

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148607.16 - phase: 0 /pseudo
(619 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
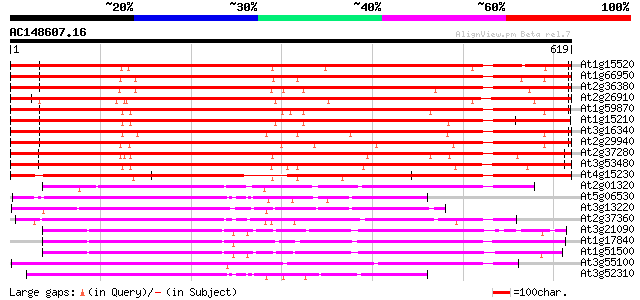
Score E
Sequences producing significant alignments: (bits) Value
At1g15520 hypothetical protein 944 0.0
At1g66950 ABC transporter, putative 837 0.0
At2g36380 putative ABC transporter 829 0.0
At2g26910 putative ABC transporter 808 0.0
At1g59870 putative ABC transporter (At1g59870) 795 0.0
At1g15210 putative ABC transporter 789 0.0
At3g16340 putative ABC transporter 788 0.0
At2g29940 putative ABC transporter 744 0.0
At2g37280 putative ABC transporter 697 0.0
At3g53480 ABC transporter - like protein 696 0.0
At4g15230 ABC transporter like protein 580 e-165
At2g01320 putative ABC transporter 202 3e-52
At5g06530 ABC transporter like protein 190 2e-48
At3g13220 ABC transporter 184 2e-46
At2g37360 ABC transporter like protein 184 2e-46
At3g21090 ABC transporter, putative 181 8e-46
At1g17840 putative ABC transporter (At1g17840) 181 8e-46
At1g51500 ATP-dependent transmembrane transporter, putative 181 1e-45
At3g55100 ABC transporter - like protein 179 3e-45
At3g52310 ABC transporter-like protein 173 2e-43
>At1g15520 hypothetical protein
Length = 1423
Score = 944 bits (2441), Expect = 0.0
Identities = 449/619 (72%), Positives = 530/619 (85%), Gaps = 12/619 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPFEPHSITFD V YSVDMPQEM +G ED+LVLLKG++GAFRPGVLTALMGV+GAG
Sbjct: 816 MVLPFEPHSITFDNVVYSVDMPQEMIEQGTQEDRLVLLKGVNGAFRPGVLTALMGVSGAG 875
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI GNITISGYPK Q+TFARISGYCEQTDIHSPHVTVYESL+YSA
Sbjct: 876 KTTLMDVLAGRKTGGYIDGNITISGYPKNQQTFARISGYCEQTDIHSPHVTVYESLVYSA 935
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
WLRL +++ RK+FIEEVMELVEL PLR ALVGLPG SGLSTEQRKRLT+AVELVANP
Sbjct: 936 WLRLPKEVDKNKRKIFIEEVMELVELTPLRQALVGLPGESGLSTEQRKRLTIAVELVANP 995
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFE+FDEL LLK+GG+
Sbjct: 996 SIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFEAFDELFLLKRGGE 1055
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
EIYVGPLGH S++LINYFE +QG++KI +GYNPATWMLEV+T+S+E L +D+A+VYKNS
Sbjct: 1056 EIYVGPLGHESTHLINYFESIQGINKITEGYNPATWMLEVSTTSQEAALGVDFAQVYKNS 1115
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
ELY+RNK LIKELS PAP SKDLYFP++YS+SF TQC+A LWKQHWSYWRNP Y A+RFL
Sbjct: 1116 ELYKRNKELIKELSQPAPGSKDLYFPTQYSQSFLTQCMASLWKQHWSYWRNPPYTAVRFL 1175
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ +A++ G+MFW+LG K + QDL NAMGSMY AV+ +G N+ SVQPVV VERTVFY
Sbjct: 1176 FTIGIALMFGTMFWDLGGKTKTRQDLSNAMGSMYTAVLFLGLQNAASVQPVVNVERTVFY 1235
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RE+AA MYSA PYA AQV I++PYV VQA+VYG++VY MIGFEWT VK W LFFMY +F
Sbjct: 1236 REQAAGMYSAMPYAFAQVFIEIPYVLVQAIVYGLIVYAMIGFEWTAVKFFWYLFFMYGSF 1295
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
L FTFYGMM+VAMTPN+HI+ +VSSAFY +WNLFSGF++PRP S+PVWW W
Sbjct: 1296 LTFTFYGMMAVAMTPNHHIASVVSSAFYGIWNLFSGFLIPRP----------SMPVWWEW 1345
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVNIAF 600
Y W PVAW+LYGL+ SQ+GD+ + + +D +V+ F+R ++G++ FLGVVA +N+ F
Sbjct: 1346 YYWLCPVAWTLYGLIASQFGDITEPM--ADSNMSVKQFIREFYGYREGFLGVVAAMNVIF 1403
Query: 601 PIVFALVFAIAIKMFNFQR 619
P++FA++FAI IK FNFQ+
Sbjct: 1404 PLLFAVIFAIGIKSFNFQK 1422
Score = 133 bits (335), Expect = 3e-31
Identities = 145/643 (22%), Positives = 284/643 (43%), Gaps = 91/643 (14%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGYIG-GNITISGYPKKQET 92
K +L +SG +PG + L+G +GKTTL+ L+G+ G +T +G+ +
Sbjct: 166 KFTILNDVSGIVKPGRMALLLGPPSSGKTTLLLALAGKLDQELKQTGRVTYNGHGMNEFV 225
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSAW----------------------LRLSPDIN- 129
R + Y Q D+H +TV E+ Y+A ++ PDI+
Sbjct: 226 PQRTAAYIGQNDVHIGEMTVRETFAYAARFQGVGSRYDMLTELARREKEANIKPDPDIDI 285
Query: 130 --------AETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
E + + +++++ L+ +VG + G+S Q+KR+T LV
Sbjct: 286 FMKAMSTAGEKTNVMTDYILKILGLEVCADTMVGDDMLRGISGGQKKRVTTGEMLVGPSR 345
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVDT-GRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ ++ ++RN V T + ++ QP+ + F FD+++L+ + G+
Sbjct: 346 ALFMDEISTGLDSSTTYQIVNSLRNYVHIFNGTALISLLQPAPETFNLFDDIILIAE-GE 404
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVE-----------L 289
IY GP H ++ +FE + + G A ++ EVT+ +++ +
Sbjct: 405 IIYEGPRDH----VVEFFETMGFKCPPRKGV--ADFLQEVTSKKDQMQYWARRDEPYRFI 458
Query: 290 RI-DYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWS- 347
R+ ++AE +++ + RR + EL+ P +K P+ + + I L K +S
Sbjct: 459 RVREFAEAFQSFHVGRR---IGDELALPFDKTKS--HPAALTTKKYGVGIKELVKTSFSR 513
Query: 348 ----YWRNPEYNAIRFLYSTAVAVLLGSMFW--NLGSKIEKDQDLFNAMGSMYAAVILIG 401
RN +F +A L ++F+ + K E D L+ G+++ ++++
Sbjct: 514 EYLLMKRNSFVYYFKFGQLLVMAFLTMTLFFRTEMQKKTEVDGSLYT--GALFFILMML- 570
Query: 402 AMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIG 461
N S + + VFY++R Y A+ Y+L ++++P F++A + + Y +IG
Sbjct: 571 MFNGMSELSMTIAKLPVFYKQRDLLFYPAWVYSLPPWLLKIPISFMEAALTTFITYYVIG 630
Query: 462 FEWTLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYS----VWNLFSGF 517
F+ + + LF Y + M ++IV++ F + V+ G
Sbjct: 631 FDPNVGR----LFKQYILLVLMNQMASALFKMVAALGRNMIVANTFGAFAMLVFFALGGV 686
Query: 518 VLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTV-E 576
VL R I WW W W +P+ + ++ +++ + + +T+
Sbjct: 687 VLSRD----------DIKKWWIWGYWISPIMYGQNAILANEFFGHSWSRAVENSSETLGV 736
Query: 577 DFLRNYFGFKHDF---LGVVALVNIAFPIVFALVFAIAIKMFN 616
FL++ H + +G AL + F ++F F +A+ N
Sbjct: 737 TFLKSRGFLPHAYWYWIGTGAL--LGFVVLFNFGFTLALTFLN 777
>At1g66950 ABC transporter, putative
Length = 1434
Score = 837 bits (2161), Expect = 0.0
Identities = 399/620 (64%), Positives = 496/620 (79%), Gaps = 11/620 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF+P S+ F+ V+Y VDMP EM+ +GV D+L LL+ + GAFRPG+LTAL+GV+GAG
Sbjct: 824 MVLPFQPLSLAFNNVNYYVDMPSEMKAQGVEGDRLQLLRDVGGAFRPGILTALVGVSGAG 883
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI G+I+ISGYPK Q TFAR+SGYCEQ DIHSPHVTVYESL+YSA
Sbjct: 884 KTTLMDVLAGRKTGGYIEGSISISGYPKNQTTFARVSGYCEQNDIHSPHVTVYESLIYSA 943
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
WLRLS DI+ +TR++F+EEVMELVELKPLR ++VGLPGV GLSTEQRKRLT+AVELVANP
Sbjct: 944 WLRLSTDIDIKTRELFVEEVMELVELKPLRNSIVGLPGVDGLSTEQRKRLTIAVELVANP 1003
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFESFDELLL+K+GGQ
Sbjct: 1004 SIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFESFDELLLMKRGGQ 1063
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY G LGH+S L+ YFE V+GV KI DGYNPATWML+VTT S E ++ +D+A+++ NS
Sbjct: 1064 VIYAGSLGHHSQKLVEYFEAVEGVPKINDGYNPATWMLDVTTPSMESQMSLDFAQIFSNS 1123
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
LYRRN+ LIK+LS P P SKD+YF ++Y++SF TQ AC WKQ+WSYWR+P+YNAIRFL
Sbjct: 1124 SLYRRNQELIKDLSTPPPGSKDVYFKTKYAQSFSTQTKACFWKQYWSYWRHPQYNAIRFL 1183
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
+ + VL G +FW +G+K E +QDL N G+MYAAV+ +GA+N+ +VQP + +ERTVFY
Sbjct: 1184 MTVVIGVLFGLIFWQIGTKTENEQDLNNFFGAMYAAVLFLGALNAATVQPAIAIERTVFY 1243
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RE+AA MYSA PYA++QV +++ Y +Q VY +++Y MIG WT+ K +W ++M +F
Sbjct: 1244 REKAAGMYSAIPYAISQVAVEIMYNTIQTGVYTLILYSMIGCNWTMAKFLWFYYYMLTSF 1303
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
+YFT YGMM +A+TPN I+ I S F S+WNLFSGF++PRP IP+WWRW
Sbjct: 1304 IYFTLYGMMLMALTPNYQIAGICMSFFLSLWNLFSGFLIPRP----------QIPIWWRW 1353
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSD-GRQTVEDFLRNYFGFKHDFLGVVALVNIA 599
Y WA PVAW+LYGL+TSQ GD + S G ++ L+ FGF+HDFL VVA+V+IA
Sbjct: 1354 YYWATPVAWTLYGLITSQVGDKDSMVHISGIGDIDLKTLLKEGFGFEHDFLPVVAVVHIA 1413
Query: 600 FPIVFALVFAIAIKMFNFQR 619
+ ++F VFA IK NFQR
Sbjct: 1414 WILLFLFVFAYGIKFLNFQR 1433
Score = 113 bits (282), Expect = 4e-25
Identities = 135/636 (21%), Positives = 264/636 (41%), Gaps = 94/636 (14%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
K+ +LK +SG +P +T L+G +GKTTL+ L+G+ + G IT G+ ++
Sbjct: 187 KIQILKDISGIVKPSRMTLLLGPPSSGKTTLLQALAGKLDDTLQMSGRITYCGHEFREFV 246
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSA----------------------WLRLSPDINA 130
+ Y Q D+H +TV E L +S ++ P I+A
Sbjct: 247 PQKTCAYISQHDLHFGEMTVREILDFSGRCLGVGSRYQLMSELSRREKEEGIKPDPKIDA 306
Query: 131 ETRKMFI---------EEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
+ + I + V++++ L L G G+S Q+KRLT LV
Sbjct: 307 FMKSIAISGQETSLVTDYVLKILGLDICADILAGDVMRRGISGGQKKRLTTGEMLVGPAR 366
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ + + +R V + T++ ++ QP+ + FE FD+++LL + GQ
Sbjct: 367 ALFMDEISTGLDSSTTFQICKFMRQLVHISDVTMIISLLQPAPETFELFDDIILLSE-GQ 425
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVEL---------RI 291
+Y GP N++ +FE + G A ++ EVT+ + + +
Sbjct: 426 IVYQGP----RDNVLEFFEYFGFQCPERKGV--ADFLQEVTSKKDQEQYWNKREQPYNYV 479
Query: 292 DYAEVYKNSELYRRNKALIKELSAP---APCSKDLYFPSRYSRSFFTQCIACLWKQHWSY 348
++ + + L E P A +Y S + AC ++
Sbjct: 480 SVSDFSSGFSTFHTGQKLTSEFRVPYDKAKTHSAALVTQKYGISNWELFKACFDREWLLM 539
Query: 349 WRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSV 408
RN + + T ++++ +++ + +D G+M+ + LI M +
Sbjct: 540 KRNSFVYVFKTVQITIMSLITMTVYLRTEMHVGTVRDGQKFYGAMFFS--LINVMFNGLA 597
Query: 409 QPVVGVER-TVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLV 467
+ V R VFY++R Y + +AL ++++P +++ ++ + Y IGF +
Sbjct: 598 ELAFTVMRLPVFYKQRDFLFYPPWAFALPAWLLKIPLSLIESGIWIGLTYYTIGFAPSAA 657
Query: 468 KVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**L 527
+ F G + +N I +++ GF++ +
Sbjct: 658 R----------------FLGAIGRTEVISNSIGTFTLLIVFTL----GGFIIAKD----- 692
Query: 528 ISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVK---QNIETSDGRQTVEDFLRNYFG 584
I W W + +P+ + +V +++ D + N +T +TV + L G
Sbjct: 693 -----DIRPWMTWAYYMSPMMYGQTAIVMNEFLDERWSSPNYDTRINAKTVGEVLLKSRG 747
Query: 585 FKHD----FLGVVALVNIAFPIVFALVFAIAIKMFN 616
F + ++ +VAL + F ++F L + +A+ N
Sbjct: 748 FFTEPYWFWICIVAL--LGFSLLFNLFYILALMYLN 781
>At2g36380 putative ABC transporter
Length = 1450
Score = 829 bits (2141), Expect = 0.0
Identities = 399/620 (64%), Positives = 491/620 (78%), Gaps = 11/620 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF+P S+ F+ V+Y VDMP EM+ +GV D+L LL+ + GAFRPGVLTAL+GV+GAG
Sbjct: 840 MVLPFQPLSLAFNNVNYYVDMPAEMKAQGVEGDRLQLLRDVGGAFRPGVLTALVGVSGAG 899
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGY+ G+I ISGYPK Q TFAR+SGYCEQ DIHSPHVTVYESL+YSA
Sbjct: 900 KTTLMDVLAGRKTGGYVEGSINISGYPKNQATFARVSGYCEQNDIHSPHVTVYESLIYSA 959
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
WLRLS DI+ +TR+MF+EEVMELVELKPLR ++VGLPGV GLSTEQRKRLT+AVELVANP
Sbjct: 960 WLRLSADIDTKTREMFVEEVMELVELKPLRNSIVGLPGVDGLSTEQRKRLTIAVELVANP 1019
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFESFDELLL+K+GGQ
Sbjct: 1020 SIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFESFDELLLMKRGGQ 1079
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY G LGH+S L+ YFE ++GV KIKDGYNPATWML+VTT S E ++ +D+A+++ NS
Sbjct: 1080 VIYAGTLGHHSQKLVEYFEAIEGVPKIKDGYNPATWMLDVTTPSMESQMSVDFAQIFVNS 1139
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
+ RRN+ LIKELS P P S DLYF ++Y++ F TQ AC WK +WS WR P+YNAIRFL
Sbjct: 1140 SVNRRNQELIKELSTPPPGSNDLYFRTKYAQPFSTQTKACFWKMYWSNWRYPQYNAIRFL 1199
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
+ + VL G +FW G+KIEK+QDL N G+MYAAV+ +GA N+ +VQP V +ERTVFY
Sbjct: 1200 MTVVIGVLFGLLFWQTGTKIEKEQDLNNFFGAMYAAVLFLGATNAATVQPAVAIERTVFY 1259
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RE+AA MYSA PYA++QV +++ Y +Q VY +++Y MIG++WT+VK W ++M F
Sbjct: 1260 REKAAGMYSAIPYAISQVAVEIMYNTIQTGVYTLILYSMIGYDWTVVKFFWFYYYMLTCF 1319
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
+YFT YGMM VA+TPN I+ I S F S WNLFSGF++PRP IP+WWRW
Sbjct: 1320 VYFTLYGMMLVALTPNYQIAGICLSFFLSFWNLFSGFLIPRP----------QIPIWWRW 1369
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIE-TSDGRQTVEDFLRNYFGFKHDFLGVVALVNIA 599
Y WA+PVAW+LYG++TSQ GD + T G +++ L+N FGF +DFL VVA+V+IA
Sbjct: 1370 YYWASPVAWTLYGIITSQVGDRDSIVHITGVGDMSLKTLLKNGFGFDYDFLPVVAVVHIA 1429
Query: 600 FPIVFALVFAIAIKMFNFQR 619
+ ++F FA IK NFQR
Sbjct: 1430 WILIFLFAFAYGIKFLNFQR 1449
Score = 127 bits (320), Expect = 1e-29
Identities = 145/641 (22%), Positives = 274/641 (42%), Gaps = 84/641 (13%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
K+ +LK +SG +P +T L+G +GKTTL+ L+G+ + G IT G+ ++
Sbjct: 185 KIEILKDISGIIKPSRMTLLLGPPSSGKTTLLQALAGKLDDTLQMSGRITYCGHEFREFV 244
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYS----------------------AWLRLSPDINA 130
+ Y Q D+H +TV ESL +S A ++ P+I+A
Sbjct: 245 PQKTCAYISQHDLHFGEMTVRESLDFSGRCLGVGTRYQLLTELSRREREAGIKPDPEIDA 304
Query: 131 ETRKMFI---------EEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
+ + I + V++L+ L LVG G+S QRKRLT LV +
Sbjct: 305 FMKSIAISGQETSLVTDYVLKLLGLDICADTLVGDVMRRGISGGQRKRLTTGEMLVGPAT 364
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVDTGR-TVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ + + +R V T+V ++ QP+ + FE FD+++LL + GQ
Sbjct: 365 ALFMDEISTGLDSSTTFQICKFMRQLVHIADVTMVISLLQPAPETFELFDDIILLSE-GQ 423
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEV------ELRIDYA 294
+Y G + N++ +FE + K + A ++ EVT+ + E Y
Sbjct: 424 IVYQG----SRDNVLEFFEYMG--FKCPERKGIADFLQEVTSKKDQEQYWNRREHPYSYV 477
Query: 295 EVYKNS---ELYRRNKALIKELSAPAPCSKD---LYFPSRYSRSFFTQCIACLWKQHWSY 348
V+ S + + L E P +K +Y S AC ++
Sbjct: 478 SVHDFSSGFNSFHAGQQLASEFRVPYDKAKTHPAALVTQKYGISNKDLFKACFDREWLLM 537
Query: 349 WRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSV 408
RN + + T ++++ ++++ + QD G+++ + LI M +
Sbjct: 538 KRNSFVYVFKTVQITIMSLIAMTVYFRTEMHVGTVQDGQKFYGALFFS--LINLMFNGMA 595
Query: 409 QPVVGVER-TVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLV 467
+ V R VF+++R Y + +AL ++++P +++V++ + Y IGF +
Sbjct: 596 ELAFTVMRLPVFFKQRDFLFYPPWAFALPGFLLKIPLSLIESVIWIALTYYTIGFAPSAA 655
Query: 468 K------VVWCLFFMYFT-FLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLP 520
+ +C+ M + F + G V ++++V + GF++
Sbjct: 656 RFFRQLLAYFCVNQMALSLFRFLGALGRTEVIANSGGTLALLVVF-------VLGGFIIS 708
Query: 521 RPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGR---QTVED 577
+ IP W W + +P+ + LV +++ D + +D R +TV +
Sbjct: 709 KD----------DIPSWLTWCYYTSPMMYGQTALVINEFLDERWGSPNNDTRINAKTVGE 758
Query: 578 FLRNYFGFKHD--FLGVVALVNIAFPIVFALVFAIAIKMFN 616
L GF + + + + F ++F + IA+ N
Sbjct: 759 VLLKSRGFFTEPYWFWICIGALLGFTVLFNFCYIIALMYLN 799
>At2g26910 putative ABC transporter
Length = 1420
Score = 808 bits (2087), Expect = 0.0
Identities = 383/621 (61%), Positives = 493/621 (78%), Gaps = 12/621 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF+P S++F ++Y VD+P ++ +G++ED+L LL ++GAFRPGVLTAL+GV+GAG
Sbjct: 809 MVLPFQPLSLSFSNINYYVDVPLGLKEQGILEDRLQLLVNITGAFRPGVLTALVGVSGAG 868
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGG I G++ ISG+PK+QETFARISGYCEQ D+HSP +TV ESLL+SA
Sbjct: 869 KTTLMDVLAGRKTGGTIEGDVYISGFPKRQETFARISGYCEQNDVHSPCLTVVESLLFSA 928
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
LRL DI++ET++ F+ EVMELVEL L ALVGLPGV GLSTEQRKRLT+AVELVANP
Sbjct: 929 CLRLPADIDSETQRAFVHEVMELVELTSLSGALVGLPGVDGLSTEQRKRLTIAVELVANP 988
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SI+FMDEPTSGLDARAAAIVMR VRN V+TGRT+VCTIHQPSIDIFESFDELL +K+GG+
Sbjct: 989 SIVFMDEPTSGLDARAAAIVMRTVRNIVNTGRTIVCTIHQPSIDIFESFDELLFMKRGGE 1048
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG S LI YFE ++GV KIK G+NPA WML+VT S++E L +D+AE+Y+NS
Sbjct: 1049 LIYAGPLGQKSCELIKYFESIEGVQKIKPGHNPAAWMLDVTASTEEHRLGVDFAEIYRNS 1108
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
L +RNK LI+ LS P+ +K++ FP+RYS+S ++Q +ACLWKQ+ SYWRNP+Y A+RF
Sbjct: 1109 NLCQRNKELIEVLSKPSNIAKEIEFPTRYSQSLYSQFVACLWKQNLSYWRNPQYTAVRFF 1168
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
Y+ ++++LG++ W GSK + Q LFNAMGSMYAAV+ IG N+ + QPVV +ER V Y
Sbjct: 1169 YTVVISLMLGTICWKFGSKRDTQQQLFNAMGSMYAAVLFIGITNATAAQPVVSIERFVSY 1228
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RERAA MYSA P+A AQV I+ PYV Q+ +Y + Y M FEW+ VK +W LFFMYF+
Sbjct: 1229 RERAAGMYSALPFAFAQVFIEFPYVLAQSTIYSTIFYAMAAFEWSAVKFLWYLFFMYFSI 1288
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
+YFTFYGMM+ A+TPN++++ I+++ FY +WNLFSGF++P IP+WWRW
Sbjct: 1289 MYFTFYGMMTTAITPNHNVASIIAAPFYMLWNLFSGFMIP----------YKRIPLWWRW 1338
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDG--RQTVEDFLRNYFGFKHDFLGVVALVNI 598
Y WANPVAW+LYGL+ SQYGD +++++ SDG + V+ L + G+KHDFLGV A++ +
Sbjct: 1339 YYWANPVAWTLYGLLVSQYGDDERSVKLSDGIHQVMVKQLLEDVMGYKHDFLGVSAIMVV 1398
Query: 599 AFPIVFALVFAIAIKMFNFQR 619
AF + F+LVFA AIK FNFQR
Sbjct: 1399 AFCVFFSLVFAFAIKAFNFQR 1419
Score = 152 bits (385), Expect = 4e-37
Identities = 166/660 (25%), Positives = 288/660 (43%), Gaps = 103/660 (15%)
Query: 25 MRNRGVI---EDKLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRK-TGGYIGGN 80
+RN VI +KL +L G+SG RP LT L+G +GKTTL+ L+GR T G
Sbjct: 135 LRNIHVIGGKRNKLTILDGISGVIRPSRLTLLLGPPSSGKTTLLLALAGRLGTNLQTSGK 194
Query: 81 ITISGYPKKQETFARISGYCEQTDIHSPHVTVYESL-----------LYSAWLRLS---- 125
IT +GY K+ R S Y Q D H +TV ++L Y L L+
Sbjct: 195 ITYNGYDLKEIIAPRTSAYVSQQDWHVAEMTVRQTLEFAGRCQGVGFKYDMLLELARREK 254
Query: 126 -----PD-----------INAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKR 169
PD + + +E VM+++ L LVG + G+S Q+KR
Sbjct: 255 LAGIVPDEDLDIFMKSLALGGMETSLVVEYVMKILGLDTCADTLVGDEMIKGISGGQKKR 314
Query: 170 LTVAVELVANPSIIFMDEPTSGLDARAA-AIVMRAVRNTVDTGRTVVCTIHQPSIDIFES 228
LT LV ++FMDE ++GLD+ I+M +T T V ++ QPS + +E
Sbjct: 315 LTTGELLVGPARVLFMDEISNGLDSSTTHQIIMYMRHSTHALEGTTVISLLQPSPETYEL 374
Query: 229 FDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVE 288
FD+++L+ + GQ IY GP ++++F + D N A ++ EVT+ + +
Sbjct: 375 FDDVILMSE-GQIIYQGP----RDEVLDFFSSLG--FTCPDRKNVADFLQEVTSKKDQQQ 427
Query: 289 LRI------------DYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQ 336
+AE +++ Y K L K+L P K + S S +
Sbjct: 428 YWSVPFRPYRYVPPGKFAEAFRS---YPTGKKLAKKLE--VPFDKRFNHSAALSTSQYGV 482
Query: 337 CIACLWKQHWSYWR-----NPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMG 391
+ L K ++++ + N +F+ VA++ ++F D +G
Sbjct: 483 KKSELLKINFAWQKQLMKQNAFIYVFKFVQLLLVALITMTVFCRTTMHHNTIDDGNIYLG 542
Query: 392 SMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVV 451
S+Y ++++I N + P++ + V Y+ R Y ++ Y L ++ +P +++
Sbjct: 543 SLYFSMVII-LFNGFTEVPMLVAKLPVLYKHRDLHFYPSWAYTLPSWLLSIPTSIIESAT 601
Query: 452 YGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYS-- 509
+ V Y IG++ L F +YF+ L+ G+ V + H +IV++ F S
Sbjct: 602 WVAVTYYTIGYD-PLFSRFLQQFLLYFS-LHQMSLGLFRVMGSLGRH--MIVANTFGSFA 657
Query: 510 --VWNLFSGFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIE 567
V GF++ R SIP WW W W +P+ ++ +++ + N +
Sbjct: 658 MLVVMTLGGFIISRD----------SIPSWWIWGYWISPLMYAQNAASVNEF--LGHNWQ 705
Query: 568 TSDGRQTVED-----------FLRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAIKMFN 616
+ G T + F NY+ ++GV AL + + ++F ++F + + N
Sbjct: 706 KTAGNHTSDSLGLALLKERSLFSGNYW----YWIGVAAL--LGYTVLFNILFTLFLAHLN 759
>At1g59870 putative ABC transporter (At1g59870)
Length = 1469
Score = 795 bits (2054), Expect = 0.0
Identities = 377/620 (60%), Positives = 488/620 (77%), Gaps = 12/620 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF P +++FD+V Y VDMP EMR++GV E +L LLKG++GAFRPGVLTALMGV+GAG
Sbjct: 858 MVLPFTPLAMSFDDVKYFVDMPGEMRDQGVTETRLQLLKGVTGAFRPGVLTALMGVSGAG 917
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI G++ ISG+PK QETFARISGYCEQTDIHSP VTV ESL++SA
Sbjct: 918 KTTLMDVLAGRKTGGYIEGDVRISGFPKVQETFARISGYCEQTDIHSPQVTVRESLIFSA 977
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
+LRL ++ + + MF+++VMELVEL LR ++VGLPGV+GLSTEQRKRLT+AVELVANP
Sbjct: 978 FLRLPKEVGKDEKMMFVDQVMELVELDSLRDSIVGLPGVTGLSTEQRKRLTIAVELVANP 1037
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFE+FDEL+L+K+GGQ
Sbjct: 1038 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDELMLMKRGGQ 1097
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG NS ++ YFE GVSKI + YNPATWMLE ++ + E++L +D+AE+Y S
Sbjct: 1098 VIYAGPLGQNSHKVVEYFESFPGVSKIPEKYNPATWMLEASSLAAELKLSVDFAELYNQS 1157
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
L++RNKAL+KELS P + DLYF +++S++ + Q +CLWKQ W+YWR+P+YN +RF+
Sbjct: 1158 ALHQRNKALVKELSVPPAGASDLYFATQFSQNTWGQFKSCLWKQWWTYWRSPDYNLVRFI 1217
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ A ++L+G++FW +G DL +G++YAA+I +G N ++VQP+V VERTVFY
Sbjct: 1218 FTLATSLLIGTVFWQIGGNRSNAGDLTMVIGALYAAIIFVGINNCSTVQPMVAVERTVFY 1277
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RERAA MYSA PYA++QV +LPYV +Q V Y ++VY M+GFEW K W +F YF+F
Sbjct: 1278 RERAAGMYSAMPYAISQVTCELPYVLIQTVYYSLIVYAMVGFEWKAEKFFWFVFVSYFSF 1337
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
LY+T+YGMM+V++TPN ++ I +SAFY ++NLFSGF +PRP IP WW W
Sbjct: 1338 LYWTYYGMMTVSLTPNQQVASIFASAFYGIFNLFSGFFIPRP----------KIPKWWIW 1387
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQ--TVEDFLRNYFGFKHDFLGVVALVNI 598
Y W PVAW++YGL+ SQYGDV+ I+ G TV+ ++ +++GF+ DF+G VA V I
Sbjct: 1388 YYWICPVAWTVYGLIVSQYGDVETRIQVLGGAPDLTVKQYIEDHYGFQSDFMGPVAAVLI 1447
Query: 599 AFPIVFALVFAIAIKMFNFQ 618
AF + FA +FA I+ NFQ
Sbjct: 1448 AFTVFFAFIFAFCIRTLNFQ 1467
Score = 143 bits (360), Expect = 3e-34
Identities = 147/645 (22%), Positives = 284/645 (43%), Gaps = 93/645 (14%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
+L +LK +SG +PG +T L+G +GKTTL+ L+G+ + G+IT +GY +
Sbjct: 183 QLTILKDISGVIKPGRMTLLLGPPSSGKTTLLLALAGKLDKSLQVSGDITYNGYQLDEFV 242
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSAWLR--------------------LSPDINAET 132
+ S Y Q D+H +TV E+L +SA + + P+ + +
Sbjct: 243 PRKTSAYISQNDLHVGIMTVKETLDFSARCQGVGTRYDLLNELARREKDAGIFPEADVDL 302
Query: 133 -----------RKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
+ + ++++ L + +VG + G+S Q+KR+T +V
Sbjct: 303 FMKASAAQGVKNSLVTDYTLKILGLDICKDTIVGDDMMRGISGGQKKRVTTGEMIVGPTK 362
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVDTGR-TVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ +++ ++ V TV+ ++ QP+ + F+ FD+++L+ + GQ
Sbjct: 363 TLFMDEISTGLDSSTTFQIVKCLQQIVHLNEATVLMSLLQPAPETFDLFDDIILVSE-GQ 421
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKN- 299
+Y GP N++ +FE K + A ++ EVT+ + + ++ Y
Sbjct: 422 IVYQGP----RDNILEFFESFG--FKCPERKGTADFLQEVTSKKDQEQYWVNPNRPYHYI 475
Query: 300 --SELYRRNKA------LIKELSAPAPCSKD----LYFPSRYSRSFFTQCIACLWKQHWS 347
SE R K+ + EL+ P S+ L F +YS S +C W + W
Sbjct: 476 PVSEFASRYKSFHVGTKMSNELAVPFDKSRGHKAALVF-DKYSVSKRELLKSC-WDKEWL 533
Query: 348 YW-RNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSN 406
RN + + + +A + ++F + D +G++ +I I N
Sbjct: 534 LMQRNAFFYVFKTVQIVIIAAITSTLFLRTEMNTRNEGDANLYIGALLFGMI-INMFNGF 592
Query: 407 SVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGF---- 462
+ ++ VFY++R Y ++ ++L ++ +P +++ + +V Y IGF
Sbjct: 593 AEMAMMVSRLPVFYKQRDLLFYPSWTFSLPTFLLGIPSSILESTAWMVVTYYSIGFAPDA 652
Query: 463 -----EWTLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGF 517
++ LV ++ + F + MM +A T ++V L GF
Sbjct: 653 SRFFKQFLLVFLIQQMAASLFRLIASVCRTMM-IANTGGALTLLLVF--------LLGGF 703
Query: 518 VLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVE- 576
+LP+ GK IP WW W W +P+ ++ GLV ++ + + + T++
Sbjct: 704 LLPK-GK---------IPDWWGWAYWVSPLTYAFNGLVVNEMFAPRWMNKMASSNSTIKL 753
Query: 577 -DFLRNYFGFKHD----FLGVVALVNIAFPIVFALVFAIAIKMFN 616
+ N + H ++ V AL + F +F ++F +A+ N
Sbjct: 754 GTMVLNTWDVYHQKNWYWISVGAL--LCFTALFNILFTLALTYLN 796
>At1g15210 putative ABC transporter
Length = 1442
Score = 789 bits (2037), Expect = 0.0
Identities = 375/620 (60%), Positives = 488/620 (78%), Gaps = 12/620 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF P +++FD+V Y VDMP EMR +GV E +L LLKG++ AFRPGVLTALMGV+GAG
Sbjct: 831 MVLPFTPLAMSFDDVKYFVDMPAEMREQGVQETRLQLLKGVTSAFRPGVLTALMGVSGAG 890
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI G++ +SG+PKKQETFARISGYCEQTDIHSP VTV ESL++SA
Sbjct: 891 KTTLMDVLAGRKTGGYIEGDVRVSGFPKKQETFARISGYCEQTDIHSPQVTVRESLIFSA 950
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
+LRL+ +++ E + MF+++VMELVEL LR A+VGLPGV+GLSTEQRKRLT+AVELVANP
Sbjct: 951 FLRLAKEVSKEDKLMFVDQVMELVELVDLRDAIVGLPGVTGLSTEQRKRLTIAVELVANP 1010
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFE+FDELLL+K+GG
Sbjct: 1011 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFEAFDELLLMKRGGH 1070
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG NS ++ YFE GV KI + YNPATWMLE ++ + E++L +D+AE+YK S
Sbjct: 1071 VIYSGPLGRNSHKVVEYFESFPGVPKIPEKYNPATWMLEASSLAAELKLGVDFAELYKAS 1130
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
L +RNKAL++ELS P + DLYF +++S++ + Q +CLWKQ W+YWR+P+YN +RF+
Sbjct: 1131 ALCQRNKALVQELSVPPQGATDLYFATQFSQNTWGQFKSCLWKQWWTYWRSPDYNLVRFI 1190
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ A ++++GS+FW +G K QDL +G++YAAV+ +G N ++VQP+V VERTVFY
Sbjct: 1191 FTLATSLMIGSVFWQIGGKRSNVQDLTMVIGAIYAAVVFVGINNCSTVQPMVAVERTVFY 1250
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RE+AA MYSA PYA++QV +LPYV +Q Y +++Y M+GFEW K +W +F YF+F
Sbjct: 1251 REKAAGMYSAIPYAISQVTCELPYVLIQTTYYSLIIYSMVGFEWKASKFLWFIFINYFSF 1310
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
LY+T+YGMM+V++TPN ++ I +SAFY ++NLFSGF +PRP IP WW W
Sbjct: 1311 LYWTYYGMMTVSLTPNQQVASIFASAFYGIFNLFSGFFIPRP----------KIPKWWVW 1360
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQ--TVEDFLRNYFGFKHDFLGVVALVNI 598
Y W PVAW++YGL+TSQYGDV+ I G TV+ ++++ +GF+ D++G VA V +
Sbjct: 1361 YYWICPVAWTIYGLITSQYGDVETPIALLGGAPGLTVKQYIKDQYGFESDYMGPVAGVLV 1420
Query: 599 AFPIVFALVFAIAIKMFNFQ 618
F + FA +FA IK NFQ
Sbjct: 1421 GFTVFFAFIFAFCIKTLNFQ 1440
Score = 146 bits (368), Expect = 4e-35
Identities = 133/581 (22%), Positives = 250/581 (42%), Gaps = 85/581 (14%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
+L +LK +SG +P +T L+G +GKTTL+ L+G+ + G +T +GY +
Sbjct: 181 QLTILKDVSGIVKPSRMTLLLGPPSSGKTTLLLALAGKLDKSLDVSGEVTYNGYRLNEFV 240
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSAWLR--------------------LSPDINAET 132
+ S Y Q D+H +TV E+L +SA + + P+ + +
Sbjct: 241 PIKTSAYISQNDLHVGIMTVKETLDFSARCQGVGTRYDLLNELARREKDAGIFPEADVDL 300
Query: 133 -----------RKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
+ + ++++ L + +VG + G+S Q+KR+T +V
Sbjct: 301 FMKASAAQGVKSSLITDYTLKILGLDICKDTIVGDDMMRGISGGQKKRVTTGEMIVGPTK 360
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ +++ ++ V T TV+ ++ QP+ + F+ FD+++LL + GQ
Sbjct: 361 TLFMDEISTGLDSSTTFQIVKCLQQIVHLTEATVLISLLQPAPETFDLFDDIILLSE-GQ 419
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRID-------- 292
+Y GP H ++ +FE K + A ++ EVT+ + + +D
Sbjct: 420 IVYQGPRDH----ILEFFESFG--FKCPERKGTADFLQEVTSKKDQEQYWVDPNRPYRYI 473
Query: 293 -YAEVYKNSELYRRNKALIKELSAPAPCSKD----LYFPSRYSRSFFTQCIACLWKQHWS 347
+E + + + L ELS P SK L F +YS T+ + W + W
Sbjct: 474 PVSEFASSFKKFHVGSKLSNELSVPYDKSKSHKAALMF-DKYSIK-KTELLKSCWDKEWM 531
Query: 348 YW-RNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSN 406
RN + + + +A + +++ + D +GS+ A+I + N
Sbjct: 532 LMKRNSFFYVFKTVQIIIIAAITSTLYLRTEMHTRNEIDANIYVGSLLFAMI-VNMFNGL 590
Query: 407 SVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTL 466
+ + VFY++R + + Y L ++ +P ++ + +V Y IG+
Sbjct: 591 AEMAMTIQRLPVFYKQRDLLFHPPWTYTLPTFLLGIPISIFESTAWMVVTYYSIGYAPDA 650
Query: 467 VKVVWCLFFMYFTFLY---------FTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGF 517
+ FF F ++ F F MT N ++V + L GF
Sbjct: 651 ER-----FFKQFLIIFLIQQMAAGIFRFIASTCRTMTIANTGGVLVLLVVF----LTGGF 701
Query: 518 VLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQ 558
+LPR IPVWWRW W +P++++ + ++
Sbjct: 702 LLPRS----------EIPVWWRWAYWISPLSYAFNAITVNE 732
>At3g16340 putative ABC transporter
Length = 1416
Score = 788 bits (2034), Expect = 0.0
Identities = 379/621 (61%), Positives = 480/621 (77%), Gaps = 12/621 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF P +++FD V+Y VDMP+EM+ +GV +DKL LLK ++G FRPGVLTALMGV+GAG
Sbjct: 805 MVLPFTPLTMSFDNVNYYVDMPKEMKEQGVSKDKLQLLKEVTGVFRPGVLTALMGVSGAG 864
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGYI G+I ISG+PK+QETFARISGYCEQ DIHSP VTV ESL+YSA
Sbjct: 865 KTTLMDVLAGRKTGGYIEGDIRISGFPKRQETFARISGYCEQNDIHSPQVTVKESLIYSA 924
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
+LRL ++ + F++EVMELVEL+ L+ A+VGLPG++GLSTEQRKRLT+AVELVANP
Sbjct: 925 FLRLPKEVTKYEKMRFVDEVMELVELESLKDAVVGLPGITGLSTEQRKRLTIAVELVANP 984
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFE+FDELLLLK+GGQ
Sbjct: 985 SIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFEAFDELLLLKRGGQ 1044
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG NS +I YF+ + GV KIK+ YNPATWMLEV++ + E +L ID+AE YK S
Sbjct: 1045 VIYAGPLGQNSHKIIEYFQAIHGVPKIKEKYNPATWMLEVSSMAAEAKLEIDFAEHYKTS 1104
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
LY++NK L+KELS P + DLYF +R+S+S Q +CLWKQ +YWR P+YN RF
Sbjct: 1105 SLYQQNKNLVKELSTPPQGASDLYFSTRFSQSLLGQFKSCLWKQWITYWRTPDYNLARFF 1164
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ A AV+LGS+FW +G+K E DL +G+MYAAV+ +G NS+SVQP++ VER+VFY
Sbjct: 1165 FTLAAAVMLGSIFWKVGTKRENANDLTKVIGAMYAAVLFVGVNNSSSVQPLIAVERSVFY 1224
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RERAA MYSA PYALAQVV ++PYV +Q Y +++Y M+ FEWTL K W F + +F
Sbjct: 1225 RERAAEMYSALPYALAQVVCEIPYVLIQTTYYTLIIYAMMCFEWTLAKFFWFYFVSFMSF 1284
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
LYFT+YGMM+VA+TPN ++ + + AFY ++NLFSGFV+PRP IP WW W
Sbjct: 1285 LYFTYYGMMTVALTPNQQVAAVFAGAFYGLFNLFSGFVIPRP----------RIPKWWIW 1334
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSD--GRQTVEDFLRNYFGFKHDFLGVVALVNI 598
Y W PVAW++YGL+ SQYGDV+ I+ T++ ++ N++G+ DF+ +A V +
Sbjct: 1335 YYWICPVAWTVYGLIVSQYGDVEDTIKVPGMANDPTIKWYIENHYGYDADFMIPIATVLV 1394
Query: 599 AFPIVFALVFAIAIKMFNFQR 619
F + FA +FA I+ NFQ+
Sbjct: 1395 GFTLFFAFMFAFGIRTLNFQQ 1415
Score = 137 bits (346), Expect = 1e-32
Identities = 148/643 (23%), Positives = 275/643 (42%), Gaps = 90/643 (13%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
K+ +L+ +SG +P +T L+G +GKTTL+ L+G+ + G +T +G+ ++
Sbjct: 159 KVTILRDVSGIIKPSRMTLLLGPPSSGKTTLLLALAGKLDQSLKVTGRVTYNGHGLEEFV 218
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSAWLR--------LSPDINAETRKMFIEE----- 139
+ S Y Q D+H +TV E+L +SA + LS + E + E
Sbjct: 219 PQKTSAYISQNDVHVGVMTVQETLDFSARCQGVGTRYDLLSELVRREKDAGILPEPEVDL 278
Query: 140 ------------------VMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPS 181
+ ++ L + +VG + G+S Q+KR+T +V
Sbjct: 279 FMKSIAAGNVKSSLITDYTLRILGLDICKDTVVGDEMIRGISGGQKKRVTTGEMIVGPTK 338
Query: 182 IIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
+FMDE ++GLD+ +++ ++ V T TV+ ++ QP+ + FE FD+++LL + GQ
Sbjct: 339 TLFMDEISTGLDSSTTYQIVKCLQEIVRFTDATVLMSLLQPAPETFELFDDIILLSE-GQ 397
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTT---------SSKEVELRI 291
+Y GP H ++ +FE K D A ++ EVT+ SK+ I
Sbjct: 398 IVYQGPRDH----VLTFFETCG--FKCPDRKGTADFLQEVTSRKDQEQYWADSKKPYSYI 451
Query: 292 DYAEVYKNSELYRRNKALIKELSAP------APCSKDLYFPSRYSRSFFTQCIACLWKQH 345
+E K + L K+LS P P S S F C W +
Sbjct: 452 SVSEFSKRFRTFHVGANLEKDLSVPYDRFKSHPASLVFKKHSVPKSQLFKVC----WDRE 507
Query: 346 WSYWRNPEYNAIRFLYSTAVAVLLGSMFW---NLGSKIEKDQDLFNAMGSMYAAVILIGA 402
+ + I + L+ S + +G+K E D ++ +G++ ++I +
Sbjct: 508 LLLMKRNAFFYITKTVQIIIMALIASTVYLRTEMGTKNESDGAVY--IGALMFSMI-VNM 564
Query: 403 MNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGF 462
N + ++ VFY++R + + ++L ++ +P ++VV+ + Y MIGF
Sbjct: 565 FNGFAELALMIQRLPVFYKQRDLLFHPPWTFSLPTFLLGIPISIFESVVWVTITYYMIGF 624
Query: 463 EWTLVKVVWCLFFMYFTFL----YFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFV 518
L + + L ++ T F F +M N +V + L GF+
Sbjct: 625 APELSRFLKHLLVIFLTQQMAGGIFRFIAATCRSMILANTGGALVILLLF----LLGGFI 680
Query: 519 LPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQ-NIETSDGRQTVED 577
+PR IP WW+W W +P+A++ L ++ + N +SD ++
Sbjct: 681 VPRG----------EIPKWWKWAYWVSPMAYTYDALTVNEMLAPRWINQPSSDNSTSLGL 730
Query: 578 FLRNYFGFKHD----FLGVVALVNIAFPIVFALVFAIAIKMFN 616
+ F D ++GV + + F ++F ++ +A+ N
Sbjct: 731 AVLEIFDIFTDPNWYWIGVGGI--LGFTVLFNILVTLALTFLN 771
>At2g29940 putative ABC transporter
Length = 1443
Score = 744 bits (1920), Expect = 0.0
Identities = 361/619 (58%), Positives = 469/619 (75%), Gaps = 10/619 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
M+LPF+P ++TF V+Y VDMP+EMR++GV E +L LL +SG F PGVLTAL+G +GAG
Sbjct: 834 MILPFKPLTMTFHNVNYYVDMPKEMRSQGVPETRLQLLSNVSGVFSPGVLTALVGSSGAG 893
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTLMDVL+GRKTGGY G+I ISG+PK+Q+TFARISGY EQ DIHSP VTV ESL +SA
Sbjct: 894 KTTLMDVLAGRKTGGYTEGDIRISGHPKEQQTFARISGYVEQNDIHSPQVTVEESLWFSA 953
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
LRL +I E +K F+E+VM LVEL LRYALVGLPG +GLSTEQRKRLT+AVELVANP
Sbjct: 954 SLRLPKEITKEQKKEFVEQVMRLVELDTLRYALVGLPGTTGLSTEQRKRLTIAVELVANP 1013
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPTSGLDARAAAIVMR VRNTVDTGRTVVCTIHQPSIDIFE+FDELLL+K+GGQ
Sbjct: 1014 SIIFMDEPTSGLDARAAAIVMRTVRNTVDTGRTVVCTIHQPSIDIFEAFDELLLMKRGGQ 1073
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY G LG +S L++YF+G+ GV I GYNPATWMLEVTT + E + +++A++YK S
Sbjct: 1074 VIYGGKLGTHSQVLVDYFQGINGVPPISSGYNPATWMLEVTTPALEEKYNMEFADLYKKS 1133
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
+ +R +A IK+LS P S+ + F SRYS++ +Q + CLWKQ+ YWR+PEYN +R +
Sbjct: 1134 DQFREVEANIKQLSVPPEGSEPISFTSRYSQNQLSQFLLCLWKQNLVYWRSPEYNLVRLV 1193
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++T A +LG++FW++GSK QDL MG++Y+A + +G N++SVQP+V +ERTVFY
Sbjct: 1194 FTTIAAFILGTVFWDIGSKRTSSQDLITVMGALYSACLFLGVSNASSVQPIVSIERTVFY 1253
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RE+AA MY+ PYA AQ ++++PY+ Q ++YG++ Y IGFE T K V L FM+ TF
Sbjct: 1254 REKAAGMYAPIPYAAAQGLVEIPYILTQTILYGVITYFTIGFERTFSKFVLYLVFMFLTF 1313
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
YFTFYGMM+V +TPN H++ ++SSAFYS+WNL SGF++ +P IPVWW W
Sbjct: 1314 TYFTFYGMMAVGLTPNQHLAAVISSAFYSLWNLLSGFLVQKP----------LIPVWWIW 1363
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVNIAF 600
+ + PVAW+L G++ SQ GDV+ I TV++F+ YFG+K + +GV A V + F
Sbjct: 1364 FYYICPVAWTLQGVILSQLGDVESMINEPLFHGTVKEFIEYYFGYKPNMIGVSAAVLVGF 1423
Query: 601 PIVFALVFAIAIKMFNFQR 619
+F FA+++K NFQR
Sbjct: 1424 CALFFSAFALSVKYLNFQR 1442
Score = 143 bits (360), Expect = 3e-34
Identities = 154/643 (23%), Positives = 282/643 (42%), Gaps = 90/643 (13%)
Query: 34 KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQET 92
KL +LK +SG +PG +T L+G G+GK+TL+ L+G+ GNIT +G +
Sbjct: 189 KLNILKDISGIIKPGRMTLLLGPPGSGKSTLLLALAGKLDKSLKKTGNITYNGENLNKFH 248
Query: 93 FARISGYCEQTDIHSPHVTVYESLLYSA-----------------------WLRLSPDIN 129
R S Y QTD H +TV E+L ++A +R S +I+
Sbjct: 249 VKRTSAYISQTDNHIAELTVRETLDFAARCQGASEGFAGYMKDLTRLEKERGIRPSSEID 308
Query: 130 A---------ETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
A E + + V++++ L +VG + G+S QRKR+T V
Sbjct: 309 AFMKAASVKGEKHSVSTDYVLKVLGLDVCSDTMVGNDMMRGVSGGQRKRVTTGEMTVGPR 368
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQGG 239
+FMDE ++GLD+ +++ +RN V TV+ + QP+ + F+ FD+L+LL + G
Sbjct: 369 KTLFMDEISTGLDSSTTFQIVKCIRNFVHLMDATVLMALLQPAPETFDLFDDLILLSE-G 427
Query: 240 QEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYK- 298
+Y GP ++I +FE + ++ A ++ EVT+ + + D ++ Y+
Sbjct: 428 YMVYQGP----REDVIAFFESLG--FRLPPRKGVADFLQEVTSKKDQAQYWADPSKPYQF 481
Query: 299 -----NSELYRRNK-ALIKELSAPAPCSKDLYFPSRYSRSFFT-----QCIACLWKQHWS 347
+ +R +K + AP K PS R+ F C ++
Sbjct: 482 IPVSDIAAAFRNSKYGHAADSKLAAPFDKKSADPSALCRTKFAISGWENLKVCFVRELLL 541
Query: 348 YWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAM---- 403
R+ R V ++ ++F L +++ + F G+ Y + + G +
Sbjct: 542 IKRHKFLYTFRTCQVGFVGLVTATVF--LKTRLHPTSEQF---GNEYLSCLFFGLVHMMF 596
Query: 404 NSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFE 463
N S P++ VFY++R + A+ +++A ++++PY ++AVV+ VVY +G
Sbjct: 597 NGFSELPLMISRLPVFYKQRDNSFHPAWSWSIASWLLRVPYSVLEAVVWSGVVYFTVGLA 656
Query: 464 WTLVKVVWCLFFMYFTFLYFTFYGM------MSVAMTPNNHISIIVSSAFYSVWNLFSGF 517
+ + FF Y L F+ + M M ++ + I+ SA + L GF
Sbjct: 657 PSAGR-----FFRYM-LLLFSVHQMALGLFRMMASLARDMVIANTFGSAAILIVFLLGGF 710
Query: 518 VLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVED 577
V+P+ I WW W W +P+++ + +++ + ++ T+
Sbjct: 711 VIPK----------ADIKPWWVWGFWVSPLSYGQRAIAVNEFTATRWMTPSAISDTTIGL 760
Query: 578 FLRNYFGFKHD----FLGVVALVNIAFPIVFALVFAIAIKMFN 616
L F + ++G+ L I + I+F V +A+ N
Sbjct: 761 NLLKLRSFPTNDYWYWIGIAVL--IGYAILFNNVVTLALAYLN 801
>At2g37280 putative ABC transporter
Length = 1413
Score = 697 bits (1798), Expect = 0.0
Identities = 336/619 (54%), Positives = 451/619 (72%), Gaps = 10/619 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
M+LPF+P +ITF +++Y VD+P EM+ +G E KL LL ++GAFRPGVLTALMG++GAG
Sbjct: 804 MILPFKPLTITFQDLNYYVDVPVEMKGQGYNEKKLQLLSEITGAFRPGVLTALMGISGAG 863
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTL+DVL+GRKT GYI G I ISG+ K QETFAR+SGYCEQTDIHSP +TV ESL+YSA
Sbjct: 864 KTTLLDVLAGRKTSGYIEGEIRISGFLKVQETFARVSGYCEQTDIHSPSITVEESLIYSA 923
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
WLRL P+IN +T+ F+++V+E +EL+ ++ ALVG+ GVSGLSTEQRKRLTVAVELVANP
Sbjct: 924 WLRLVPEINPQTKIRFVKQVLETIELEEIKDALVGVAGVSGLSTEQRKRLTVAVELVANP 983
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPT+GLDARAAAIVMRAV+N +TGRT+VCTIHQPSI IFE+FDEL+LLK+GG+
Sbjct: 984 SIIFMDEPTTGLDARAAAIVMRAVKNVAETGRTIVCTIHQPSIHIFEAFDELVLLKRGGR 1043
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG +SS +I YF+ + GV+KI+D YNPATWMLEVT+ S E EL +D+A++Y S
Sbjct: 1044 MIYSGPLGQHSSCVIEYFQNIPGVAKIRDKYNPATWMLEVTSESVETELDMDFAKIYNES 1103
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
+LY+ N L+KELS P S DL+F ++++++ Q +CLWK SYWR+P YN +R
Sbjct: 1104 DLYKNNSELVKELSKPDHGSSDLHFKRTFAQNWWEQFKSCLWKMSLSYWRSPSYNLMRIG 1163
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ + + G +FWN G KI+ Q+LF +G++Y V+ +G N S ER V Y
Sbjct: 1164 HTFISSFIFGLLFWNQGKKIDTQQNLFTVLGAIYGLVLFVGINNCTSALQYFETERNVMY 1223
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RER A MYSAF YALAQVV ++PY+F+Q+ + IV+Y MIGF + KV W L+ M+
Sbjct: 1224 RERFAGMYSAFAYALAQVVTEIPYIFIQSAEFVIVIYPMIGFYASFSKVFWSLYAMFCNL 1283
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
L F + M +++TPN ++ I+ S F++ +N+F+GF++P+P IP WW W
Sbjct: 1284 LCFNYLAMFLISITPNFMVAAILQSLFFTTFNIFAGFLIPKP----------QIPKWWVW 1333
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVNIAF 600
+ + P +W+L +SQYGD+ Q I +TV FL +YFGF HD L + A++ IAF
Sbjct: 1334 FYYITPTSWTLNLFFSSQYGDIHQKINAFGETKTVASFLEDYFGFHHDRLMITAIILIAF 1393
Query: 601 PIVFALVFAIAIKMFNFQR 619
PI A ++A + NFQ+
Sbjct: 1394 PIALATMYAFFVAKLNFQK 1412
Score = 136 bits (343), Expect = 3e-32
Identities = 144/642 (22%), Positives = 285/642 (43%), Gaps = 95/642 (14%)
Query: 32 EDKLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQ 90
E + +L +SG PG LT L+G G GKTTL+ LSG G I+ +G+ +
Sbjct: 149 EANIKILTDVSGIISPGRLTLLLGPPGCGKTTLLKALSGNLENNLKCYGEISYNGHGLNE 208
Query: 91 ETFARISGYCEQTDIHSPHVTVYESLLYSA-----------WLRLS-----------PDI 128
+ S Y Q D+H +T E++ +SA + +S P+I
Sbjct: 209 VVPQKTSAYISQHDLHIAEMTTRETIDFSARCQGVGSRTDIMMEVSKREKDGGIIPDPEI 268
Query: 129 NAE---------TRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVAN 179
+A R + + +++++ L LVG G+S Q+KRLT A +V
Sbjct: 269 DAYMKAISVKGLKRSLQTDYILKILGLDICAETLVGNAMKRGISGGQKKRLTTAEMIVGP 328
Query: 180 PSIIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQG 238
+FMDE T+GLD+ A ++++++ T TV ++ QP+ + ++ FD+++L+ +
Sbjct: 329 TKALFMDEITNGLDSSTAFQIIKSLQQVAHITNATVFVSLLQPAPESYDLFDDIVLMAE- 387
Query: 239 GQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVE---------- 288
G+ +Y GP +++ +FE + G A ++ EV + + +
Sbjct: 388 GKIVYHGP----RDDVLKFFEECGFQCPERKGV--ADFLQEVISKKDQGQYWLHQNLPHS 441
Query: 289 -LRID-YAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHW 346
+ +D ++ +K+ E+ R+ + + + + KD + YS + AC+ ++
Sbjct: 442 FVSVDTLSKRFKDLEIGRKIEEALSKPYDISKTHKDALSFNVYSLPKWELFRACISREFL 501
Query: 347 SYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSM----YAAVILIGA 402
R N +L+ T VL + + + D D+ + M +A V+L+
Sbjct: 502 LMKR----NYFVYLFKTFQLVLAAIITMTVFIRTRMDIDIIHGNSYMSCLFFATVVLL-- 555
Query: 403 MNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGF 462
++ + +VFY+++ Y A+ YA+ V+++P F +++V+ + Y +IG+
Sbjct: 556 VDGIPELSMTVQRLSVFYKQKQLCFYPAWAYAIPATVLKIPLSFFESLVWTCLTYYVIGY 615
Query: 463 ---EWTLVKVVWCLFFMYFTFLYF-----TFYGMMSVAMTPNNHISIIVSSAFYSVWNLF 514
+ + LF ++FT + + AMT + + +I +F
Sbjct: 616 TPEPYRFFRQFMILFAVHFTSISMFRCIAAIFQTGVAAMTAGSFVMLITF--------VF 667
Query: 515 SGFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQY----GDVKQNIETSD 570
+GF +P +P W +W W NP++++ GL +++ Q +
Sbjct: 668 AGFAIP----------YTDMPGWLKWGFWVNPISYAEIGLSVNEFLAPRWQKMQPTNVTL 717
Query: 571 GRQTVEDFLRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAI 612
GR +E NY + + ++ + AL+ + I+F +F +A+
Sbjct: 718 GRTILESRGLNYDDYMY-WVSLSALLGLT--IIFNTIFTLAL 756
>At3g53480 ABC transporter - like protein
Length = 1450
Score = 696 bits (1795), Expect = 0.0
Identities = 334/619 (53%), Positives = 451/619 (71%), Gaps = 10/619 (1%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
MVLPF+P ++TF +++Y VDMP EMR++G + KL LL ++GAFRPG+LTALMGV+GAG
Sbjct: 841 MVLPFKPLTVTFQDLNYFVDMPVEMRDQGYDQKKLQLLSDITGAFRPGILTALMGVSGAG 900
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTL+DVL+GRKT GYI G+I ISG+PK QETFAR+SGYCEQTDIHSP++TV ES++YSA
Sbjct: 901 KTTLLDVLAGRKTSGYIEGDIRISGFPKVQETFARVSGYCEQTDIHSPNITVEESVIYSA 960
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
WLRL+P+I+A T+ F+++V+E +EL ++ +LVG+ GVSGLSTEQRKRLT+AVELVANP
Sbjct: 961 WLRLAPEIDATTKTKFVKQVLETIELDEIKDSLVGVTGVSGLSTEQRKRLTIAVELVANP 1020
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQ 240
SIIFMDEPT+GLDARAAAIVMRAV+N DTGRT+VCTIHQPSIDIFE+FDEL+LLK+GG+
Sbjct: 1021 SIIFMDEPTTGLDARAAAIVMRAVKNVADTGRTIVCTIHQPSIDIFEAFDELVLLKRGGR 1080
Query: 241 EIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNS 300
IY GPLG +S ++I YFE V + KIKD +NPATWML+V++ S E+EL +D+A++Y +S
Sbjct: 1081 MIYTGPLGQHSRHIIEYFESVPEIPKIKDNHNPATWMLDVSSQSVEIELGVDFAKIYHDS 1140
Query: 301 ELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFL 360
LY+RN L+K+LS P S D+ F +++S++ Q + LWK + SYWR+P YN +R +
Sbjct: 1141 ALYKRNSELVKQLSQPDSGSSDIQFKRTFAQSWWGQFKSILWKMNLSYWRSPSYNLMRMM 1200
Query: 361 YSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFY 420
++ +++ G++FW G ++ Q +F G++Y V+ +G N S ER V Y
Sbjct: 1201 HTLVSSLIFGALFWKQGQNLDTQQSMFTVFGAIYGLVLFLGINNCASALQYFETERNVMY 1260
Query: 421 RERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTF 480
RER A MYSA YAL QVV ++PY+F+QA + IV Y MIGF + KV W L+ M+ +
Sbjct: 1261 RERFAGMYSATAYALGQVVTEIPYIFIQAAEFVIVTYPMIGFYPSAYKVFWSLYSMFCSL 1320
Query: 481 LYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRW 540
L F + M V++TPN ++ I+ S FY +NLFSGF++P+ +P WW W
Sbjct: 1321 LTFNYLAMFLVSITPNFMVAAILQSLFYVGFNLFSGFLIPQ----------TQVPGWWIW 1370
Query: 541 YSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFKHDFLGVVALVNIAF 600
+ P +W+L G ++SQYGD+ + I TV FL++YFGF HD L V A+V IAF
Sbjct: 1371 LYYLTPTSWTLNGFISSQYGDIHEEINVFGQSTTVARFLKDYFGFHHDLLAVTAVVQIAF 1430
Query: 601 PIVFALVFAIAIKMFNFQR 619
PI A +FA + NFQR
Sbjct: 1431 PIALASMFAFFVGKLNFQR 1449
Score = 149 bits (377), Expect = 3e-36
Identities = 146/638 (22%), Positives = 294/638 (45%), Gaps = 87/638 (13%)
Query: 32 EDKLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGY-IGGNITISGYPKKQ 90
E K+ ++ ++G +PG LT L+G GKTTL+ LSG G I+ +G+ +
Sbjct: 184 EAKINIINDVNGIIKPGRLTLLLGPPSCGKTTLLKALSGNLENNLKCSGEISYNGHRLDE 243
Query: 91 ETFARISGYCEQTDIHSPHVTVYESLLYSAWLR--------------------LSPDINA 130
+ S Y Q D+H +TV E++ +SA + + PD
Sbjct: 244 FVPQKTSAYISQYDLHIAEMTVRETVDFSARCQGVGSRTDIMMEVSKREKEKGIIPDTEV 303
Query: 131 ET-----------RKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVAN 179
+ R + + +++++ L L+G G+S Q+KRLT A +V
Sbjct: 304 DAYMKAISVEGLQRSLQTDYILKILGLDICAEILIGDVMRRGISGGQKKRLTTAEMIVGP 363
Query: 180 PSIIFMDEPTSGLDARAAAIVMRAVRNTVD-TGRTVVCTIHQPSIDIFESFDELLLLKQG 238
+FMDE T+GLD+ A ++++++ + TV+ ++ QP+ + ++ FD+++L+ +
Sbjct: 364 TKALFMDEITNGLDSSTAFQIVKSLQQFAHISSATVLVSLLQPAPESYDLFDDIMLMAK- 422
Query: 239 GQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKEV------ELRID 292
G+ +Y GP G ++N+FE + G A ++ EV + + +L
Sbjct: 423 GRIVYHGPRG----EVLNFFEDCGFRCPERKGV--ADFLQEVISKKDQAQYWWHEDLPYS 476
Query: 293 YAEVYKNSELYRR---NKALIKELSAP---APCSKDLYFPSRYSRSFFTQCIACLWKQHW 346
+ V S+ ++ K + LS P + KD S YS + IAC+ +++
Sbjct: 477 FVSVEMLSKKFKDLSIGKKIEDTLSKPYDRSKSHKDALSFSVYSLPNWELFIACISREYL 536
Query: 347 SYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNA---MGSMYAAVILIGAM 403
R N +++ TA V+ + + + D+ + M +++ A+I++ +
Sbjct: 537 LMKR----NYFVYIFKTAQLVMAAFITMTVFIRTRMGIDIIHGNSYMSALFFALIIL-LV 591
Query: 404 NSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFE 463
+ + VFY+++ Y A+ YA+ V+++P F +++V+ + Y +IG+
Sbjct: 592 DGFPELSMTAQRLAVFYKQKQLCFYPAWAYAIPATVLKVPLSFFESLVWTCLSYYVIGYT 651
Query: 464 WTLVKVVWCLFFMYFTFLYFTFYGMMS----VAMTPNNHISIIVSSAFYSVWN-LFSGFV 518
+ FF F L+ + +S +A ++ I + +F ++ +F+GFV
Sbjct: 652 PEASR-----FFKQFILLFAVHFTSISMFRCLAAIFQTVVASITAGSFGILFTFVFAGFV 706
Query: 519 LPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSD----GRQT 574
+P P S+P W +W WANP+++ GL +++ + N + GR
Sbjct: 707 IPPP----------SMPAWLKWGFWANPLSYGEIGLSVNEFLAPRWNQMQPNNFTLGRTI 756
Query: 575 VEDFLRNYFGFKHDFLGVVALVNIAFPIVFALVFAIAI 612
++ +Y G+ + ++ + AL + F ++F ++F +A+
Sbjct: 757 LQTRGMDYNGYMY-WVSLCAL--LGFTVLFNIIFTLAL 791
>At4g15230 ABC transporter like protein
Length = 979
Score = 580 bits (1494), Expect = e-165
Identities = 298/633 (47%), Positives = 411/633 (64%), Gaps = 78/633 (12%)
Query: 1 MVLPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAG 60
++LPF+P ++TF V Y ++ PQ + LL ++GA +PGVLT+LMGV+GAG
Sbjct: 410 IILPFKPLTVTFQNVQYYIETPQGKTRQ--------LLSDITGALKPGVLTSLMGVSGAG 461
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
KTTL+DVLSGRKT G I G I + GYPK QETFAR+SGYCEQ DIHSP++TV ESL YSA
Sbjct: 462 KTTLLDVLSGRKTRGIIKGEIKVGGYPKVQETFARVSGYCEQFDIHSPNITVEESLKYSA 521
Query: 121 WLRLSPDINAETRKM--------------FIEEVMELVELKPLRYALVGLPGVSGLSTEQ 166
WLRL +I+++T+ + ++EV+E VEL ++ ++VGLPG+SGLS EQ
Sbjct: 522 WLRLPYNIDSKTKNVRNYTLKTNRLKEIELVKEVLETVELDDIKDSVVGLPGISGLSIEQ 581
Query: 167 RKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIF 226
RKRLT+AVELVANPSIIFMDEPT+GLDARAAAIVMRAV+N +TGRTVVCTIHQPSIDIF
Sbjct: 582 RKRLTIAVELVANPSIIFMDEPTTGLDARAAAIVMRAVKNVAETGRTVVCTIHQPSIDIF 641
Query: 227 ESFDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSSKE 286
E+FDEL+L+K GGQ +Y GP G NSS +I YFE
Sbjct: 642 ETFDELILMKNGGQLVYYGPPGQNSSKVIEYFE--------------------------- 674
Query: 287 VELRIDYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHW 346
NK ++++LS+ + S+ L FPS++S++ + Q ACLWKQH+
Sbjct: 675 -------------------NKMVVEQLSSASLGSEALRFPSQFSQTAWVQLKACLWKQHY 715
Query: 347 SYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSN 406
SYWRNP +N R ++ + L G +FW I QDL + GSMY V+ G N
Sbjct: 716 SYWRNPSHNITRIVFILLDSTLCGLLFWQKAEDINNQQDLISIFGSMYTLVVFPGMNNCA 775
Query: 407 SVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTL 466
+V + ER VFYRER ARMYS++ Y+ +QV+I++PY +Q+++ I+VY IG+ ++
Sbjct: 776 AVINFIAAERNVFYRERFARMYSSWAYSFSQVLIEVPYSLLQSLLCTIIVYPTIGYHMSV 835
Query: 467 VKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK** 526
K+ W L+ ++ + L F + GM+ VA+TPN H+++ + S+F+S+ NLF+GFV+P+
Sbjct: 836 YKMFWSLYSIFCSLLIFNYSGMLMVALTPNIHMAVTLRSSFFSMLNLFAGFVIPKQ---- 891
Query: 527 LISVILSIPVWWRWYSWANPVAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFK 586
IP WW W + +P +W L GL++SQYGDV + I ++ V FL +YFG+K
Sbjct: 892 ------KIPKWWIWMYYLSPTSWVLEGLLSSQYGDVDKEILVFGEKKRVSAFLEDYFGYK 945
Query: 587 HDFLGVVALVNIAFPIVFALVFAIAIKMFNFQR 619
H+ L VVA V IA+PI+ A +FA + +FQ+
Sbjct: 946 HESLAVVAFVLIAYPIIVATLFAFFMSKLSFQK 978
Score = 56.6 bits (135), Expect = 4e-08
Identities = 66/307 (21%), Positives = 143/307 (46%), Gaps = 39/307 (12%)
Query: 157 PGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGR-TVV 215
PG+SG +++RLT +V + +FMDE ++GLD+ ++ ++ T++
Sbjct: 42 PGISG---GEKRRLTTGELVVGPATTLFMDEISNGLDSSTTFQIVSCLQQLAHIAEATIL 98
Query: 216 CTIHQPSIDIFESFDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPAT 275
++ QP+ + FE FD+++L+ + G+ IY P +++ +FE + K + A
Sbjct: 99 ISLLQPAPETFELFDDVILMGE-GKIIYHAP----RADICRFFE--EFGFKCPERKGVAD 151
Query: 276 WMLEVTTSSKEVE-----------LRID-YAEVYKNSELYRRNKALIKELSAP---APCS 320
++ E+ + + + + +D + +K S L L +ELS P +
Sbjct: 152 FLQEIMSKKDQEQYWCHRDKPYSYISVDSFINKFKESNL---GLLLKEELSKPFNKSQTR 208
Query: 321 KDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAV----AVLLGSMFWNL 376
KD +YS + AC ++ R N+ +L+ +A+ A++ ++F +
Sbjct: 209 KDGLCYKKYSLGKWEMLKACSRREFLLMKR----NSFIYLFKSALLVFNALVTMTVFLQV 264
Query: 377 GSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALA 436
G+ + + MGS++ A+ + A + + VF +++ Y A+ YA+
Sbjct: 265 GATTDSLHGNY-LMGSLFTALFRLLADGLPELTLTIS-RLGVFCKQKDLYFYPAWAYAIP 322
Query: 437 QVVIQLP 443
+++++P
Sbjct: 323 SIILKIP 329
>At2g01320 putative ABC transporter
Length = 725
Score = 202 bits (515), Expect = 3e-52
Identities = 157/560 (28%), Positives = 266/560 (47%), Gaps = 43/560 (7%)
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGG---YIGGNITISGYPKKQETF 93
LLK +SG +PG L A+MG +G+GKTTL++VL+G+ + ++ G + ++G P + +
Sbjct: 90 LLKNVSGEAKPGRLLAIMGPSGSGKTTLLNVLAGQLSLSPRLHLSGLLEVNGKPSSSKAY 149
Query: 94 ARISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYAL 153
+ Q D+ +TV E+L ++A L+L +AE R ++ ++ + L +
Sbjct: 150 KL--AFVRQEDLFFSQLTVRETLSFAAELQLPEISSAEERDEYVNNLLLKLGLVSCADSC 207
Query: 154 VGLPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRT 213
VG V G+S ++KRL++A EL+A+PS+IF DEPT+GLDA A VM ++ G T
Sbjct: 208 VGDAKVRGISGGEKKRLSLACELIASPSVIFADEPTTGLDAFQAEKVMETLQKLAQDGHT 267
Query: 214 VVCTIHQPSIDIFESFDELLLLKQGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNP 273
V+C+IHQP ++ FD+++LL + G +Y GP G F + + NP
Sbjct: 268 VICSIHQPRGSVYAKFDDIVLLTE-GTLVYAGPAGKEPLTYFGNFGFL-----CPEHVNP 321
Query: 274 ATWMLE-----------VTTSSKEVELRIDYAEVYKNSELYRRNKALIKEL-SAPAPCSK 321
A ++ + V +S K V +D +S LY ++ +E + P K
Sbjct: 322 AEFLADLISVDYSSSETVYSSQKRVHALVDAFSQRSSSVLYATPLSMKEETKNGMRPRRK 381
Query: 322 DLY-FPSRYSRSFFTQCIACLWKQHW-SYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSK 379
+ + R FF L K+ W R+ N +R S A AV+ GS+FW +G
Sbjct: 382 AIVERTDGWWRQFF-----LLLKRAWMQASRDGPTNKVRARMSVASAVIFGSVFWRMGKS 436
Query: 380 IEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVV 439
QD MG + A I V ER + RER+ YS PY L++ +
Sbjct: 437 QTSIQD---RMGLLQVAAINTAMAALTKTVGVFPKERAIVDRERSKGSYSLGPYLLSKTI 493
Query: 440 IQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGMMSVAMTPNNHI 499
++P +++G V+Y M TL + + + G+ AM P+
Sbjct: 494 AEIPIGAAFPLMFGAVLYPMARLNPTLSRFGKFCGIVTVESFAASAMGLTVGAMVPSTEA 553
Query: 500 SIIVSSAFYSVWNLFSGFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQY 559
++ V + +V+ +F G+ + + P+ +RW A+ + W+ GL +++
Sbjct: 554 AMAVGPSLMTVFIVFGGYYVNAD----------NTPIIFRWIPRASLIRWAFQGLCINEF 603
Query: 560 GDVKQNIETSDGRQTVEDFL 579
+K + + + QT E L
Sbjct: 604 SGLKFDHQNTFDVQTGEQAL 623
>At5g06530 ABC transporter like protein
Length = 627
Score = 190 bits (482), Expect = 2e-48
Identities = 142/490 (28%), Positives = 248/490 (49%), Gaps = 51/490 (10%)
Query: 4 PFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLVLLKGLSGAFRPGVLTALMGVTGAGKTT 63
P P + F +V+Y V + + + +E ++ L G+SG+ PG + ALMG +G+GKTT
Sbjct: 27 PTLPIFLKFRDVTYKVVIKKLTSS---VEKEI--LTGISGSVNPGEVLALMGPSGSGKTT 81
Query: 64 LMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSAWLR 123
L+ +L+GR + GG++T + P + ++I G+ Q D+ PH+TV E+L Y+A LR
Sbjct: 82 LLSLLAGRISQSSTGGSVTYNDKPYSKYLKSKI-GFVTQDDVLFPHLTVKETLTYAARLR 140
Query: 124 LSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPSII 183
L + E +K +V++ + L+ + ++G V G+S +RKR+++ E++ NPS++
Sbjct: 141 LPKTLTREQKKQRALDVIQELGLERCQDTMIGGAFVRGVSGGERKRVSIGNEIIINPSLL 200
Query: 184 FMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQEIY 243
+DEPTSGLD+ A + + + + G+TV+ TIHQPS +F FD+L+LL +G +Y
Sbjct: 201 LLDEPTSGLDSTTALRTILMLHDIAEAGKTVITTIHQPSSRLFHRFDKLILLGRGSL-LY 259
Query: 244 VGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTTSS-------KEVELRIDYAEV 296
G SS ++YF + G S + NPA ++L++ + E++ R+
Sbjct: 260 FG----KSSEALDYFSSI-GCSPL-IAMNPAEFLLDLANGNINDISVPSELDDRVQVGNS 313
Query: 297 YKNSELYRRNKALI-----------------KELSAPAPCSKDLYFPS-RYSRSFFTQCI 338
+ ++ + + A + K+L P P ++ S R R + T
Sbjct: 314 GRETQTGKPSPAAVHEYLVEAYETRVAEQEKKKLLDPVPLDEEAKAKSTRLKRQWGT--- 370
Query: 339 ACLWKQHWSYW-------RNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMG 391
C W+Q+ + R+ ++ +R + AV+LG ++W S I L + G
Sbjct: 371 -CWWEQYCILFCRGLKERRHEYFSWLRVTQVLSTAVILGLLWWQ--SDIRTPMGLQDQAG 427
Query: 392 SMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVV 451
++ + G + ER + +ERAA MY Y LA+ LP F+ +
Sbjct: 428 LLFFIAVFWGFFPVFTAIFAFPQERAMLNKERAADMYRLSAYFLARTTSDLPLDFILPSL 487
Query: 452 YGIVVYVMIG 461
+ +VVY M G
Sbjct: 488 FLLVVYFMTG 497
>At3g13220 ABC transporter
Length = 647
Score = 184 bits (466), Expect = 2e-46
Identities = 138/505 (27%), Positives = 246/505 (48%), Gaps = 43/505 (8%)
Query: 3 LPFEPHSITFDEVSYSVDMPQEMRNRGVIEDKLV------LLKGLSGAFRPGVLTALMGV 56
+ F P + ++ S +D+ +E + +ED + +LKG++G+ PG + ALMG
Sbjct: 28 MEFMPQAYLRNQYSSEIDIDEEFVSTYPLEDAPLPIFLKHILKGITGSTGPGEILALMGP 87
Query: 57 TGAGKTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESL 116
+G+GKTTL+ ++ GR T + G +T + P RI G+ Q D+ P +TV E+L
Sbjct: 88 SGSGKTTLLKIMGGRLTDN-VKGKLTYNDIPYSPSVKRRI-GFVTQDDVLLPQLTVEETL 145
Query: 117 LYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVEL 176
++A+LRL ++ E + IE +++ + L+ R VG V G+S +RKR ++A E+
Sbjct: 146 AFAAFLRLPSSMSKEQKYAKIEMIIKELGLERCRRTRVGGGFVKGISGGERKRASIAYEI 205
Query: 177 VANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLK 236
+ +PS++ +DEPTSGLD+ +A ++ ++ GRTV+ TIHQPS +F FD+LLL+
Sbjct: 206 LVDPSLLLLDEPTSGLDSTSATKLLHILQGVAKAGRTVITTIHQPSSRMFHMFDKLLLIS 265
Query: 237 QGGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEVTT-------------- 282
+G Y + + YF ++ + +I NPA ++L++ T
Sbjct: 266 EGHPAFY-----GKARESMEYFSSLRILPEI--AMNPAEFLLDLATGQVSDISLPDELLA 318
Query: 283 ------SSKEVELRIDYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQ 336
S+EV L+ + YK + + + AP + ++ S++ Q
Sbjct: 319 AKTAQPDSEEVLLKY-LKQRYKTDLEPKEKEENHRNRKAPEHLQIAIQVKKDWTLSWWDQ 377
Query: 337 CIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAA 396
+ L ++ + R ++ +R + S VAV+LG ++W SK + + L + +G M+
Sbjct: 378 FL-ILSRRTFRERRRDYFDKLRLVQSLGVAVVLGLLWWK--SKTDTEAHLRDQVGLMFYI 434
Query: 397 VILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVV 456
I + + V E+ +ER A MY Y + + + + + I+V
Sbjct: 435 CIFWTSSSLFGAVYVFPFEKIYLVKERKAEMYRLSVYYVCSTLCDMVAHVLYPTFFMIIV 494
Query: 457 YVMIGFEWTLVKVVWCLFFMYFTFL 481
Y M F + + C F T L
Sbjct: 495 YFMAEFN----RNIPCFLFTVLTIL 515
>At2g37360 ABC transporter like protein
Length = 755
Score = 184 bits (466), Expect = 2e-46
Identities = 148/591 (25%), Positives = 274/591 (46%), Gaps = 64/591 (10%)
Query: 7 PHSITFDEVSYSVDMPQEMR---------NRGVIEDKLVLLKGLSGAFRPGVLTALMGVT 57
P ++F +++YSV + ++ N + K +LL G+SG R G + A++G +
Sbjct: 95 PFVLSFTDLTYSVKIQKKFNPLACCRRSGNDSSVNTK-ILLNGISGEAREGEMMAVLGAS 153
Query: 58 GAGKTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLL 117
G+GK+TL+D L+ R + G+IT++G + IS Y Q D+ P +TV E+L+
Sbjct: 154 GSGKSTLIDALANRIAKDSLRGSITLNGEVLESSMQKVISAYVMQDDLLFPMLTVEETLM 213
Query: 118 YSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELV 177
+SA RL ++ + +K ++ +++ + L+ ++G G G+S +R+R+++ +++
Sbjct: 214 FSAEFRLPRSLSKKKKKARVQALIDQLGLRSAAKTVIGDEGHRGVSGGERRRVSIGNDII 273
Query: 178 ANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQ 237
+P I+F+DEPTSGLD+ +A +V++ ++ +G V+ +IHQPS I D+L+ L +
Sbjct: 274 HDPIILFLDEPTSGLDSTSAYMVIKVLQRIAQSGSIVIMSIHQPSYRIMGLLDQLIFLSK 333
Query: 238 GGQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPATWMLEV-------TTSSKEV--- 287
G +Y G + ++L +F + I + N + L++ T +K +
Sbjct: 334 -GNTVYSG----SPTHLPQFFSEFK--HPIPENENKTEFALDLIRELEYSTEGTKPLVEF 386
Query: 288 --ELRIDYAEVYKNSELYRRNKALIKE-----------LSAPAPCSKDLYFPS--RYSRS 332
+ R A Y N+ N + +KE +S + PS ++
Sbjct: 387 HKQWRAKQAPSYNNNNKRNTNVSSLKEAITASISRGKLVSGATNNNSSNLTPSFQTFANP 446
Query: 333 FFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGS 392
F+ + I + + R PE +R ++L +MF NL + + Q+
Sbjct: 447 FWIEMIVIGKRAILNSRRQPELLGMRLGAVMVTGIILATMFTNLDNSPKGAQERL----G 502
Query: 393 MYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVY 452
+A + PV ER +F RE A Y Y L+Q +I +P + V + +
Sbjct: 503 FFAFAMSTTFYTCAEAIPVFLQERYIFMRETAYNAYRRSSYVLSQSIISIPALIVLSASF 562
Query: 453 GIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGMMSV----AMTPNNHISIIVSSAFY 508
+ +G + FF YFT L + G V + PN + V A
Sbjct: 563 AATTFWAVGLDGGANG----FFFFYFTILASFWAGSSFVTFLSGVIPNVMLGFTVVVAIL 618
Query: 509 SVWNLFSGFVLPRPGK**LISVILSIPVWWRWYSWANPVAWSLYGLVTSQY 559
+ + LFSGF + R IPV+W W+ + + V + G++ +++
Sbjct: 619 AYFLLFSGFFISRD----------RIPVYWLWFHYISLVKYPYEGVLQNEF 659
>At3g21090 ABC transporter, putative
Length = 680
Score = 181 bits (460), Expect = 8e-46
Identities = 156/614 (25%), Positives = 288/614 (46%), Gaps = 76/614 (12%)
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGYI-GGNITISGYPKKQETFAR 95
LL+ L+G PG + A+MG +G+GK+TL+D L+GR + GN+ ++G KK
Sbjct: 45 LLQRLNGYAEPGRIMAIMGPSGSGKSTLLDSLAGRLARNVVMTGNLLLNG--KKARLDYG 102
Query: 96 ISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVG 155
+ Y Q D+ +TV E++ YSA LRL D++ E +E + + L+ ++G
Sbjct: 103 LVAYVTQEDVLLGTLTVRETITYSAHLRLPSDMSKEEVSDIVEGTIMELGLQDCSDRVIG 162
Query: 156 LPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVV 215
G+S +RKR+++A+E++ P I+F+DEPTSGLD+ +A V++A+RN GRTV+
Sbjct: 163 NWHARGVSGGERKRVSIALEILTRPQILFLDEPTSGLDSASAFFVIQALRNIARDGRTVI 222
Query: 216 CTIHQPSIDIFESFDELLLLKQGGQEIYVG---------------------PLGHNSSNL 254
++HQPS ++F FD+L LL G+ +Y G P H +
Sbjct: 223 SSVHQPSSEVFALFDDLFLL-SSGESVYFGEAKSAVEFFAESGFPCPKKRNPSDHFLRCI 281
Query: 255 INYFEGV----QGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNSELYRRNKALI 310
+ F+ V +G +I++ PAT + ++ ++ R+ E YK S+ + K+ I
Sbjct: 282 NSDFDTVTATLKGSQRIQE--TPATSDPLMNLATSVIKARL--VENYKRSKYAKSAKSRI 337
Query: 311 KELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLLG 370
+ELS ++ S +++ Q + + R+ Y R + V++ +G
Sbjct: 338 RELSNIEGLEMEIRKGS--EATWWKQLRTLTARSFINMCRDVGYYWTRIISYIVVSISVG 395
Query: 371 SMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQ--PVVGVERTVFYRERAARMY 428
++F+++G + + + + G M S+ P E VFY+ER + Y
Sbjct: 396 TIFYDVG------YSYTSILARVSCGGFITGFMTFMSIGGFPSFLEEMKVFYKERLSGYY 449
Query: 429 SAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGM 488
Y L+ + P++ +V+ G + Y ++ F + ++F+ M
Sbjct: 450 GVSVYILSNYISSFPFLVAISVITGTITYNLVKFRPGFSHYAFFCLNIFFSVSVIESLMM 509
Query: 489 MSVAMTPNNHISIIVSSAFYSVWNLFSGF--VLPRPGK**LISVILSIPVWWRWYSWANP 546
+ ++ PN + +I + + + SGF +LP K ++WR+ P
Sbjct: 510 VVASVVPNFLMGLITGAGLIGIIMMTSGFFRLLPDLPK-----------IFWRY-----P 553
Query: 547 VAWSLYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFK------HDFLGVVALVNIAF 600
V++ YG Q+ + + + + T E+ + FG K D VVA++
Sbjct: 554 VSYISYGSWAIQF----EPLFPGEPKMTGEEVIEKVFGVKVTYSKWWDLAAVVAIL---- 605
Query: 601 PIVFALVFAIAIKM 614
+ + L+F + +K+
Sbjct: 606 -VCYRLLFFVVLKL 618
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 181 bits (460), Expect = 8e-46
Identities = 156/604 (25%), Positives = 276/604 (44%), Gaps = 50/604 (8%)
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGR-KTGGYIGGNITISGYPKKQETFAR 95
+L+GL+G PG LTALMG +G+GK+T++D L+ R ++ G + ++G +K +
Sbjct: 69 VLEGLTGYAEPGSLTALMGPSGSGKSTMLDALASRLAANAFLSGTVLLNG--RKTKLSFG 126
Query: 96 ISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVG 155
+ Y Q D +TV E++ YSA +RL + ++ +E + + L+ ++G
Sbjct: 127 TAAYVTQDDNLIGTLTVRETIWYSARVRLPDKMLRSEKRALVERTIIEMGLQDCADTVIG 186
Query: 156 LPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVV 215
+ G+S +++R+++A+E++ P ++F+DEPTSGLD+ +A V + +R GRTV+
Sbjct: 187 NWHLRGISGGEKRRVSIALEILMRPRLLFLDEPTSGLDSASAFFVTQTLRALSRDGRTVI 246
Query: 216 CTIHQPSIDIFESFDELLLLKQGGQEIYVG---------------------PLGHNSSNL 254
+IHQPS ++FE FD L LL GG+ +Y G P H +
Sbjct: 247 ASIHQPSSEVFELFDRLYLL-SGGKTVYFGQASDAYEFFAQAGFPCPALRNPSDHFLRCI 305
Query: 255 INYFEGVQGVSK--IKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNSELYRRNKALIKE 312
+ F+ V+ K +K + + LE T+++ + L +DY Y S+ Y KA ++E
Sbjct: 306 NSDFDKVRATLKGSMKLRFEASDDPLEKITTAEAIRLLVDY---YHTSDYYYTAKAKVEE 362
Query: 313 LSAPAPCSKDLYFPSRYSR-SFFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLLGS 371
+S K S S+ SF Q + + R+ Y +R L V V +G+
Sbjct: 363 ISQ----FKGTILDSGGSQASFLLQTYTLTKRSFINMSRDFGYYWLRLLIYILVTVCIGT 418
Query: 372 MFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQPVVGVERTVFYRERAARMYSAF 431
++ N+G+ A GS + V S P + VF RER Y
Sbjct: 419 IYLNVGT----SYSAILARGSCASFVFGFVTFMSIGGFPSFVEDMKVFQRERLNGHYGVA 474
Query: 432 PYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYGMMSV 491
+ +A + P++ + + G + Y M+G ++ + +Y + M
Sbjct: 475 AFVIANTLSATPFLIMITFISGTICYFMVGLHPGFTHYLFFVLCLYASVTVVESLMMAIA 534
Query: 492 AMTPNNHISIIVSSAFYSVWNLFSGFV-LPRPGK**LISVILSIPVWWRWYSWANPVAWS 550
++ PN + II+ + ++ L SGF LP + P W S+ + W+
Sbjct: 535 SIVPNFLMGIIIGAGIQGIFMLVSGFFRLPND---------IPKPFWRYPMSYISFHFWA 585
Query: 551 LYGLVTSQYGDVKQNIETSDGRQTVEDFLRNYFGFK-HDFLGVVALVNIAFPIVFALVFA 609
L G + + + + S + E L N F H + V ++ I++ ++F
Sbjct: 586 LQGQYQNDLRGLTFDSQGSAFKIPGEYVLENVFQIDLHRSKWINLSVILSMIIIYRIIFF 645
Query: 610 IAIK 613
I IK
Sbjct: 646 IMIK 649
>At1g51500 ATP-dependent transmembrane transporter, putative
Length = 687
Score = 181 bits (458), Expect = 1e-45
Identities = 163/612 (26%), Positives = 285/612 (45%), Gaps = 63/612 (10%)
Query: 37 LLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGYI-GGNITISGYPKKQETFAR 95
LL GL+G PG + A+MG +G+GK+TL+D L+GR I GN+ ++G KK
Sbjct: 44 LLDGLNGHAEPGRIMAIMGPSGSGKSTLLDSLAGRLARNVIMTGNLLLNG--KKARLDYG 101
Query: 96 ISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFIEEVMELVELKPLRYALVG 155
+ Y Q DI +TV E++ YSA LRLS D+ E +E + + L+ ++G
Sbjct: 102 LVAYVTQEDILMGTLTVRETITYSAHLRLSSDLTKEEVNDIVEGTIIELGLQDCADRVIG 161
Query: 156 LPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAAAIVMRAVRNTV-DTGRTV 214
G+S +RKR++VA+E++ P I+F+DEPTSGLD+ +A V++A+RN D GRTV
Sbjct: 162 NWHSRGVSGGERKRVSVALEILTRPQILFLDEPTSGLDSASAFFVIQALRNIARDGGRTV 221
Query: 215 VCTIHQPSIDIFESFDELLLLKQGGQEIYVG---------------------PLGHNSSN 253
V +IHQPS ++F FD+L LL G+ +Y G P H
Sbjct: 222 VSSIHQPSSEVFALFDDLFLL-SSGETVYFGESKFAVEFFAEAGFPCPKKRNPSDHFLRC 280
Query: 254 LINYFEGV----QGVSKIKDGYNPATWMLEVTTSSKEVELRIDYAEVYKNSELYRRNKAL 309
+ + F+ V +G +I++ PAT + ++ E++ R+ E Y+ S + K+
Sbjct: 281 INSDFDTVTATLKGSQRIRE--TPATSDPLMNLATSEIKARL--VENYRRSVYAKSAKSR 336
Query: 310 IKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWRNPEYNAIRFLYSTAVAVLL 369
I+EL A + ++F Q + + R+ Y R + V+ +
Sbjct: 337 IREL-ASIEGHHGMEVRKGSEATWFKQLRTLTKRSFVNMCRDIGYYWSRIVIYIVVSFCV 395
Query: 370 GSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQ--PVVGVERTVFYRERAARM 427
G++F+++G + + + + G M S+ P E VFY+ER +
Sbjct: 396 GTIFYDVG------HSYTSILARVSCGGFITGFMTFMSIGGFPSFIEEMKVFYKERLSGY 449
Query: 428 YSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVVWCLFFMYFTFLYFTFYG 487
Y Y ++ V P++ A++ G + Y M+ F + + ++F+
Sbjct: 450 YGVSVYIISNYVSSFPFLVAIALITGSITYNMVKFRPGVSHWAFFCLNIFFSVSVIESLM 509
Query: 488 MMSVAMTPNNHISIIVSSAFYSVWNLFSGF--VLPRPGK**LISVILSIPVWWRW-YSWA 544
M+ ++ PN + +I + + + SGF +LP K V+WR+ S+
Sbjct: 510 MVVASLVPNFLMGLITGAGIIGIIMMTSGFFRLLPDLPK-----------VFWRYPISFM 558
Query: 545 NPVAWSLYGLVTSQY-GDVKQNIETSDGRQTVEDFLRNYFGF-----KHDFLGVVALVNI 598
+ +W++ G + + G + + + T E + FG K L + L+ +
Sbjct: 559 SYGSWAIQGAYKNDFLGLEFDPMFAGEPKMTGEQVINKIFGVQVTHSKWWDLSAIVLILV 618
Query: 599 AFPIVFALVFAI 610
+ I+F +V +
Sbjct: 619 CYRILFFIVLKL 630
>At3g55100 ABC transporter - like protein
Length = 662
Score = 179 bits (455), Expect = 3e-45
Identities = 130/571 (22%), Positives = 264/571 (45%), Gaps = 29/571 (5%)
Query: 3 LPFEPHSITFDEVSYSVDMPQEMRNR-GVIEDKL-VLLKGLSGAFRPGVLTALMGVTGAG 60
+P P + F++++Y+V + Q R G K+ LL G++G + G + A++G +GAG
Sbjct: 15 IPPIPFVLAFNDLTYNVTLQQRFGLRFGHSPAKIKTLLNGITGEAKEGEILAILGASGAG 74
Query: 61 KTTLMDVLSGRKTGGYIGGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSA 120
K+TL+D L+G+ G + G +T++G + IS Y Q D+ P +TV E+L+++A
Sbjct: 75 KSTLIDALAGQIAEGSLKGTVTLNGEALQSRLLRVISAYVMQEDLLFPMLTVEETLMFAA 134
Query: 121 WLRLSPDINAETRKMFIEEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANP 180
RL ++ ++ +E +++ + L ++ ++G G G+S +R+R+++ +++ +P
Sbjct: 135 EFRLPRSLSKSKKRNRVETLIDQLGLTTVKNTVIGDEGHRGVSGGERRRVSIGTDIIHDP 194
Query: 181 SIIFMDEPTSGLDARAAAIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQG-- 238
++F+DEPTSGLD+ +A +V++ ++ +G V+ +IHQPS I E D +++L G
Sbjct: 195 IVLFLDEPTSGLDSTSAFMVVQVLKKIARSGSIVIMSIHQPSGRIMEFLDRVIVLSSGQI 254
Query: 239 -------GQEIYVGPLGHNSSNLINYFEGVQGVSKIKDGYNPAT-WMLEVTTSSKEVELR 290
++ G N E + K +G T ++E + + +LR
Sbjct: 255 VFSDSPATLPLFFSEFGSPIPEKENIAEFTLDLIKDLEGSPEGTRGLVEFNRNWQHRKLR 314
Query: 291 IDYAEVYKNSELYRRNKALIKELSAPAPCSKDLYFPSRYSRSFFTQCIACLWKQHWSYWR 350
+ + +S L +A+ +S S Y ++ + + + ++ R
Sbjct: 315 VSQEPHHNSSSL---GEAINASISRGKLVSTSYRSIPSYVNPWWVETVILAKRYMINWTR 371
Query: 351 NPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQP 410
PE R LL +++W + Q+ + A + A P
Sbjct: 372 TPELIGTRVFIVMMTGFLLATVYWKVDDSPRGVQERLSFFSFAMATMFYSCA----DGLP 427
Query: 411 VVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIGFEWTLVKVV 470
ER +F RE A Y Y ++ ++ LP++F ++ + + +G L +
Sbjct: 428 AFIQERYIFLRETAHNAYRRSSYVISHSLVTLPHLFALSIGFAATTFWFVGLNGGLAGFI 487
Query: 471 WCLFFMYFTFLYFTFYGMMSVAMTPNNHISIIVSSAFYSVWNLFSGFVLPRPGK**LISV 530
+ L ++ +F + + PN +S +V+ + S LFSGF + R
Sbjct: 488 YYLMIIFASFWSGCSFVTFVSGVIPNVMMSYMVTFGYLSYCLLFSGFYVNRD-------- 539
Query: 531 ILSIPVWWRWYSWANPVAWSLYGLVTSQYGD 561
I ++W W + + + + ++ +++ D
Sbjct: 540 --RIHLYWIWIHYISLLKYPYEAVLHNEFDD 568
>At3g52310 ABC transporter-like protein
Length = 737
Score = 173 bits (439), Expect = 2e-43
Identities = 130/472 (27%), Positives = 240/472 (50%), Gaps = 43/472 (9%)
Query: 19 VDMPQEMRNRGVIED-KLVLLKGLSGAFRPGVLTALMGVTGAGKTTLMDVLSGRKTGGYI 77
+D+ ++ +G+ + +L G+SG+ PG L ALMG +G+GKTTL++ L GR I
Sbjct: 148 IDITYKVTTKGMTSSSEKSILNGISGSAYPGELLALMGPSGSGKTTLLNALGGRFNQQNI 207
Query: 78 GGNITISGYPKKQETFARISGYCEQTDIHSPHVTVYESLLYSAWLRLSPDINAETRKMFI 137
GG+++ + P + RI G+ Q D+ PH+TV E+L Y+A LRL + + ++
Sbjct: 208 GGSVSYNDKPYSKHLKTRI-GFVTQDDVLFPHLTVKETLTYTALLRLPKTLTEQEKEQRA 266
Query: 138 EEVMELVELKPLRYALVGLPGVSGLSTEQRKRLTVAVELVANPSIIFMDEPTSGLDARAA 197
V++ + L+ + ++G V G+S +RKR+ + E++ NPS++ +DEPTS LD+ A
Sbjct: 267 ASVIQELGLERCQDTMIGGSFVRGVSGGERKRVCIGNEIMTNPSLLLLDEPTSSLDSTTA 326
Query: 198 AIVMRAVRNTVDTGRTVVCTIHQPSIDIFESFDELLLLKQGGQEIYVGPLGHNSSNLINY 257
+++ + G+T+V TIHQPS +F FD+L++L +G +Y G +S ++Y
Sbjct: 327 LKIVQMLHCIAKAGKTIVTTIHQPSSRLFHRFDKLVVLSRGSL-LYFG----KASEAMSY 381
Query: 258 FEGVQGVSKIKDGYNPATWMLEVTT---------SSKEVELRIDYAEVYKNS-------- 300
F + G S + NPA ++L++ S+ + +++I E+Y +
Sbjct: 382 FSSI-GCSPLL-AMNPAEFLLDLVNGNMNDISVPSALKEKMKIIRLELYVRNVKCDVETQ 439
Query: 301 ---ELYRRNKALIK--ELSAPAPCSKDLYF----PSR-YSRSFFTQ-CIACLWKQHWSYW 349
E Y+ A+++ +L AP P +++ P R + S++ Q C+ L +
Sbjct: 440 YLEEAYKTQIAVMEKMKLMAPVPLDEEVKLMITCPKREWGLSWWEQYCLLSL--RGIKER 497
Query: 350 RNPEYNAIRFLYSTAVAVLLGSMFWNLGSKIEKDQDLFNAMGSMYAAVILIGAMNSNSVQ 409
R+ ++ +R + A++LG ++W ++ G ++ + G +
Sbjct: 498 RHDYFSWLRVTQVLSTAIILGLLWWQSDITSQRP----TRSGLLFFIAVFWGFFPVFTAI 553
Query: 410 PVVGVERTVFYRERAARMYSAFPYALAQVVIQLPYVFVQAVVYGIVVYVMIG 461
ER + +ER + MY Y +A+ LP + V++ +VVY M G
Sbjct: 554 FTFPQERAMLSKERESNMYRLSAYFVARTTSDLPLDLILPVLFLVVVYFMAG 605
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,620,142
Number of Sequences: 26719
Number of extensions: 590928
Number of successful extensions: 2351
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1912
Number of HSP's gapped (non-prelim): 254
length of query: 619
length of database: 11,318,596
effective HSP length: 105
effective length of query: 514
effective length of database: 8,513,101
effective search space: 4375733914
effective search space used: 4375733914
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148607.16


