

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148651.4 + phase: 0 /pseudo
(764 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
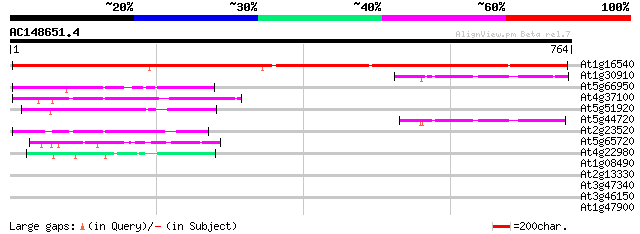
Score E
Sequences producing significant alignments: (bits) Value
At1g16540 molybdenum cofactor sulfurase 841 0.0
At1g30910 unknown protein 76 8e-14
At5g66950 unknown protein 72 9e-13
At4g37100 putative protein 72 9e-13
At5g51920 putative protein 67 3e-11
At5g44720 unknown protein 67 5e-11
At2g23520 hypothetical protein 64 3e-10
At5g65720 NifS-like aminotranfserase 50 4e-06
At4g22980 putative protein 50 6e-06
At1g08490 NIFS like protein CpNifsp precursor (NIFS) 42 0.001
At2g13330 F14O4.9 30 3.9
At3g47340 glutamine-dependent asparagine synthetase 30 5.1
At3g46150 hypothetical protein 30 6.6
At1g47900 mysoin-like protein 29 8.6
>At1g16540 molybdenum cofactor sulfurase
Length = 819
Score = 841 bits (2173), Expect = 0.0
Identities = 449/793 (56%), Positives = 555/793 (69%), Gaps = 44/793 (5%)
Query: 4 EEFLKEFGEHYGYPNAARTIDQIRATEFNRL-QDLVYLDHAGATLYSELQMESVFKDLTT 62
E FLKEFG++YGYP+ + I +IR TEF RL + +VYLDHAG+TLYSELQME +FKD T+
Sbjct: 2 EAFLKEFGDYYGYPDGPKNIQEIRDTEFKRLDKGVVYLDHAGSTLYSELQMEYIFKDFTS 61
Query: 63 NVYGNPHSQSDSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWS 122
NV+GNPHSQSD S+AT D++ DAR QVL+Y NASPEDY C+FTSGATAALKLVGE FPW+
Sbjct: 62 NVFGNPHSQSDISSATSDLIADARHQVLEYFNASPEDYSCLFTSGATAALKLVGETFPWT 121
Query: 123 CNSNFMYTMENHNSVLGIREYALGQGAAAIAVDIEDV--HPRIEGEKFPT-KISLHQEQR 179
+SNF+YTMENHNSVLGIREYAL QGA+A AVDIE+ P P+ K+ Q
Sbjct: 122 QDSNFLYTMENHNSVLGIREYALAQGASACAVDIEEAANQPGQLTNSGPSIKVKHRAVQM 181
Query: 180 RKVTGLQEEE-----------PMECNFSGLRFDLDLAKIIKEDSSKIL-GASVCKKGRWL 227
R + LQ+EE P ECNFSGLRF+LDL K++KE++ +L G+ K RW+
Sbjct: 182 RNTSKLQKEESRGNAYNLFAFPSECNFSGLRFNLDLVKLMKENTETVLQGSPFSKSKRWM 241
Query: 228 VLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGALVVRNDAAKLLKKSYFSGGT 287
VLIDAAKG AT+PPDLS+YP DFV LSFYKLFGYPTGLGAL+VRNDAAKLLKK+YFSGGT
Sbjct: 242 VLIDAAKGCATLPPDLSEYPADFVVLSFYKLFGYPTGLGALLVRNDAAKLLKKTYFSGGT 301
Query: 288 VAASIADIDFIKRREGIEELFEDGTVSFLSIASIRHGFKILNSLTVSAI-SRTYNIS--- 343
VAASIADIDF+KRRE +EE FEDG+ SFLSIA+IRHGFK+L SLT SAI T ++S
Sbjct: 302 VAASIADIDFVKRRERVEEFFEDGSASFLSIAAIRHGFKLLKSLTPSAIWMHTTSLSIYV 361
Query: 344 ---------------CPVYEENAFGPKAWQWI*RLHPLWTP*FNGPDGSWYGYREVEKLA 388
C +Y + + P T PDGSW+GY EVEKLA
Sbjct: 362 KKKLQALRHGNGAAVCVLYGSENLELSSH----KSGPTVTFNLKRPDGSWFGYLEVEKLA 417
Query: 389 SLSGIQLRTGCFCNPGACAKYLGLSHMDLISNTEAGHVCWDDQDIISGKPIGAVRVSFGY 448
SLSGIQLRTGCFCNPGACAKYL LSH +L SN EAGH+CWDD D+I+GKP GAVRVSFGY
Sbjct: 418 SLSGIQLRTGCFCNPGACAKYLELSHSELRSNVEAGHICWDDNDVINGKPTGAVRVSFGY 477
Query: 449 MSTFEDAKKFIDFVKSSFMSPQNHVDNGNQLKDIYSVIYMQE*MVSMILVIISNQLQCIR 508
MSTFEDAKKFIDF+ SSF SP NG + + + ++ + S+ L+ I
Sbjct: 478 MSTFEDAKKFIDFIISSFASPPKKTGNGTVVSGRFPQLPSED--LESKESFPSHYLKSIT 535
Query: 509 *NPVE--ASVQAVGLLATMGSLKHDREWILKSLSGEILTLKRVPEMGLISSFIDLSQGML 566
P++ A + L HDREW+++ L+GEILT K+VPEM LI +FIDL +G+L
Sbjct: 536 VYPIKSCAGFSVIRWPLCRTGLLHDREWMVQGLTGEILTQKKVPEMSLIKTFIDLEEGLL 595
Query: 567 FVESPHCKERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRY 626
VES C+++L IR++ D Y+ + + + + + N WF+ AI R C LLRY
Sbjct: 596 SVESSRCEDKLHIRIKSDSYNPRNDEFDSHANILENRNEETRINRWFTNAIGRQCKLLRY 655
Query: 627 SGSSHDFVLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETE 686
S S+ L+R K C+D S ++FANE QFLL+SEESV+DLN+RL + + D
Sbjct: 656 SSSTSKDCLNRNKSPGLCRDLESNINFANEAQFLLISEESVADLNRRLEAKDE-DYKRAH 714
Query: 687 IEINTNRFRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSK 746
++N +RFRPNLV+SGG PY ED W ++IG+ +F SLGGCNRCQ+IN++ AG V+KS
Sbjct: 715 EKLNPHRFRPNLVISGGEPYGEDKWKTVKIGDNHFTSLGGCNRCQMINISNEAGLVKKSN 774
Query: 747 EPLATLASYRRVK 759
EPL TLASYRRVK
Sbjct: 775 EPLTTLASYRRVK 787
>At1g30910 unknown protein
Length = 318
Score = 75.9 bits (185), Expect = 8e-14
Identities = 62/248 (25%), Positives = 107/248 (43%), Gaps = 34/248 (13%)
Query: 524 TMGSLKHDREWILKSLSGEILTLKRVPEMGLISSFI-----------DLSQGMLFVESPH 572
T + DR W++ + G LT + P++ LI + + S M+ V +P
Sbjct: 43 TPTGFRWDRNWLIVNSKGRGLTQRVEPKLSLIEVEMPKHAFGEDWEPEKSSNMV-VRAPG 101
Query: 573 CKERLQIRLQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHD 632
+++ L D + + E + WF+ + +PC L+R++ +
Sbjct: 102 MDA---LKVSLAKPDKIADGVSVWEWSGSALDEGEEASQWFTNFVGKPCRLVRFNSAYE- 157
Query: 633 FVLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTN 692
T+ + F++ FLL+S+ S+ LNK L V + N
Sbjct: 158 -----TRPVDPNYAPGHIAMFSDMYPFLLISQGSLDSLNKLLKEPVPI-----------N 201
Query: 693 RFRPNLVVSGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSKEPLATL 752
RFRPN+ V G P+ ED W++I I F + C+RC+V ++ G +EP+ TL
Sbjct: 202 RFRPNIFVDGCEPFAEDLWTEILINGFTFHGVKLCSRCKVPTISQETG--IGGQEPIETL 259
Query: 753 ASYRRVKV 760
++R KV
Sbjct: 260 RTFRSDKV 267
>At5g66950 unknown protein
Length = 870
Score = 72.4 bits (176), Expect = 9e-13
Identities = 73/286 (25%), Positives = 131/286 (45%), Gaps = 24/286 (8%)
Query: 4 EEFLKEFGEHYGYPNAARTIDQIRATEFNRLQ-DLVYLDHAGATLYSELQMESVFKDLTT 62
EE L F Y ++ +D++R E+ L V LD+ G L+S LQ + T
Sbjct: 112 EEALTIFLTMYPKYQSSEKVDELRNDEYFHLSLPKVCLDYCGFGLFSYLQTVHYWDTCTF 171
Query: 63 NVYGNPHSQSDSS----AATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEA 118
++ + S+ + A I D + +++DY N +Y +FT +A KL+ E+
Sbjct: 172 SLSEISANLSNHAIYGGAEKGSIEHDIKIRIMDYLNIPENEYGLVFTVSRGSAFKLLAES 231
Query: 119 FPWSCNSNFMYTMENH--NSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQ 176
+P+ N + TM +H SV + + A +GA + K+PT
Sbjct: 232 YPFHTNKKLL-TMFDHESQSVSWMGQCAKEKGAKVGSAWF----------KWPTLRLCSM 280
Query: 177 EQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDA-AKG 235
+ ++++ L +++ + + +GL F + + ++ ++ W VL+DA A G
Sbjct: 281 DLKKEI--LSKKKRKKDSATGL-FVFPVQSRVTGSKYSYQWMALAQQNNWHVLLDAGALG 337
Query: 236 SATMPP-DLSKYPVDFVALSFYKLFGY-PTGLGALVVRNDAAKLLK 279
M LS + DF+ SFY++FGY PTG G L+++ L+
Sbjct: 338 PKDMDSLGLSLFRPDFIITSFYRVFGYDPTGFGCLLIKKSVISCLQ 383
Score = 35.4 bits (80), Expect = 0.12
Identities = 29/91 (31%), Positives = 41/91 (44%), Gaps = 19/91 (20%)
Query: 384 VEKLASLSGIQLRTGCFCNPGACAKYLGLSHMDLISNTEAGHVCWDDQDIISGKPIGAVR 443
V+KLA GI L G LSH+ +I N W D G+ G +R
Sbjct: 774 VQKLAEREGISLGIGY------------LSHIKIIDNRSEDSSSWKPVDR-EGRNNGFIR 820
Query: 444 V-----SFGYMSTFEDAKKFIDFVKSSFMSP 469
V S G+++ FED + +FV + F+SP
Sbjct: 821 VEVVTASLGFLTNFEDVYRLWNFV-AKFLSP 850
>At4g37100 putative protein
Length = 896
Score = 72.4 bits (176), Expect = 9e-13
Identities = 75/327 (22%), Positives = 142/327 (42%), Gaps = 36/327 (11%)
Query: 5 EFLKEFGEHYGYPNAARTIDQIRATEFNRLQDL---VYLDHAGATLYSELQMESVF---- 57
E L +F Y A+ IDQ+R+ E++ L V LD+ G L+S +Q +
Sbjct: 117 EALTKFLSMYPKYQASEKIDQLRSDEYSHLSSSASKVCLDYCGFGLFSYVQTLHYWDTCT 176
Query: 58 ---KDLTTNVYGNP-HSQSDSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALK 113
++T N+ + + ++S HDI + +++DY N +Y +FT +A +
Sbjct: 177 FSLSEITANLSNHALYGGAESGTVEHDI----KTRIMDYLNIPENEYGLVFTVSRGSAFR 232
Query: 114 LVGEAFPWSCNSNFMYTMENH--NSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTK 171
L+ E++P+ N + TM +H SV + + A +GA A + ++ +
Sbjct: 233 LLAESYPFQSNKRLL-TMFDHESQSVNWMAQTAREKGAKAYNAWFKWPTLKLCSTDLKKR 291
Query: 172 ISLHQEQRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLID 231
+S + +++ P + +G ++ ++ ++ W VL+D
Sbjct: 292 LSYKKRKKKDSAVGLFVFPAQSRVTGTKYSYQ-------------WMALAQQNHWHVLLD 338
Query: 232 AAK-GSATMPP-DLSKYPVDFVALSFYKLFGY-PTGLGALVVRNDAAKLLKKSYFSGGTV 288
A G M LS + +F+ SFY++FG+ PTG G L+++ L+ G+
Sbjct: 339 AGSLGPKDMDSLGLSLFRPEFIITSFYRVFGHDPTGFGCLLIKKSVMGSLQSQSGKTGSG 398
Query: 289 AASIADIDFIKRREGIEELFEDGTVSF 315
I + + ++ L DG V F
Sbjct: 399 IVKITPEYPLYLSDSVDGL--DGLVGF 423
>At5g51920 putative protein
Length = 570
Score = 67.4 bits (163), Expect = 3e-11
Identities = 70/290 (24%), Positives = 121/290 (41%), Gaps = 38/290 (13%)
Query: 16 YPNAART--IDQIRATEFNRLQDLVY--LDHAGATLYSELQM---------------ESV 56
YPN + T ID++R+ + L Y LD+ G LYS Q+ ES
Sbjct: 83 YPNYSDTYKIDRLRSDHYFHLGLSHYTCLDYIGIGLYSYSQLLNYDPSTYQISSSLSESP 142
Query: 57 FKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVG 116
F ++ + + + +++++ + S EDY +FT+ T+A +LV
Sbjct: 143 FFSVSPKIGNLKEKLLNDGGQETEFEYSMKRRIMGFLKISEEDYSMVFTANRTSAFRLVA 202
Query: 117 EAFPWSCNSNFMYTME-NHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLH 175
E++P++ + + +V I + +GA A + ++ K ++
Sbjct: 203 ESYPFNSKRKLLTVYDYESEAVSEINRVSEKRGAKVAAAEFSWPRLKLCSSKLRKLVTAG 262
Query: 176 QE-QRRKVTGLQEEEPMECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDA-A 233
+ + K G+ P+ +G R+ L SV ++ W V+IDA
Sbjct: 263 KNGSKTKKKGIY-VFPLHSRVTGSRY-------------PYLWMSVAQENGWHVMIDACG 308
Query: 234 KGSATMPP-DLSKYPVDFVALSFYKLFG-YPTGLGALVVRNDAAKLLKKS 281
G M LS Y DF+ SFYK+FG P+G G L V+ +L+ S
Sbjct: 309 LGPKDMDSFGLSIYNPDFMVCSFYKVFGENPSGFGCLFVKKSTISILESS 358
>At5g44720 unknown protein
Length = 308
Score = 66.6 bits (161), Expect = 5e-11
Identities = 54/236 (22%), Positives = 101/236 (41%), Gaps = 31/236 (13%)
Query: 531 DREWILKSLSGEILTLKRVPEMGLISS------FID----LSQGMLFVESPHCKERLQIR 580
DR W++ + G T + P + L+ S F++ + +L + +P ++
Sbjct: 36 DRYWLVVNYKGRAYTQRVEPTLALVESELPKEAFLEDWEPTNDSLLVIRAPGMSP---LK 92
Query: 581 LQLDFYDSAIQDIELQGQRYKVYSYDNETNSWFSKAIERPCTLLRYSGSSHDFVLDRTKD 640
+ L S + + + + E WFS + + L+R++ + T+
Sbjct: 93 IPLTRPSSVAEGVSMWEWSGSAFDEGEEAAKWFSDYLGKQSRLVRFNKDTE------TRP 146
Query: 641 IVTCKDTNSAVSFANEGQFLLVSEESVSDLNKRLCSDVQMDMCETEIEINTNRFRPNLVV 700
+ +F + FL+ S+ S+ LN L V + NRFRPN++V
Sbjct: 147 SPPEFAAGYSTTFMDMFPFLVASQGSLDHLNTLLPEPVPI-----------NRFRPNILV 195
Query: 701 SGGRPYDEDGWSDIRIGNKYFRSLGGCNRCQVINLTLNAGQVQKSKEPLATLASYR 756
P+ ED W +I+I + F+ + C+RC+V + G + K+ EP TL +R
Sbjct: 196 DNCDPFGEDLWDEIKINDLVFQGVRLCSRCKVPTVNQETGVMGKA-EPTETLMKFR 250
>At2g23520 hypothetical protein
Length = 862
Score = 63.9 bits (154), Expect = 3e-10
Identities = 61/274 (22%), Positives = 125/274 (45%), Gaps = 33/274 (12%)
Query: 5 EFLKEFGEHYGYPNAARTIDQIRATEFNRLQDLVYLDHAGATLYSELQMESVFKDLTTNV 64
E +F Y + +DQ+R+ E+ L D L + + +S ++T N+
Sbjct: 115 EAFNKFLTMYPKFETSEKVDQLRSDEYGHLLDSKTLHYWDSCTFS-------LSEITANL 167
Query: 65 YGNP-HSQSDSSAATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSC 123
+ + ++ HD+ + +++DY N +Y +FT +A +L+ E++P+
Sbjct: 168 SNHALYGGAEIGTVEHDL----KTRIMDYLNIPESEYGLVFTGSRGSAFRLLAESYPFHT 223
Query: 124 NSNFMYTMENH--NSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRK 181
N + TM +H SV + + A +GA A + ++ ++S H+++++K
Sbjct: 224 NKRLL-TMFDHESQSVNWMAQTAREKGAKAYNAWFKWPTLKLCSTDLKKRLS-HKKRKKK 281
Query: 182 VTGL-QEEEPMECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDAAK-GSATM 239
+ + P + +G ++ + ++++ W VL+DA G M
Sbjct: 282 DSAVGLFVFPAQSRVTGSKYSYQWMALAQQNN-------------WHVLLDAGSLGPKDM 328
Query: 240 PP-DLSKYPVDFVALSFYKLFGY-PTGLGALVVR 271
LS + +F+ SFYK+FG+ PTG G L+++
Sbjct: 329 DSLGLSLFRPEFIITSFYKVFGHDPTGFGCLLIK 362
>At5g65720 NifS-like aminotranfserase
Length = 453
Score = 50.4 bits (119), Expect = 4e-06
Identities = 65/274 (23%), Positives = 116/274 (41%), Gaps = 39/274 (14%)
Query: 28 ATEFNRLQDLVYLDH---AGATLYSELQMES-----VFKDLTTNV---YGNPHSQSDSSA 76
ATE N + + + +G LY ++Q + VF + + YGNPHS++
Sbjct: 33 ATEVNYEDESIMMKGVRISGRPLYLDMQATTPIDPRVFDAMNASQIHEYGNPHSRTHLYG 92
Query: 77 -ATHDIVRDARQQVLDYCNASPEDYKCIFTSGATAALKLVGEA---FPWSCNSNFMYTME 132
+ V +AR QV ASP++ +F SGAT A + + F + + T
Sbjct: 93 WEAENAVENARNQVAKLIEASPKEI--VFVSGATEANNMAVKGVMHFYKDTKKHVITTQT 150
Query: 133 NHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPME 192
H VL + +G + + + +G + + +E R TGL
Sbjct: 151 EHKCVLDSCRHLQQEGFEVTYLPV-----KTDGL---VDLEMLREAIRPDTGL------- 195
Query: 193 CNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVA 252
+ + ++ + + ++E +CK+ DAA+ +P D+ K+ V ++
Sbjct: 196 VSIMAVNNEIGVVQPMEEIGM------ICKEHNVPFHTDAAQAIGKIPVDVKKWNVALMS 249
Query: 253 LSFYKLFGYPTGLGALVVRNDAAKLLKKSYFSGG 286
+S +K++G P G+GAL VR L+ GG
Sbjct: 250 MSAHKIYG-PKGVGALYVRRRPRIRLEPLMNGGG 282
>At4g22980 putative protein
Length = 559
Score = 49.7 bits (117), Expect = 6e-06
Identities = 62/274 (22%), Positives = 108/274 (38%), Gaps = 36/274 (13%)
Query: 24 DQIRATEFNRLQDLVYLDHAGATLYSELQMESVFK---DLTTNVYGNPHSQSDSSAATHD 80
D +R+TE+ L ++ L+S Q + + DL ++ Q S
Sbjct: 82 DHLRSTEYQNLSSSSHVFGQQQPLFSYSQFREISESESDLNHSLLTLSCKQVSSGKELLS 141
Query: 81 IVRDARQQ------VLDYCNASPEDYKCIFTSGATAALKLVGEAFPWSCNSNFM----YT 130
++R Q + + N +Y I T ++A K+V E + + N N + Y
Sbjct: 142 FEEESRFQSRIRKRITSFMNLEESEYHMILTQDRSSAFKIVAELYSFKTNPNLLTVYNYE 201
Query: 131 MENHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEP 190
E ++ I E +G + + I EK +I+ + +RR GL
Sbjct: 202 DEAVEEMIRISE---KKGIKPQSAEFSWPSTEILSEKLKRRIT--RSKRRGKRGL----- 251
Query: 191 MECNFSGLRFDLDLAKIIKEDSSKILGASVCKKGRWLVLID-AAKGSATMPP-DLSKYPV 248
F L ++ S S+ ++ W VL+D +A GS M LS +
Sbjct: 252 ---------FVFPLQSLVTGASYSYSWMSLARESEWHVLLDTSALGSKDMETLGLSLFQP 302
Query: 249 DFVALSFYKLFGY--PTGLGALVVRNDAAKLLKK 280
DF+ SF ++ G P+G G L V+ ++ L +
Sbjct: 303 DFLICSFTEVLGQDDPSGFGCLFVKKSSSTALSE 336
>At1g08490 NIFS like protein CpNifsp precursor (NIFS)
Length = 463
Score = 42.4 bits (98), Expect = 0.001
Identities = 59/265 (22%), Positives = 116/265 (43%), Gaps = 38/265 (14%)
Query: 37 LVYLDHAGATLYSELQMESV---FKDLTTNVYGNPHSQSDSSAATHDIVRDARQQVLDYC 93
LVYLD A + ++++ ++ +NV+ H S + ++ AR++V +
Sbjct: 73 LVYLDSAATSQKPAAVLDALQNYYEFYNSNVHRGIHYLSAKATDEFEL---ARKKVARFI 129
Query: 94 NASPEDYKCIFTSGATAALKLVGEAFPWSCNS-----NFMYTMENHNSVLGIREYALGQG 148
NAS + + +FT AT A+ LV A+ W ++ + T+ H+S + + + Q
Sbjct: 130 NAS-DSREIVFTRNATEAINLV--AYSWGLSNLKPGDEVILTVAEHHSCI-VPWQIVSQK 185
Query: 149 AAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQEEEPMECNFSGLRFDLD-LAKI 207
A+ KF ++L++++ + L+E + + + LA
Sbjct: 186 TGAVL-------------KF---VTLNEDEVPDINKLRELISPKTKLVAVHHVSNVLASS 229
Query: 208 IKEDSSKILGASVCKKGRWLVLIDAAKGSATMPPDLSKYPVDFVALSFYKLFGYPTGLGA 267
+ + + V K VL+DA + M D+ K DF+ S +K+ G PTG+G
Sbjct: 230 LPIEEIVVWAHDVGAK----VLVDACQSVPHMVVDVQKLNADFLVASSHKMCG-PTGIGF 284
Query: 268 LVVRNDAAKLLKKSYFSGGTVAASI 292
L ++D + + GG + + +
Sbjct: 285 LYGKSDLLHSM-PPFLGGGEMISDV 308
>At2g13330 F14O4.9
Length = 889
Score = 30.4 bits (67), Expect = 3.9
Identities = 30/109 (27%), Positives = 43/109 (38%), Gaps = 23/109 (21%)
Query: 185 LQEEEPMECNFSGLRFDL----DLAKIIKED-------SSKILGASVCKKGRWLVLIDAA 233
+ E +P C G DL DL +KE+ S+ + G S+ K G+W V ID
Sbjct: 242 IDESDPKRCVGIGHDLDLTVREDLITFLKENKDSFAWSSANLQGISLEKNGKWRVCIDFR 301
Query: 234 KGSATMPPDLSKYP---------VDFVALSFYKLFGYPTGLGALVVRND 273
+ P D P V+ LSF F G +++R D
Sbjct: 302 DLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAF---YGYNQILMRRD 347
>At3g47340 glutamine-dependent asparagine synthetase
Length = 584
Score = 30.0 bits (66), Expect = 5.1
Identities = 22/76 (28%), Positives = 31/76 (39%), Gaps = 6/76 (7%)
Query: 128 MYTMENHNSVLGIREYALGQGAAAIAVDIEDVHPRIEGEKFPTKISLHQEQRRKVTGLQE 187
M+ M LG++ G+GA I H K P K HQE RK+ L +
Sbjct: 324 MFLMSRKIKSLGVKMVLSGEGADEIFGGYLYFH------KAPNKKEFHQETCRKIKALHK 377
Query: 188 EEPMECNFSGLRFDLD 203
+ + N S F L+
Sbjct: 378 YDCLRANKSTSAFGLE 393
>At3g46150 hypothetical protein
Length = 142
Score = 29.6 bits (65), Expect = 6.6
Identities = 20/60 (33%), Positives = 27/60 (44%), Gaps = 4/60 (6%)
Query: 262 PTGLGALVVR----NDAAKLLKKSYFSGGTVAASIADIDFIKRREGIEELFEDGTVSFLS 317
PTG G +V D K++ +F VA + D RRE + LF D T + LS
Sbjct: 18 PTGFGIMVTSLDLLRDHQKVIHTLWFIDDVVARELGGFDGFCRRERVLLLFVDSTNTTLS 77
>At1g47900 mysoin-like protein
Length = 1054
Score = 29.3 bits (64), Expect = 8.6
Identities = 25/130 (19%), Positives = 60/130 (45%), Gaps = 9/130 (6%)
Query: 554 LISSFIDLSQGMLFVESPHCKERLQIRLQLDFYDSAIQ------DIELQGQRYKVYSYDN 607
+I S D S + E+ + +QI ++ + + ++ D++++G +V +Y+N
Sbjct: 19 VIDSAADASHSQIDKEAIKKPKYVQISVEQYTHFTGLEEQIKSYDVQIKGYDVQVKTYEN 78
Query: 608 ETNSWFSKAIERPCTLLRYSGSSHDFVLDRTKDIVTCKDTNSAVSFANEGQFLLVSEESV 667
+ S+ + + + Y H++ K +D N +S ANE +V++E++
Sbjct: 79 QVESYEEQVKDFEEQIDAYDEKVHEYEEQVQKLNEDVEDLNEKLSVANEE---IVTKEAL 135
Query: 668 SDLNKRLCSD 677
+ ++ D
Sbjct: 136 VKQHSKVAED 145
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,836,125
Number of Sequences: 26719
Number of extensions: 718281
Number of successful extensions: 1729
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1703
Number of HSP's gapped (non-prelim): 21
length of query: 764
length of database: 11,318,596
effective HSP length: 107
effective length of query: 657
effective length of database: 8,459,663
effective search space: 5557998591
effective search space used: 5557998591
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC148651.4


