.1) 70 2e-13
At3g12490 unknown protein 67 2e-12
At5g05110 cysteine proteinase inhibitor-like protein 65 4e-12
At5g05060 unknown protein 42 5e-05
At2g31980 putative cysteine proteinase inhibitor B (cystatin B) 40 1e-04
At5g05040 putative protein 37 0.002
At5g56920 unknown protein 31 0.087
At4g20810 putative protein 30 0.25
At5g56910 unknown protein 29 0.43
At5g60390 translation elongation factor eEF-1 alpha chain (gene A4) 28 0.96
At1g07940 elongation factor 1-alpha 28 0.96
At1g07930 elongation factor 1-alpha 28 0.96
At1g07920 elongation factor 1-alpha 28 0.96
At1g16260 putative wall-associated kinase 27 2.1
At5g23820 putative protein 26 2.8
At3g10930 unknown protein 26 2.8
At1g78770 hypothetical protein 26 2.8
>At5g47550 unknown protein
Length = 122
Score = 125 bits (313), Expect = 4e-30
Identities = 64/112 (57%), Positives = 84/112 (74%), Gaps = 3/112 (2%)
Query: 6 LLLVLLFVVVLFSYAAARNQFAPGGWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEK 65
LLL+ L VV+L YA+A + GGWSPI ++ DP V EI FAV+EY++RS + LKFE
Sbjct: 8 LLLLSLVVVLLPLYASAAARV--GGWSPISNVTDPQVVEIGEFAVSEYNKRSESGLKFET 65
Query: 66 VINGESQVVAGTNYRLTLSASDG-SYSKNYEAVVWEKIWQHFRNLTSFVPVH 116
V++GE+QVV+GTNYRL ++A+DG SKNY A+VW+K W FRNLTSF P +
Sbjct: 66 VVSGETQVVSGTNYRLKVAANDGDGVSKNYLAIVWDKPWMKFRNLTSFEPAN 117
>At4g16500 cysteine proteinase inhibitor like protein
Length = 117
Score = 82.0 bits (201), Expect = 4e-17
Identities = 40/85 (47%), Positives = 57/85 (67%), Gaps = 1/85 (1%)
Query: 29 GGWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDG 88
G PI +++DP V +A +A+ E+++ S KL F KV+ G +QVV+GT Y L ++A DG
Sbjct: 30 GSRKPIKNVSDPDVVAVAKYAIEEHNKESKEKLVFVKVVEGTTQVVSGTKYDLKIAAKDG 89
Query: 89 SYS-KNYEAVVWEKIWQHFRNLTSF 112
KNYEAVV EK+W H ++L SF
Sbjct: 90 GGKIKNYEAVVVEKLWLHSKSLESF 114
>At2g40880 putative cysteine proteinase inhibitor B (cystatin B)
Length = 125
Score = 72.8 bits (177), Expect = 3e-14
Identities = 31/75 (41%), Positives = 47/75 (62%)
Query: 38 NDPHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDGSYSKNYEAV 97
N + +A FA+ E++++ L+F+K++ QVVAGT Y LTL A +G +KN+EA
Sbjct: 46 NSGEIESLARFAIQEHNKQQNKILEFKKIVKAREQVVAGTMYHLTLEAKEGDQTKNFEAK 105
Query: 98 VWEKIWQHFRNLTSF 112
VW K W +F+ L F
Sbjct: 106 VWVKPWMNFKQLQEF 120
>At5g12140 cystatin (emb|CAA03929.1)
Length = 101
Score = 70.1 bits (170), Expect = 2e-13
Identities = 35/89 (39%), Positives = 56/89 (62%), Gaps = 1/89 (1%)
Query: 29 GGWSPID-DINDPHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASD 87
GG ID + ND V +A FAV E+++ L++++++ ++QVVAGT + LT+ +D
Sbjct: 11 GGVRDIDANANDLQVESLARFAVDEHNKNENLTLEYKRLLGAKTQVVAGTMHHLTVEVAD 70
Query: 88 GSYSKNYEAVVWEKIWQHFRNLTSFVPVH 116
G +K YEA V EK W++ + L SF +H
Sbjct: 71 GETNKVYEAKVLEKAWENLKQLESFNHLH 99
>At3g12490 unknown protein
Length = 201
Score = 66.6 bits (161), Expect = 2e-12
Identities = 32/77 (41%), Positives = 45/77 (57%)
Query: 38 NDPHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDGSYSKNYEAV 97
N V +A FAV E++++ A L+F +V+ + QVVAGT + LTL + K YEA
Sbjct: 15 NSGEVESLARFAVDEHNKKENALLEFARVVKAKEQVVAGTLHHLTLEILEAGQKKLYEAK 74
Query: 98 VWEKIWQHFRNLTSFVP 114
VW K W +F+ L F P
Sbjct: 75 VWVKPWLNFKELQEFKP 91
>At5g05110 cysteine proteinase inhibitor-like protein
Length = 232
Score = 65.5 bits (158), Expect = 4e-12
Identities = 38/88 (43%), Positives = 49/88 (55%), Gaps = 6/88 (6%)
Query: 29 GGWSPIDDIND----PHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLS 84
GG+S D ND + +IA FAV E++RR A L+ +V+ QVVAG YRLTL
Sbjct: 46 GGFS--DSKNDWNGGKEIDDIALFAVQEHNRRENAVLELARVLKATEQVVAGKLYRLTLE 103
Query: 85 ASDGSYSKNYEAVVWEKIWQHFRNLTSF 112
+ K YEA VW K W +F+ L F
Sbjct: 104 VIEAGEKKIYEAKVWVKPWMNFKQLQEF 131
>At5g05060 unknown protein
Length = 172
Score = 42.0 bits (97), Expect = 5e-05
Identities = 27/91 (29%), Positives = 47/91 (50%), Gaps = 11/91 (12%)
Query: 19 YAAARNQFAPGGWSPIDD--INDPHVTE------IANFAVTEYDRRSGAKLKFEKVINGE 70
++ R++F G DD +++PH T ++N A++ Y+ ++G KL+ KV+
Sbjct: 63 FSKVRHRFGRGAVHLDDDNMVSEPHETNRDLLNRLSNMAISFYNDKTGIKLELVKVLRAN 122
Query: 71 SQVVAGTNYRLTLSA---SDGSYSKNYEAVV 98
A +T A SDG+ +K Y+AVV
Sbjct: 123 FHPSAAITLYITFEANDPSDGNQTKRYQAVV 153
>At2g31980 putative cysteine proteinase inhibitor B (cystatin B)
Length = 147
Score = 40.4 bits (93), Expect = 1e-04
Identities = 35/140 (25%), Positives = 70/140 (50%), Gaps = 26/140 (18%)
Query: 1 MRNPTLLLVLL-FVVVLFSYAAARNQFAP----GGWSPIDDIN-DPHVTEIANFAVTEYD 54
M +L+L LL F+V+ +A N F GG S + +I + + ++ + V +++
Sbjct: 4 MLKVSLVLSLLGFLVIAVVTPSAANPFRKSVVLGGKSGVPNIRTNREIQQLGRYCVEQFN 63
Query: 55 RRSGAK-----------------LKFEKVINGESQVVAGTNY--RLTLSASDGSYSKNYE 95
+++ + L+F +V++ + QVVAG Y R+ ++ +GS ++ ++
Sbjct: 64 QQAQNEQGNIGSIAKTDTAISNPLQFSRVVSAQKQVVAGLKYYLRIEVTQPNGS-TRMFD 122
Query: 96 AVVWEKIWQHFRNLTSFVPV 115
+VV + W H + L F PV
Sbjct: 123 SVVVIQPWLHSKQLLGFTPV 142
>At5g05040 putative protein
Length = 171
Score = 36.6 bits (83), Expect = 0.002
Identities = 24/91 (26%), Positives = 46/91 (50%), Gaps = 11/91 (12%)
Query: 19 YAAARNQFAPGGWSPIDD--INDPHVTE------IANFAVTEYDRRSGAKLKFEKVINGE 70
++ R++F G DD +++PH T ++N A++ Y+ ++ + L+ KV+
Sbjct: 63 FSKVRHRFGCGAVHLDDDNMVSEPHETNRDLLNRLSNMAISFYNDKTDSSLELVKVLRAN 122
Query: 71 SQVVAGTNYRLTLSAS---DGSYSKNYEAVV 98
A +T A+ DG+ +K Y+AVV
Sbjct: 123 FHPSAAITLYITFEANDPKDGNQTKRYQAVV 153
>At5g56920 unknown protein
Length = 227
Score = 31.2 bits (69), Expect = 0.087
Identities = 16/72 (22%), Positives = 33/72 (45%)
Query: 42 VTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDGSYSKNYEAVVWEK 101
+ E+ A+ +++ G KL F + + Q+ G LT A+D + S +
Sbjct: 96 IREMTLTAIDKHNEAHGTKLVFVEHVEANYQLTRGLTCWLTFWATDMASSPPQSKIYQAH 155
Query: 102 IWQHFRNLTSFV 113
+W+ +N +F+
Sbjct: 156 VWRRGQNFHTFI 167
>At4g20810 putative protein
Length = 416
Score = 29.6 bits (65), Expect = 0.25
Identities = 17/60 (28%), Positives = 28/60 (46%)
Query: 42 VTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDGSYSKNYEAVVWEK 101
VT A + DR+ G K K E+ E+ V A NY++ L+ ++ V W++
Sbjct: 356 VTTDIELATSSSDRQVGMKSKREEEEEEEASVAANGNYKVDLNVEAEEAEQDENDVDWQE 415
>At5g56910 unknown protein
Length = 224
Score = 28.9 bits (63), Expect = 0.43
Identities = 18/74 (24%), Positives = 36/74 (48%), Gaps = 2/74 (2%)
Query: 33 PIDDI--NDPHVTEIANFAVTEYDRRSGAKLKFEKVINGESQVVAGTNYRLTLSASDGSY 90
PI DI N + ++ FA+ +++ G+KL F + ++ + G LT A+D +
Sbjct: 82 PISDIRTNADVIRDVTLFAIEKHNEAHGSKLVFVEHVSANFKFANGLLCWLTFWATDMAS 141
Query: 91 SKNYEAVVWEKIWQ 104
S + ++W+
Sbjct: 142 SAPTSQIYQVELWR 155
>At5g60390 translation elongation factor eEF-1 alpha chain (gene A4)
Length = 449
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query: 30 GWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEK--VINGESQVVAGT 77
G++P+ D + H+ + +T+ DRRSG +++ E + NG++ +V T
Sbjct: 344 GYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
>At1g07940 elongation factor 1-alpha
Length = 449
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query: 30 GWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEK--VINGESQVVAGT 77
G++P+ D + H+ + +T+ DRRSG +++ E + NG++ +V T
Sbjct: 344 GYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
>At1g07930 elongation factor 1-alpha
Length = 449
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query: 30 GWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEK--VINGESQVVAGT 77
G++P+ D + H+ + +T+ DRRSG +++ E + NG++ +V T
Sbjct: 344 GYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
>At1g07920 elongation factor 1-alpha
Length = 449
Score = 27.7 bits (60), Expect = 0.96
Identities = 15/50 (30%), Positives = 30/50 (60%), Gaps = 2/50 (4%)
Query: 30 GWSPIDDINDPHVTEIANFAVTEYDRRSGAKLKFEK--VINGESQVVAGT 77
G++P+ D + H+ + +T+ DRRSG +++ E + NG++ +V T
Sbjct: 344 GYAPVLDCHTSHIAVKFSEILTKIDRRSGKEIEKEPKFLKNGDAGMVKMT 393
>At1g16260 putative wall-associated kinase
Length = 720
Score = 26.6 bits (57), Expect = 2.1
Identities = 9/21 (42%), Positives = 13/21 (61%)
Query: 24 NQFAPGGWSPIDDINDPHVTE 44
N + PGG ID+ DPH+ +
Sbjct: 275 NPYIPGGCQDIDECRDPHLNK 295
>At5g23820 putative protein
Length = 164
Score = 26.2 bits (56), Expect = 2.8
Identities = 10/17 (58%), Positives = 14/17 (81%)
Query: 68 NGESQVVAGTNYRLTLS 84
N E+++ AGTN+ LTLS
Sbjct: 101 NNEAEIEAGTNFELTLS 117
>At3g10930 unknown protein
Length = 97
Score = 26.2 bits (56), Expect = 2.8
Identities = 24/83 (28%), Positives = 38/83 (44%), Gaps = 9/83 (10%)
Query: 3 NPTLLLVLLFVVVLFSYAAARNQFAPGGWSPIDDINDPHVTEIANFAVTEYDRRSGAKLK 62
N +LLL+LLF+ V S A + PG + P+ I + ++ AV K +
Sbjct: 4 NRSLLLILLFISVSLSTA----RILPGEFVPV--IFSGEIPPVSKSAVVGCGGEQETKTE 57
Query: 63 FEKVINGESQVVAGTNYRLTLSA 85
+ + +VVAG L L+A
Sbjct: 58 YSSFV---PEVVAGKFGSLVLNA 77
>At1g78770 hypothetical protein
Length = 542
Score = 26.2 bits (56), Expect = 2.8
Identities = 11/29 (37%), Positives = 20/29 (68%)
Query: 50 VTEYDRRSGAKLKFEKVINGESQVVAGTN 78
+ +YD+ S +LKF+K+ N S V+G++
Sbjct: 204 IKKYDKESTVELKFKKLENETSGSVSGSS 232
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,590,346
Number of Sequences: 26719
Number of extensions: 92870
Number of successful extensions: 254
Number of sequences better than 10.0: 30
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 229
Number of HSP's gapped (non-prelim): 30
length of query: 117
length of database: 11,318,596
effective HSP length: 93
effective length of query: 24
effective length of database: 8,833,729
effective search space: 212009496
effective search space used: 212009496
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 52 (24.6 bits)
Medicago: description of AC148653.2


