

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148532.8 + phase: 0
(380 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
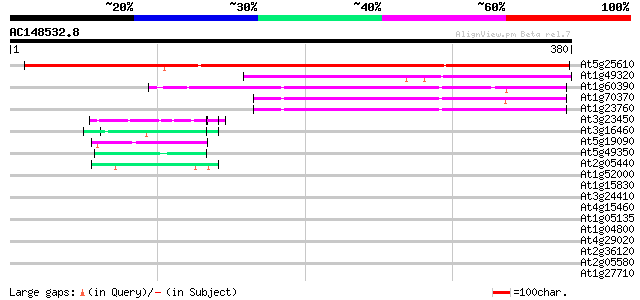
Score E
Sequences producing significant alignments: (bits) Value
At5g25610 dehydration-induced protein RD22 412 e-115
At1g49320 unknown protein 164 1e-40
At1g60390 140 1e-33
At1g70370 unknown protein 139 2e-33
At1g23760 putative polygalacuronase isoenzyme 1 beta subunit 130 1e-30
At3g23450 hypothetical protein 52 7e-07
At3g16460 putative jasmonate inducible protein (MDC8.9/AT3g16460) 43 3e-04
At5g19090 unknown protein 43 3e-04
At5g49350 unknown protein 42 7e-04
At2g05440 putative glycine-rich protein 42 7e-04
At1g52000 40 0.002
At1g15830 hypothetical protein 39 0.004
At3g24410 hypothetical protein 38 0.011
At4g15460 glycine-rich protein like, predicted GPI-anchored protein 37 0.018
At1g05135 unknown protein 37 0.023
At1g04800 unknown protein 36 0.031
At4g29020 glycine-rich protein like 36 0.040
At2g36120 unknown protein 36 0.040
At2g05580 unknown protein 36 0.040
At1g27710 unknown protein 36 0.040
>At5g25610 dehydration-induced protein RD22
Length = 392
Score = 412 bits (1058), Expect = e-115
Identities = 217/378 (57%), Positives = 272/378 (71%), Gaps = 11/378 (2%)
Query: 11 LVATHAALPPEVYWKSKLPTTQMPKAITDLLHLDWTEEKSTSVAVGKGGVNVDSGKTKPG 70
+VA A L PE YW + LP T +P ++ +LL D+T+EKST+V VGKGGVNV++ K K G
Sbjct: 16 VVAIAADLTPERYWSTALPNTPIPNSLHNLLTFDFTDEKSTNVQVGKGGVNVNTHKGKTG 75
Query: 71 -GTSVNVGKGGVHVDAGKTKPGG-TAVNVGKGGVH---VNAGKGKPGGGTRVNVGKGGVN 125
GT+VNVGKGGV VD GK KPGG T V+VG G H V GKPG T V VGKGGV
Sbjct: 76 SGTAVNVGKGGVRVDTGKGKPGGGTHVSVGSGKGHGGGVAVHTGKPGKRTDVGVGKGGVT 135
Query: 126 VHAGKGRGKPVHVSVG-NKSPFLYNYAASETQLHDKPNVALFFLEKDLHQGTKLNLQFTK 184
VH + +G+P++V V +PF+YNYAA ETQLHD PN ALFFLEKDL +G ++N++F
Sbjct: 136 VHT-RHKGRPIYVGVKPGANPFVYNYAAKETQLHDDPNAALFFLEKDLVRGKEMNVRFNA 194
Query: 185 TNS-NDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEESEIVKNTISECEENGIKGE 243
+ FLP+ A ++PF S K L FS++ G+EE+E++K TI ECE + GE
Sbjct: 195 EDGYGGKTAFLPRGEAETVPFGSEKFSETLKRFSVEAGSEEAEMMKKTIEECEARKVSGE 254
Query: 244 EKLCVTSLESMVDFTTSKLGN-NVEAVSTEVKKESSDLQEY-VMAKGVKKLGEKNKAVVC 301
EK C TSLESMVDF+ SKLG +V AVSTEV K+++ +Q+Y + A GVKKL + +K+VVC
Sbjct: 255 EKYCATSLESMVDFSVSKLGKYHVRAVSTEVAKKNAPMQKYKIAAAGVKKLSD-DKSVVC 313
Query: 302 HKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRVKAVAVCHTDTSQWNPKHLAFQVLNVQP 361
HK+ YP+AVFYCHK T VY+VPLEG +G R KAVAVCH +TS WNP HLAF+VL V+P
Sbjct: 314 HKQKYPFAVFYCHKAMMTTVYAVPLEGENGMRAKAVAVCHKNTSAWNPNHLAFKVLKVKP 373
Query: 362 GTVPVCHFLPQDHVVWVS 379
GTVPVCHFLP+ HVVW S
Sbjct: 374 GTVPVCHFLPETHVVWFS 391
>At1g49320 unknown protein
Length = 280
Score = 164 bits (414), Expect = 1e-40
Identities = 88/231 (38%), Positives = 136/231 (58%), Gaps = 10/231 (4%)
Query: 159 DKPNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSI 218
D P++ ++F DL GTKL + F K + L ++ A+ IPF+ SK++ +L+ FSI
Sbjct: 51 DDPSLYMYFTLNDLKLGTKLLIYFYKNDLQKLPPLLTRQQADLIPFTKSKLDFLLDHFSI 110
Query: 219 KEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVE--AVSTEVKKE 276
+ + + + +K T+ C+ I+GE K C TSLES++D +G NV+ ++T+V
Sbjct: 111 TKDSPQGKAIKETLGHCDAKAIEGEHKFCGTSLESLIDLVKKTMGYNVDLKVMTTKVMVP 170
Query: 277 SSD-----LQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCH-KTDSTKVYSVPLEGVD 330
+ + L Y + K+L K + CH+ YPYAV+YCH ++V+ V L D
Sbjct: 171 AQNSISYALHNYTFVEAPKEL-VGIKMLGCHRMPYPYAVYYCHGHKGGSRVFEVNLVTDD 229
Query: 331 G-SRVKAVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVWVSK 380
G RV AVCH DTS W+ H+AF+VL ++P + PVCHF P D++VWV+K
Sbjct: 230 GRQRVVGPAVCHMDTSTWDADHVAFKVLKMEPRSAPVCHFFPLDNIVWVTK 280
>At1g60390
Length = 624
Score = 140 bits (353), Expect = 1e-33
Identities = 95/289 (32%), Positives = 138/289 (46%), Gaps = 14/289 (4%)
Query: 95 VNVGKGGVHVNAGKGKPGGGTRVNVGKGGVNVHA-GKGRGKPVHVSVGNKSPFLYNYAAS 153
VN GK N G G + NVG G V+ G+G+ V+ G N +S
Sbjct: 340 VNYGKS---FNLGSDNFTGYGQDNVG-GNVSFKTYGQGQSFKVYTKDGVVFARYSNNVSS 395
Query: 154 ETQLHDK-PNVALFFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENI 212
+ +K FF E L +GT + + K + FLP+ + ++PFSSS + I
Sbjct: 396 NGKTVNKWVEEGKFFREAMLKEGTLMQMPDIK-DKMPKRTFLPRNIVKNLPFSSSTIGEI 454
Query: 213 LNLFSIKEGTEESEIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTE 272
+F E + + I+ + +SECE GE K CV S E M+DF TS LG V +TE
Sbjct: 455 WRVFGAGENSSMAGIISSAVSECERPASHGETKRCVGSAEDMIDFATSVLGRGVVVRTTE 514
Query: 273 VKKESSDLQEYVMAKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGS 332
S G+ G+ +AV CH+ YPY ++YCH +VY L +D
Sbjct: 515 NVVGSKKKVVIGKVNGING-GDVTRAVSCHQSLYPYLLYYCHSVPRVRVYETDL--LDPK 571
Query: 333 RVK----AVAVCHTDTSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVW 377
++ VA+CH DTS W+P H AF L PG + VCH++ ++ + W
Sbjct: 572 SLEKINHGVAICHIDTSAWSPSHGAFLALGSGPGQIEVCHWIFENDMTW 620
>At1g70370 unknown protein
Length = 626
Score = 139 bits (351), Expect = 2e-33
Identities = 74/214 (34%), Positives = 111/214 (51%), Gaps = 4/214 (1%)
Query: 166 FFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEES 225
FF E L +GT + + K + FLP+ + +PFS+SK+ I +F E +
Sbjct: 411 FFRESSLKEGTVIPMPDIK-DKMPKRSFLPRSIITKLPFSTSKLGEIKRIFHAVENSTMG 469
Query: 226 EIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVM 285
I+ + ++ECE GE K CV S E M+DF TS LG +V +TE S +
Sbjct: 470 GIITDAVTECERPPSVGETKRCVGSAEDMIDFATSVLGRSVVLRTTENVAGSKEKVVIGK 529
Query: 286 AKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVYSVPLEGVDGSRV--KAVAVCHTD 343
G+ G+ KAV CH+ YPY ++YCH +VY L ++ + +A+CH D
Sbjct: 530 VNGING-GKLTKAVSCHQSLYPYLLYYCHSVPKVRVYEADLLELNSKKKINHGIAICHMD 588
Query: 344 TSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVW 377
TS W P H AF L +PG + VCH++ ++ + W
Sbjct: 589 TSSWGPSHGAFLALGSKPGRIEVCHWIFENDMNW 622
>At1g23760 putative polygalacuronase isoenzyme 1 beta subunit
Length = 622
Score = 130 bits (327), Expect = 1e-30
Identities = 71/214 (33%), Positives = 111/214 (51%), Gaps = 4/214 (1%)
Query: 166 FFLEKDLHQGTKLNLQFTKTNSNDDAKFLPKEVANSIPFSSSKVENILNLFSIKEGTEES 225
FF E L +GT + + K + FLP+ + + +PFS+SK+ I +F + +
Sbjct: 407 FFRESMLKEGTLIWMPDIK-DKMPKRSFLPRSIVSKLPFSTSKIAEIKRVFHANDNSTME 465
Query: 226 EIVKNTISECEENGIKGEEKLCVTSLESMVDFTTSKLGNNVEAVSTEVKKESSDLQEYVM 285
I+ + + ECE E K CV S E M+DF TS LG +V +TE S +
Sbjct: 466 GIITDAVRECERPPTVSETKRCVGSAEDMIDFATSVLGRSVVLRTTESVAGSKEKVMIGK 525
Query: 286 AKGVKKLGEKNKAVVCHKESYPYAVFYCHKTDSTKVY-SVPLEGVDGSRVK-AVAVCHTD 343
G+ G K+V CH+ YPY ++YCH +VY S L+ +++ +A+CH D
Sbjct: 526 VNGING-GRVTKSVSCHQSLYPYLLYYCHSVPKVRVYESDLLDPKSKAKINHGIAICHMD 584
Query: 344 TSQWNPKHLAFQVLNVQPGTVPVCHFLPQDHVVW 377
TS W H AF +L +PG + VCH++ ++ + W
Sbjct: 585 TSAWGANHGAFMLLGSRPGQIEVCHWIFENDMNW 618
>At3g23450 hypothetical protein
Length = 452
Score = 51.6 bits (122), Expect = 7e-07
Identities = 36/87 (41%), Positives = 45/87 (51%), Gaps = 5/87 (5%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGG 114
+GKGG V G K GG +GKGG V G K GG +GKGG + G GK GGG
Sbjct: 204 IGKGG-GVGGGFGKGGGVGGGIGKGG-GVGGGFGKGGGVGGGIGKGG-GIGGGIGK-GGG 259
Query: 115 TRVNVGKGGVNVHAGKGRGKPVHVSVG 141
+GKGG + G G+G + +G
Sbjct: 260 IGGGIGKGG-GIGGGIGKGGGIGGGIG 285
Score = 51.6 bits (122), Expect = 7e-07
Identities = 36/87 (41%), Positives = 45/87 (51%), Gaps = 5/87 (5%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGG 114
+GKGG + G K GG +GKGG V G K GG +GKGG V G GK GGG
Sbjct: 184 IGKGG-GIGGGIGKGGGIGGGIGKGG-GVGGGFGKGGGVGGGIGKGG-GVGGGFGK-GGG 239
Query: 115 TRVNVGKGGVNVHAGKGRGKPVHVSVG 141
+GKGG + G G+G + +G
Sbjct: 240 VGGGIGKGG-GIGGGIGKGGGIGGGIG 265
Score = 49.7 bits (117), Expect = 3e-06
Identities = 35/79 (44%), Positives = 42/79 (52%), Gaps = 5/79 (6%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGG 114
+GKGG V G K GG +GKGG V G K GG +GKGG + G GK GGG
Sbjct: 134 IGKGG-GVGGGIGKGGGIGGGIGKGG-GVGGGIGKGGGIGGGIGKGG-GIGGGIGK-GGG 189
Query: 115 TRVNVGKGGVNVHAGKGRG 133
+GKGG + G G+G
Sbjct: 190 IGGGIGKGG-GIGGGIGKG 207
Score = 48.9 bits (115), Expect = 5e-06
Identities = 33/93 (35%), Positives = 43/93 (45%), Gaps = 3/93 (3%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGG-VHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG 113
+GKGG + G K GG +GKGG + G K GG +GKGG G GG
Sbjct: 294 IGKGG-GIGGGIGKGGGIGGGIGKGGGIGGGGGFGKGGGIGGGIGKGGGIGGGGGFGKGG 352
Query: 114 GTRVNVGKGGVNVHAGKGRGKPVHVSVGNKSPF 146
G +GKGG + G G+G + +G F
Sbjct: 353 GIGGGIGKGG-GIGGGFGKGGGIGGGIGGGGGF 384
Score = 46.2 bits (108), Expect = 3e-05
Identities = 33/80 (41%), Positives = 41/80 (51%), Gaps = 5/80 (6%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG- 113
+GKGG + G K GG +GKGG + G K GG +GKGG + G GK GG
Sbjct: 264 IGKGG-GIGGGIGKGGGIGGGIGKGG-GIGGGIGKGGGIGGGIGKGG-GIGGGIGKGGGI 320
Query: 114 GTRVNVGKGGVNVHAGKGRG 133
G GKGG + G G+G
Sbjct: 321 GGGGGFGKGG-GIGGGIGKG 339
Score = 43.9 bits (102), Expect = 1e-04
Identities = 31/81 (38%), Positives = 37/81 (45%), Gaps = 3/81 (3%)
Query: 55 VGKGG-VNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG 113
+GKGG + G K GG +GKGG + G K GG +G GG G GG
Sbjct: 336 IGKGGGIGGGGGFGKGGGIGGGIGKGG-GIGGGFGKGGGIGGGIGGGGGFGGGGGFGKGG 394
Query: 114 GTRVNVGKGGVNVHAGKGRGK 134
G +GKGG G G GK
Sbjct: 395 GIGGGIGKGG-GFGGGGGFGK 414
Score = 36.6 bits (83), Expect = 0.023
Identities = 32/82 (39%), Positives = 36/82 (43%), Gaps = 4/82 (4%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKT-KPGGTAVNVGK-GGVHVNAGKGKPG 112
+GKGG + G K GG +G GG G K GG +GK GG G GK G
Sbjct: 358 IGKGG-GIGGGFGKGGGIGGGIGGGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGG 416
Query: 113 G-GTRVNVGKGGVNVHAGKGRG 133
G G GKGG G G G
Sbjct: 417 GIGGGGGFGKGGGFGGGGFGGG 438
Score = 34.3 bits (77), Expect = 0.12
Identities = 32/89 (35%), Positives = 36/89 (39%), Gaps = 14/89 (15%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVH---VNAGKGKPG 112
G GG G GG G GG H G V G GG H V G GK G
Sbjct: 78 GGGGGGGGGGGGGGGGFGGGGGFGGGH---------GGGVGGGVGGGHGGGVGGGFGK-G 127
Query: 113 GGTRVNVGKGGVNVHAGKGRGKPVHVSVG 141
GG +GKGG V G G+G + +G
Sbjct: 128 GGIGGGIGKGG-GVGGGIGKGGGIGGGIG 155
Score = 34.3 bits (77), Expect = 0.12
Identities = 27/75 (36%), Positives = 29/75 (38%), Gaps = 5/75 (6%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPG---GTAVNVGKGGVHVNAGKGKPG 112
G GG G K GG +GKGG G G G GKGG G G G
Sbjct: 380 GGGGFGGGGGFGKGGGIGGGIGKGGGFGGGGGFGKGGGIGGGGGFGKGGGFGGGGFGGGG 439
Query: 113 GGTRVNVGKGGVNVH 127
GG G GG+ H
Sbjct: 440 GGG--GGGGGGIGHH 452
Score = 30.8 bits (68), Expect = 1.3
Identities = 28/77 (36%), Positives = 30/77 (38%), Gaps = 8/77 (10%)
Query: 47 EEKSTSVAVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNA 106
E++ T V GKGG G GG VG G AG GG G GG
Sbjct: 34 EDEKTLVGGGKGG---GFGGGFGGGAGGGVGGG-----AGGGFGGGAGGGFGGGGGGGGG 85
Query: 107 GKGKPGGGTRVNVGKGG 123
G G GGG G GG
Sbjct: 86 GGGGGGGGFGGGGGFGG 102
Score = 30.0 bits (66), Expect = 2.2
Identities = 25/73 (34%), Positives = 28/73 (38%)
Query: 61 NVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVG 120
+V SG+ + T V GKGG GG GG AG G GGG G
Sbjct: 27 DVSSGRDEDEKTLVGGGKGGGFGGGFGGGAGGGVGGGAGGGFGGGAGGGFGGGGGGGGGG 86
Query: 121 KGGVNVHAGKGRG 133
GG G G G
Sbjct: 87 GGGGGGGFGGGGG 99
Score = 29.6 bits (65), Expect = 2.9
Identities = 30/90 (33%), Positives = 34/90 (37%), Gaps = 8/90 (8%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGK------GGVHVNAGKGKP 111
GGV +G GG G GG G GG G GGV G G
Sbjct: 58 GGVGGGAGGGFGGGAGGGFGGGGGGGGGGGGGGGGGFGGGGGFGGGHGGGVGGGVGGGH- 116
Query: 112 GGGTRVNVGKGGVNVHAGKGRGKPVHVSVG 141
GGG GKGG + G G+G V +G
Sbjct: 117 GGGVGGGFGKGG-GIGGGIGKGGGVGGGIG 145
>At3g16460 putative jasmonate inducible protein (MDC8.9/AT3g16460)
Length = 705
Score = 43.1 bits (100), Expect = 3e-04
Identities = 30/87 (34%), Positives = 31/87 (35%), Gaps = 5/87 (5%)
Query: 51 TSVAVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGK-TKPG---GTAVNVGKGGVHVNA 106
T G GG +G T GGT G GG G T G GT G GG
Sbjct: 160 TGTGTGTGGTGTGTG-TGTGGTGTGTGTGGTGTGTGTGTGTGTGTGTGTGTGTGGTGTGT 218
Query: 107 GKGKPGGGTRVNVGKGGVNVHAGKGRG 133
G G G GT G G G G G
Sbjct: 219 GTGGTGTGTGTGTGTGTGTGGTGTGTG 245
Score = 41.6 bits (96), Expect = 7e-04
Identities = 26/81 (32%), Positives = 28/81 (34%), Gaps = 3/81 (3%)
Query: 62 VDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVG 120
+ +G T GGT G G G G GT G GG G G GGT G
Sbjct: 128 IPTGGTGTGGTGTGTGTGTGTGTGTGTGTGTGTGTGTGTGGTGTGTGTGT--GGTGTGTG 185
Query: 121 KGGVNVHAGKGRGKPVHVSVG 141
GG G G G G
Sbjct: 186 TGGTGTGTGTGTGTGTGTGTG 206
Score = 35.4 bits (80), Expect = 0.052
Identities = 26/80 (32%), Positives = 27/80 (33%), Gaps = 5/80 (6%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGG 114
G GG +G GT G G G G GGT G G G G G G
Sbjct: 134 GTGGTGTGTGTGTGTGTGTGTGTGTGTGTGTGT---GGTGTGTGTGTGGTGTGTGTGGTG 190
Query: 115 TRVNVGKG-GVNVHAGKGRG 133
T G G G G G G
Sbjct: 191 TGTGTGTGTGTGTGTGTGTG 210
Score = 31.2 bits (69), Expect = 0.98
Identities = 21/68 (30%), Positives = 24/68 (34%), Gaps = 6/68 (8%)
Query: 51 TSVAVGKG-GVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKG 109
T G G G +G T GGT G GG G GT G GG G G
Sbjct: 193 TGTGTGTGTGTGTGTG-TGTGGTGTGTGTGGTGTGTGT----GTGTGTGTGGTGTGTGTG 247
Query: 110 KPGGGTRV 117
G ++
Sbjct: 248 TGSGAQKL 255
>At5g19090 unknown protein
Length = 587
Score = 42.7 bits (99), Expect = 3e-04
Identities = 34/84 (40%), Positives = 39/84 (45%), Gaps = 7/84 (8%)
Query: 56 GKG---GVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGV-HVNAGK-GK 110
GKG V + G PGG G GG ++ G GG N GKGG H GK G
Sbjct: 303 GKGMPFPVQMGGGGGGPGGKKGGPGGGGGNM--GNQNQGGGGKNGGKGGGGHPLDGKMGG 360
Query: 111 PGGGTRVNVGKGGVNVHAGKGRGK 134
GGG N G GGV ++ G GK
Sbjct: 361 GGGGPNGNKGGGGVQMNGGPNGGK 384
Score = 38.5 bits (88), Expect = 0.006
Identities = 28/69 (40%), Positives = 30/69 (42%), Gaps = 2/69 (2%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGV-HVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGG 114
G GG + G GG N GKGG H GK GG N KGG V G P GG
Sbjct: 325 GPGGGGGNMGNQNQGGGGKNGGKGGGGHPLDGKMGGGGGGPNGNKGGGGVQM-NGGPNGG 383
Query: 115 TRVNVGKGG 123
+ G GG
Sbjct: 384 KKGGGGGGG 392
Score = 29.3 bits (64), Expect = 3.7
Identities = 23/65 (35%), Positives = 27/65 (41%), Gaps = 11/65 (16%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G GG +D GK GG N KGG GG +N G G G G GGG
Sbjct: 348 GGGGHPLD-GKMGGGGGGPNGNKGG----------GGVQMNGGPNGGKKGGGGGGGGGGG 396
Query: 116 RVNVG 120
++ G
Sbjct: 397 PMSGG 401
>At5g49350 unknown protein
Length = 302
Score = 41.6 bits (96), Expect = 7e-04
Identities = 25/76 (32%), Positives = 31/76 (39%), Gaps = 4/76 (5%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
G + ++ GG +G GG G T GG G GG+ G G GGG
Sbjct: 27 GNTYSEEDDSEVGGEGGGIGGGGTGFGGGGTGVGGGGTGFGGGGL----GAGGSGGGGAG 82
Query: 118 NVGKGGVNVHAGKGRG 133
+G GG V G G G
Sbjct: 83 GLGSGGTGVGGGGGLG 98
Score = 40.4 bits (93), Expect = 0.002
Identities = 32/93 (34%), Positives = 40/93 (42%), Gaps = 10/93 (10%)
Query: 45 WTEEKSTSVAVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGT----AVNVGKG 100
++EE + V G+GG + G T GG VG GG G GG+ A +G G
Sbjct: 30 YSEEDDSEVG-GEGG-GIGGGGTGFGGGGTGVGGGGTGFGGGGLGAGGSGGGGAGGLGSG 87
Query: 101 GVHVNA----GKGKPGGGTRVNVGKGGVNVHAG 129
G V G G GGG +G GG V G
Sbjct: 88 GTGVGGGGGLGAGGSGGGGAGGLGGGGTGVGGG 120
Score = 33.9 bits (76), Expect = 0.15
Identities = 30/83 (36%), Positives = 34/83 (40%), Gaps = 7/83 (8%)
Query: 55 VGKGGVNVDSG-----KTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKG 109
+G GG V G GG + +G GG V G T GGT N G G G G
Sbjct: 84 LGSGGTGVGGGGGLGAGGSGGGGAGGLGGGGTGVGGGGT-GGGTGFNGGGTGFGPFGGGG 142
Query: 110 -KPGGGTRVNVGKGGVNVHAGKG 131
GGG +G GG N G G
Sbjct: 143 LGTGGGGAGGLGGGGGNGGFGGG 165
Score = 29.3 bits (64), Expect = 3.7
Identities = 26/70 (37%), Positives = 28/70 (39%), Gaps = 6/70 (8%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVG-KGGVHVNAGKGKPGG 113
+G GG V G T GGT N G G G GG G GG+ G G GG
Sbjct: 110 LGGGGTGVGGGGTG-GGTGFNGGGTGFGPFGG----GGLGTGGGGAGGLGGGGGNGGFGG 164
Query: 114 GTRVNVGKGG 123
G G GG
Sbjct: 165 GGLGGGGLGG 174
>At2g05440 putative glycine-rich protein
Length = 154
Score = 41.6 bits (96), Expect = 7e-04
Identities = 33/94 (35%), Positives = 36/94 (38%), Gaps = 8/94 (8%)
Query: 56 GKGGVNVDSGKTKPG--GTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG 113
G GG N G G G + G GG H G GG + G GG H G G GG
Sbjct: 60 GHGGHNGGGGHGLDGYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGHYGGGGGGHGG 119
Query: 114 GTRVNVGKGGV----NVHAGKGRG--KPVHVSVG 141
G G GG H G G G +PV G
Sbjct: 120 GGHYGGGGGGYGGGGGHHGGGGHGLNEPVQTKPG 153
>At1g52000
Length = 730
Score = 40.4 bits (93), Expect = 0.002
Identities = 28/98 (28%), Positives = 38/98 (38%), Gaps = 2/98 (2%)
Query: 50 STSVAVGKGGVNVDSGKTKPGGTSVNV-GKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGK 108
ST G G+ DSG GT+ G +AG +KP + G V G+
Sbjct: 132 STGAKPGASGIGSDSGSIGSAGTNPGADGTRETEKNAGGSKPSSGSAGTNPGASAVGNGE 191
Query: 109 -GKPGGGTRVNVGKGGVNVHAGKGRGKPVHVSVGNKSP 145
K GG++ + G G N A G +VG P
Sbjct: 192 TEKNAGGSKPSSGSAGTNPGASAGGNGETEKNVGGSKP 229
Score = 37.7 bits (86), Expect = 0.011
Identities = 29/91 (31%), Positives = 39/91 (41%), Gaps = 5/91 (5%)
Query: 54 AVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKP--GGTAVNVGKGGVHVNAGKGKP 111
AVG G ++G +KP S G G+T+ GG+ + GK G + A G
Sbjct: 186 AVGNGETEKNAGGSKPSSGSAGTNPGASAGGNGETEKNVGGSKPSSGKAGTNPGANAG-G 244
Query: 112 GGGTRVNVGKGGVNVHAGKGRGKPVHVSVGN 142
GGT N GG +G R P + GN
Sbjct: 245 NGGTEKNA--GGSKSSSGSARTNPGASAGGN 273
Score = 35.8 bits (81), Expect = 0.040
Identities = 25/86 (29%), Positives = 30/86 (34%), Gaps = 12/86 (13%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGK------------GGVH 103
GK G N + GGT N G + +T PG +A G+ GG
Sbjct: 232 GKAGTNPGANAGGNGGTEKNAGGSKSSSGSARTNPGASAGGNGETVSNIGDTESNAGGSK 291
Query: 104 VNAGKGKPGGGTRVNVGKGGVNVHAG 129
N G G N G G N AG
Sbjct: 292 SNDGANNGASGIESNAGSTGTNFGAG 317
Score = 35.0 bits (79), Expect = 0.068
Identities = 28/100 (28%), Positives = 41/100 (41%), Gaps = 18/100 (18%)
Query: 50 STSVAVGKGGVNV----------DSGKTKPGGTSVNVGKGGVHVDAGKTK--PGGTAVNV 97
S S ++G G N ++G +KP S G V G+T+ GG+ +
Sbjct: 144 SDSGSIGSAGTNPGADGTRETEKNAGGSKPSSGSAGTNPGASAVGNGETEKNAGGSKPSS 203
Query: 98 GKGGVHVNAGKGKPG------GGTRVNVGKGGVNVHAGKG 131
G G + A G G GG++ + GK G N A G
Sbjct: 204 GSAGTNPGASAGGNGETEKNVGGSKPSSGKAGTNPGANAG 243
Score = 32.3 bits (72), Expect = 0.44
Identities = 18/65 (27%), Positives = 26/65 (39%), Gaps = 1/65 (1%)
Query: 61 NVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVG 120
N+ ++ GG+ N G ++ G T N G GG GG++ N G
Sbjct: 279 NIGDTESNAGGSKSNDGANN-GASGIESNAGSTGTNFGAGGTGGIGDTESDAGGSKTNSG 337
Query: 121 KGGVN 125
GG N
Sbjct: 338 NGGTN 342
Score = 32.0 bits (71), Expect = 0.58
Identities = 25/93 (26%), Positives = 34/93 (35%), Gaps = 7/93 (7%)
Query: 46 TEEKSTSVAVGKGG---VNVDSGKTKPGGTSVNVGKGGVHVDAGK--TKPGGTAVNVGKG 100
+ + ST G GG N++ + GG N G G+ G T P GT N
Sbjct: 350 SNDGSTGTNPGAGGGTDSNIEGTENNVGGKETNPGASGIGNSDGSTGTSPEGTESNA--D 407
Query: 101 GVHVNAGKGKPGGGTRVNVGKGGVNVHAGKGRG 133
G N G + G+ N + A G G
Sbjct: 408 GTKTNTGGKESNTGSESNTNSSPQKLEAQGGNG 440
>At1g15830 hypothetical protein
Length = 483
Score = 39.3 bits (90), Expect = 0.004
Identities = 32/86 (37%), Positives = 38/86 (43%), Gaps = 9/86 (10%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPG--GGT 115
GGVNVD GG+ V G GG + T GG G V + G+ G GG
Sbjct: 3 GGVNVD---WSIGGSMVTSGGGGCPM----TISGGRLFTGGGDPVIITGGRLSTGAAGGV 55
Query: 116 RVNVGKGGVNVHAGKGRGKPVHVSVG 141
V+ KGG G G G PV +S G
Sbjct: 56 NVDRSKGGGRRKTGDGGGDPVVISGG 81
Score = 32.3 bits (72), Expect = 0.44
Identities = 22/70 (31%), Positives = 28/70 (39%), Gaps = 10/70 (14%)
Query: 54 AVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG 113
A G G + G T GG + N G G G + VG G+ G G+ G
Sbjct: 297 AEGGGRGSTGEGVTDGGGRTGNKGGNG----------GSIKIGVGTNGITGGTGGGEAGA 346
Query: 114 GTRVNVGKGG 123
G +V G GG
Sbjct: 347 GMQVMQGWGG 356
Score = 30.4 bits (67), Expect = 1.7
Identities = 27/86 (31%), Positives = 37/86 (42%), Gaps = 12/86 (13%)
Query: 58 GGVNVDSG-KTKP----GGTSVNVGKGGVHVD----AGKTKPGGTAVNVGKG---GVHVN 105
GGV V+ G +T P GG G+GG + G+ GG + G+G G
Sbjct: 257 GGVIVNGGCETVPPGRGGGGDKTNGRGGEGREEDNGGGRGAEGGGRGSTGEGVTDGGGRT 316
Query: 106 AGKGKPGGGTRVNVGKGGVNVHAGKG 131
KG GG ++ VG G+ G G
Sbjct: 317 GNKGGNGGSIKIGVGTNGITGGTGGG 342
>At3g24410 hypothetical protein
Length = 240
Score = 37.7 bits (86), Expect = 0.011
Identities = 29/78 (37%), Positives = 37/78 (47%), Gaps = 2/78 (2%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKP-GGTAVNVGKGGV-HVNAGKGKPGGGT 115
GGV V+ G GG G+GG + K + GG V G+G + G+G G G
Sbjct: 82 GGVGVEGGGGDLGGVEGVAGEGGGGEEGLKGEDGGGEGVKGGEGMLGDGGGGEGVVGDGG 141
Query: 116 RVNVGKGGVNVHAGKGRG 133
R + KGG V AG G G
Sbjct: 142 RGDGVKGGEGVMAGGGEG 159
Score = 34.7 bits (78), Expect = 0.089
Identities = 26/74 (35%), Positives = 31/74 (41%), Gaps = 7/74 (9%)
Query: 70 GGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGK-------G 122
GG + GGV V+ G GG G+GG KG+ GGG V G+ G
Sbjct: 73 GGLRLPTRGGGVGVEGGGGDLGGVEGVAGEGGGGEEGLKGEDGGGEGVKGGEGMLGDGGG 132
Query: 123 GVNVHAGKGRGKPV 136
G V GRG V
Sbjct: 133 GEGVVGDGGRGDGV 146
Score = 33.9 bits (76), Expect = 0.15
Identities = 29/77 (37%), Positives = 33/77 (42%), Gaps = 10/77 (12%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKG-----GVHVDAGKTKP----GGTAVNVGKGGVHVNA 106
G GV G+ GG V VG+G GV D G+ + GG V G GG V
Sbjct: 148 GGEGVMAGGGEGVKGGEGV-VGEGVKGGEGVVGDGGRGEGVKGGGGEGVEGGIGGTVVGG 206
Query: 107 GKGKPGGGTRVNVGKGG 123
G G GG V G GG
Sbjct: 207 GGGGRAGGRAVGGGGGG 223
Score = 29.6 bits (65), Expect = 2.9
Identities = 26/74 (35%), Positives = 28/74 (37%), Gaps = 4/74 (5%)
Query: 59 GVNVDSGKTKPGGTSVNV-GKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GV G GG V G GG V+ G GGT V G GG G GGG
Sbjct: 170 GVKGGEGVVGDGGRGEGVKGGGGEGVEGGI---GGTVVGGGGGGRAGGRAVGGGGGGRAG 226
Query: 118 NVGKGGVNVHAGKG 131
GG G+G
Sbjct: 227 GRAVGGGGGVTGRG 240
>At4g15460 glycine-rich protein like, predicted GPI-anchored protein
Length = 148
Score = 37.0 bits (84), Expect = 0.018
Identities = 27/74 (36%), Positives = 31/74 (41%), Gaps = 4/74 (5%)
Query: 60 VNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNV 119
VN + K GG + G GGVHV G + GG H + G G GG V
Sbjct: 29 VNCTTVVAKGGGGGAH-GGGGVHVSVGGAHASVGGGHASGGGGHASVGGGHASGGGGHAV 87
Query: 120 GKGGVNVHAGKGRG 133
GG HAG G G
Sbjct: 88 EGGG---HAGGGGG 98
Score = 36.6 bits (83), Expect = 0.023
Identities = 28/85 (32%), Positives = 32/85 (36%), Gaps = 5/85 (5%)
Query: 50 STSVAVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKG 109
+T VA G GG G GG V+VG V G GG +VG G G
Sbjct: 32 TTVVAKGGGG-----GAHGGGGVHVSVGGAHASVGGGHASGGGGHASVGGGHASGGGGHA 86
Query: 110 KPGGGTRVNVGKGGVNVHAGKGRGK 134
GGG G G G G G+
Sbjct: 87 VEGGGHAGGGGGGHGEEEGGHGIGR 111
Score = 29.3 bits (64), Expect = 3.7
Identities = 18/45 (40%), Positives = 20/45 (44%), Gaps = 4/45 (8%)
Query: 97 VGKGGVHVNAGKGKPGGGTRVNVGKGGVNVHAGKGRGKPVHVSVG 141
V KGG G GGG V+VG +V G G H SVG
Sbjct: 35 VAKGG----GGGAHGGGGVHVSVGGAHASVGGGHASGGGGHASVG 75
>At1g05135 unknown protein
Length = 384
Score = 36.6 bits (83), Expect = 0.023
Identities = 32/87 (36%), Positives = 33/87 (37%), Gaps = 10/87 (11%)
Query: 54 AVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKP-- 111
A G GG V G GG G GG AG GG A G GGV +G G
Sbjct: 138 AGGGGGGGVGGGSGHGGGFGAGGGVGG---GAGGIGGGGGAGGGGGGGVGGGSGHGSGFG 194
Query: 112 -----GGGTRVNVGKGGVNVHAGKGRG 133
GGG VG GG G G G
Sbjct: 195 AGGGIGGGAGGGVGGGGGGGGGGGGGG 221
Score = 36.2 bits (82), Expect = 0.031
Identities = 27/76 (35%), Positives = 33/76 (42%), Gaps = 5/76 (6%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GGV +G GG + G GGV +G G+ G GG+ AG G GGG
Sbjct: 160 GGVGGGAGGIGGGGGAGGGGGGGVGGGSGH----GSGFGAG-GGIGGGAGGGVGGGGGGG 214
Query: 118 NVGKGGVNVHAGKGRG 133
G GG + G G G
Sbjct: 215 GGGGGGGGANGGSGHG 230
Score = 35.8 bits (81), Expect = 0.040
Identities = 28/76 (36%), Positives = 31/76 (39%), Gaps = 4/76 (5%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GGV +G GG G GG + G GG G GG AG G GGG
Sbjct: 84 GGVGGGAGGGLGGGGGAGGGGGG-GIGGGSGHGGGFGAGGGVGG---GAGGGIGGGGGAG 139
Query: 118 NVGKGGVNVHAGKGRG 133
G GGV +G G G
Sbjct: 140 GGGGGGVGGGSGHGGG 155
Score = 35.0 bits (79), Expect = 0.068
Identities = 29/81 (35%), Positives = 32/81 (38%), Gaps = 3/81 (3%)
Query: 54 AVGKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKP-G 112
A G GG + G GG G GG G GG A G GGV +G G G
Sbjct: 100 AGGGGGGGIGGGSGHGGGFGAGGGVGGGA--GGGIGGGGGAGGGGGGGVGGGSGHGGGFG 157
Query: 113 GGTRVNVGKGGVNVHAGKGRG 133
G V G GG+ G G G
Sbjct: 158 AGGGVGGGAGGIGGGGGAGGG 178
Score = 33.9 bits (76), Expect = 0.15
Identities = 30/82 (36%), Positives = 32/82 (38%), Gaps = 12/82 (14%)
Query: 53 VAVGKGGVNVDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKP 111
V G GG+ G GG V G G G AG GG VG GG G G
Sbjct: 162 VGGGAGGIGGGGGAGGGGGGGVGGGSGHGSGFGAGGGIGGGAGGGVGGGG-----GGGGG 216
Query: 112 GGGTRVNVGKGGVNVHAGKGRG 133
GG G GG N +G G G
Sbjct: 217 GG------GGGGANGGSGHGSG 232
Score = 33.5 bits (75), Expect = 0.20
Identities = 30/85 (35%), Positives = 31/85 (36%), Gaps = 10/85 (11%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGK---------GGVHVNAGK 108
GG SG GG VG GG G GG N G GGV G
Sbjct: 225 GGSGHGSGFGAGGGVGGGVG-GGAGGGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGG 283
Query: 109 GKPGGGTRVNVGKGGVNVHAGKGRG 133
G GGG G GGV +G G G
Sbjct: 284 GAGGGGGGGGGGGGGVGGGSGHGGG 308
Score = 33.5 bits (75), Expect = 0.20
Identities = 29/76 (38%), Positives = 32/76 (41%), Gaps = 10/76 (13%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GGV G GG G GG + +G G VG GGV AG G GGG
Sbjct: 205 GGVGGGGGGGGGGG-----GGGGANGGSGHGSGFGAGGGVG-GGVGGGAGGGGGGGGG-- 256
Query: 118 NVGKGGVNVHAGKGRG 133
G GG N +G G G
Sbjct: 257 --GGGGANGGSGHGSG 270
Score = 33.1 bits (74), Expect = 0.26
Identities = 31/93 (33%), Positives = 34/93 (36%), Gaps = 10/93 (10%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGK---------GGVHVNAGK 108
GG SG GG VG GG G GG V G GG+ AG
Sbjct: 263 GGSGHGSGFGAGGGVGGGVG-GGAGGGGGGGGGGGGGVGGGSGHGGGFGAGGGLGGGAGG 321
Query: 109 GKPGGGTRVNVGKGGVNVHAGKGRGKPVHVSVG 141
G GGG G GG+ G G G V +G
Sbjct: 322 GLGGGGGAGGGGGGGLGHGGGVGGGHGGGVGIG 354
Score = 33.1 bits (74), Expect = 0.26
Identities = 27/84 (32%), Positives = 33/84 (39%), Gaps = 4/84 (4%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GGV +G GG G GG G GG GG+ G G GGG
Sbjct: 279 GGVGGGAGGGGGGGGGGGGGVGGGSGHGGGFGAGGGLGGGAGGGLGGGGGAGGGGGG--- 335
Query: 118 NVGKGGVNVHAGKGRGKPVHVSVG 141
+G GG V G G G + + +G
Sbjct: 336 GLGHGG-GVGGGHGGGVGIGIGIG 358
Score = 32.7 bits (73), Expect = 0.34
Identities = 28/79 (35%), Positives = 32/79 (40%), Gaps = 4/79 (5%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG G+ GG G+GG AG GG +G GG G G GGG+
Sbjct: 56 GGGGFGGGRGSGGGIGGGGGQGG-GFGAGGGVGGGAGGGLGGGGGAGGGGGGGIGGGSGH 114
Query: 118 NVG---KGGVNVHAGKGRG 133
G GGV AG G G
Sbjct: 115 GGGFGAGGGVGGGAGGGIG 133
Score = 32.7 bits (73), Expect = 0.34
Identities = 31/88 (35%), Positives = 33/88 (37%), Gaps = 4/88 (4%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTR 116
GG G GG V G G G AG GG +G GG G G G G
Sbjct: 283 GGAGGGGGGGGGGGGGVGGGSGHGGGFGAGGGLGGGAGGGLGGGGGAGGGGGGGLGHGGG 342
Query: 117 VNVGKGGVNVHAGKGRGKPVHVSVGNKS 144
V G GG G G G + V VG S
Sbjct: 343 VGGGHGG---GVGIGIGIGIGVGVGGGS 367
Score = 32.3 bits (72), Expect = 0.44
Identities = 29/76 (38%), Positives = 31/76 (40%), Gaps = 5/76 (6%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GGV +G GG G GG V G GG G GG AG G GGG
Sbjct: 122 GGVGGGAGGGIGGGGGAGGGGGG-GVGGGSGHGGGFGAGGGVGG---GAG-GIGGGGGAG 176
Query: 118 NVGKGGVNVHAGKGRG 133
G GGV +G G G
Sbjct: 177 GGGGGGVGGGSGHGSG 192
Score = 32.0 bits (71), Expect = 0.58
Identities = 30/88 (34%), Positives = 32/88 (36%), Gaps = 11/88 (12%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKG---GVHVDAGKTKPGGTAVNVGKG-------GVHVN 105
G GG+ SG G VG G G+ G GG V G G G V
Sbjct: 104 GGGGIGGGSGHGGGFGAGGGVGGGAGGGIGGGGGAGGGGGGGVGGGSGHGGGFGAGGGVG 163
Query: 106 AGKGKPGGGTRVNVGKGGVNVHAGKGRG 133
G G GGG G GG V G G G
Sbjct: 164 GGAGGIGGGGGAG-GGGGGGVGGGSGHG 190
Score = 32.0 bits (71), Expect = 0.58
Identities = 24/69 (34%), Positives = 29/69 (41%), Gaps = 4/69 (5%)
Query: 65 GKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGKGGV 124
G+ GG G GG + G + GG G GG AG G GGG G GG+
Sbjct: 53 GRGGGGGFGGGRGSGG-GIGGGGGQGGGFGAGGGVGG---GAGGGLGGGGGAGGGGGGGI 108
Query: 125 NVHAGKGRG 133
+G G G
Sbjct: 109 GGGSGHGGG 117
Score = 32.0 bits (71), Expect = 0.58
Identities = 28/80 (35%), Positives = 30/80 (37%), Gaps = 4/80 (5%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTR 116
GG G GG N G G G AG GG G GG G G GGG+
Sbjct: 245 GGAGGGGGGGGGGGGGANGGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGGGVGGGSG 304
Query: 117 VNVG---KGGVNVHAGKGRG 133
G GG+ AG G G
Sbjct: 305 HGGGFGAGGGLGGGAGGGLG 324
Score = 30.8 bits (68), Expect = 1.3
Identities = 31/81 (38%), Positives = 33/81 (40%), Gaps = 8/81 (9%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKG---KPG 112
G GG N SG G VG GGV AG GG G GGV +G G G
Sbjct: 257 GGGGANGGSGHGSGFGAGGGVG-GGVGGGAGGGGGGGGG---GGGGVGGGSGHGGGFGAG 312
Query: 113 GGTRVNVGKGGVNVHAGKGRG 133
GG G GG+ G G G
Sbjct: 313 GGLGGGAG-GGLGGGGGAGGG 332
Score = 29.3 bits (64), Expect = 3.7
Identities = 29/81 (35%), Positives = 30/81 (36%), Gaps = 5/81 (6%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVGKG-GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG 113
VG GG G GG N G G G AG GG G GG G G G
Sbjct: 207 VGGGGGGGGGGG---GGGGANGGSGHGSGFGAGGGVGGGVGGGAGGGGGGGGGGGGGANG 263
Query: 114 GTRVNVGKG-GVNVHAGKGRG 133
G+ G G G V G G G
Sbjct: 264 GSGHGSGFGAGGGVGGGVGGG 284
Score = 28.9 bits (63), Expect = 4.9
Identities = 27/85 (31%), Positives = 28/85 (32%), Gaps = 11/85 (12%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG-- 113
G GGV SG G +G G G GG G GG N G G G
Sbjct: 179 GGGGVGGGSGHGSGFGAGGGIGGGA----GGGVGGGGGGGGGGGGGGGANGGSGHGSGFG 234
Query: 114 -----GTRVNVGKGGVNVHAGKGRG 133
G V G GG G G G
Sbjct: 235 AGGGVGGGVGGGAGGGGGGGGGGGG 259
Score = 28.1 bits (61), Expect = 8.3
Identities = 32/91 (35%), Positives = 37/91 (40%), Gaps = 9/91 (9%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G+GG +G GG +G GG AG GG G GG AG G GGG
Sbjct: 75 GQGG-GFGAGGGVGGGAGGGLGGGG---GAGGGGGGGIGGGSGHGG-GFGAGGGV-GGGA 128
Query: 116 RVNVGKGGVNVHAGKGRGKPVHVSVGNKSPF 146
+G GG AG G G V G+ F
Sbjct: 129 GGGIGGGG---GAGGGGGGGVGGGSGHGGGF 156
>At1g04800 unknown protein
Length = 200
Score = 36.2 bits (82), Expect = 0.031
Identities = 32/81 (39%), Positives = 36/81 (43%), Gaps = 8/81 (9%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKG---GVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPG 112
G GG +GK GG V G GV AGK GG VGKG V AGKG G
Sbjct: 91 GGGGFGGGAGKGVDGGFGKGVDGGAGKGVDGGAGKGFDGG----VGKG-VDGGAGKGFDG 145
Query: 113 GGTRVNVGKGGVNVHAGKGRG 133
G + G G + G G+G
Sbjct: 146 GVGKGFEGGIGKGIEGGVGKG 166
Score = 33.5 bits (75), Expect = 0.20
Identities = 26/64 (40%), Positives = 28/64 (43%), Gaps = 5/64 (7%)
Query: 70 GGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGKGGVNVHAG 129
GG G GG AGK GG GKG V AGKG GG + G G V G
Sbjct: 84 GGIGGGFGGGGFGGGAGKGVDGG----FGKG-VDGGAGKGVDGGAGKGFDGGVGKGVDGG 138
Query: 130 KGRG 133
G+G
Sbjct: 139 AGKG 142
Score = 31.6 bits (70), Expect = 0.75
Identities = 29/78 (37%), Positives = 34/78 (43%), Gaps = 11/78 (14%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGG-G 114
G GG++ GG G G + G GG G GG AGKG GG G
Sbjct: 58 GGGGIS--------GGGGFGAGGGWIGGSVGGFG-GGIGGGFGGGGFGGGAGKGVDGGFG 108
Query: 115 TRVNVGKG-GVNVHAGKG 131
V+ G G GV+ AGKG
Sbjct: 109 KGVDGGAGKGVDGGAGKG 126
Score = 28.1 bits (61), Expect = 8.3
Identities = 21/64 (32%), Positives = 25/64 (38%), Gaps = 5/64 (7%)
Query: 75 NVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGKG-----GVNVHAG 129
++G GG G GG + GG G G GGG GKG G V G
Sbjct: 55 DLGGGGGISGGGGFGAGGGWIGGSVGGFGGGIGGGFGGGGFGGGAGKGVDGGFGKGVDGG 114
Query: 130 KGRG 133
G+G
Sbjct: 115 AGKG 118
>At4g29020 glycine-rich protein like
Length = 158
Score = 35.8 bits (81), Expect = 0.040
Identities = 29/83 (34%), Positives = 35/83 (41%), Gaps = 4/83 (4%)
Query: 55 VGKGGVNVDSGKTKPGGTSVNVG--KGGVHVDAGKTKPGGTAVNVGK--GGVHVNAGKGK 110
V GV +G GG +G GGV AG GG +G GGV G G
Sbjct: 41 VAFAGVGGAAGIGGAGGVGAGLGGVAGGVGGVAGVLPVGGVGGGIGGLGGGVGGLGGLGG 100
Query: 111 PGGGTRVNVGKGGVNVHAGKGRG 133
GGG+ + G GG+ G G G
Sbjct: 101 LGGGSGLGHGVGGIGGDPGIGSG 123
>At2g36120 unknown protein
Length = 255
Score = 35.8 bits (81), Expect = 0.040
Identities = 26/78 (33%), Positives = 28/78 (35%), Gaps = 3/78 (3%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G GG G + GG G+ G H G GG G GG H G G G
Sbjct: 99 GPGG-GYGGGSGEGGGAGYGGGEAGGHGGGGGGGAGGGGG--GGGGAHGGGYGGGQGAGA 155
Query: 116 RVNVGKGGVNVHAGKGRG 133
G GG H G G G
Sbjct: 156 GGGYGGGGAGGHGGGGGG 173
Score = 33.5 bits (75), Expect = 0.20
Identities = 25/74 (33%), Positives = 27/74 (35%), Gaps = 10/74 (13%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G GG G GG G GG G + GG G+ G H G G GGG
Sbjct: 82 GGGGGGHGGGAGGGGGGGPGGGYGG-----GSGEGGGAGYGGGEAGGHGGGGGGGAGGG- 135
Query: 116 RVNVGKGGVNVHAG 129
G GG H G
Sbjct: 136 ----GGGGGGAHGG 145
Score = 32.0 bits (71), Expect = 0.58
Identities = 24/73 (32%), Positives = 31/73 (41%), Gaps = 2/73 (2%)
Query: 62 VDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRVNVGK 121
V+SG + G + G GG G + GG A G+G H+ G G GG G
Sbjct: 40 VNSGLSAGLGVGIGGGPGGGSGYGGGSGEGGGAGGHGEG--HIGGGGGGGHGGGAGGGGG 97
Query: 122 GGVNVHAGKGRGK 134
GG G G G+
Sbjct: 98 GGPGGGYGGGSGE 110
Score = 32.0 bits (71), Expect = 0.58
Identities = 25/76 (32%), Positives = 28/76 (35%), Gaps = 6/76 (7%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG +G GG + G GG G GG G+GG H G G GG
Sbjct: 149 GGQGAGAGGGYGGGGAGGHGGGG-----GGGNGGGGGGGSGEGGAH-GGGYGAGGGAGEG 202
Query: 118 NVGKGGVNVHAGKGRG 133
G G H G G G
Sbjct: 203 YGGGAGAGGHGGGGGG 218
Score = 30.8 bits (68), Expect = 1.3
Identities = 26/76 (34%), Positives = 27/76 (35%), Gaps = 5/76 (6%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG G GG G+GG G GG A G GG G G GGG
Sbjct: 90 GGAGGGGGGGPGGGYGGGSGEGG-----GAGYGGGEAGGHGGGGGGGAGGGGGGGGGAHG 144
Query: 118 NVGKGGVNVHAGKGRG 133
GG AG G G
Sbjct: 145 GGYGGGQGAGAGGGYG 160
Score = 30.0 bits (66), Expect = 2.2
Identities = 25/86 (29%), Positives = 28/86 (32%), Gaps = 7/86 (8%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVD-------AGKTKPGGTAVNVGKGGVHVNAGK 108
G GG G GG G+GG H AG+ GG GG G
Sbjct: 163 GAGGHGGGGGGGNGGGGGGGSGEGGAHGGGYGAGGGAGEGYGGGAGAGGHGGGGGGGGGS 222
Query: 109 GKPGGGTRVNVGKGGVNVHAGKGRGK 134
G GGG G G G G+
Sbjct: 223 GGGGGGGGGYAAASGYGHGGGAGGGE 248
Score = 28.9 bits (63), Expect = 4.9
Identities = 28/82 (34%), Positives = 31/82 (37%), Gaps = 9/82 (10%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G GG G GG GG AG GG G GG H G G GGG
Sbjct: 126 GGGGGGAGGGGGGGGGAHGGGYGGGQGAGAGGGYGGG-----GAGG-HGGGGGGGNGGGG 179
Query: 116 RVNVGKGGVN---VHAGKGRGK 134
G+GG + AG G G+
Sbjct: 180 GGGSGEGGAHGGGYGAGGGAGE 201
Score = 28.5 bits (62), Expect = 6.4
Identities = 25/82 (30%), Positives = 29/82 (34%), Gaps = 5/82 (6%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G+GG G+ GG GG G GG G+GG G G GG
Sbjct: 67 GEGGGAGGHGEGHIGGGGGGGHGGGAGGGGGGGPGGGYGGGSGEGG-----GAGYGGGEA 121
Query: 116 RVNVGKGGVNVHAGKGRGKPVH 137
+ G GG G G G H
Sbjct: 122 GGHGGGGGGGAGGGGGGGGGAH 143
Score = 28.1 bits (61), Expect = 8.3
Identities = 24/76 (31%), Positives = 26/76 (33%), Gaps = 5/76 (6%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG G+ G G GG G GG G G AG G GGG
Sbjct: 112 GGAGYGGGEAGGHGGGGGGGAGGGGGGGGGAHGGGYGGGQGAG-----AGGGYGGGGAGG 166
Query: 118 NVGKGGVNVHAGKGRG 133
+ G GG G G G
Sbjct: 167 HGGGGGGGNGGGGGGG 182
>At2g05580 unknown protein
Length = 302
Score = 35.8 bits (81), Expect = 0.040
Identities = 25/78 (32%), Positives = 29/78 (37%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G GG G GG G+GG G+ + G G G H G G GG
Sbjct: 164 GGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKGGGGGGGQGGH 223
Query: 116 RVNVGKGGVNVHAGKGRG 133
+ G GG H G G G
Sbjct: 224 KGGGGGGGQGGHKGGGGG 241
Score = 35.8 bits (81), Expect = 0.040
Identities = 30/81 (37%), Positives = 31/81 (38%), Gaps = 4/81 (4%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAV---NVGKGGVHVNAGKGKPG 112
G GG G GG + G GG G K GG G GG HV G G
Sbjct: 216 GGGGQGGHKGGGGGGGQGGHKGGGGGGQGGGGHKGGGGGQGGHKGGGGGGHVGGGGRGGG 275
Query: 113 GGTRVNVGKGGVNVHAGKGRG 133
GG R G GG G GRG
Sbjct: 276 GGGRGGGGSGG-GGGGGSGRG 295
Score = 35.0 bits (79), Expect = 0.068
Identities = 26/77 (33%), Positives = 28/77 (35%)
Query: 57 KGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTR 116
KGG G K GG G+ G G+ GG G GG G G G G
Sbjct: 141 KGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGM 200
Query: 117 VNVGKGGVNVHAGKGRG 133
G GG H G G G
Sbjct: 201 KGGGGGGQGGHKGGGGG 217
Score = 33.9 bits (76), Expect = 0.15
Identities = 28/79 (35%), Positives = 33/79 (41%), Gaps = 2/79 (2%)
Query: 57 KGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVG-KGGVHVNAGKGKPG-GG 114
KGG G+ GG KGG G K GG G KGG KG G GG
Sbjct: 120 KGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGG 179
Query: 115 TRVNVGKGGVNVHAGKGRG 133
+ G+GG G+G+G
Sbjct: 180 QKGGGGQGGQKGGGGRGQG 198
Score = 33.9 bits (76), Expect = 0.15
Identities = 27/77 (35%), Positives = 28/77 (36%)
Query: 57 KGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTR 116
KGG G K GG KGG G+ GG KGG G K GGG
Sbjct: 99 KGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGG 158
Query: 117 VNVGKGGVNVHAGKGRG 133
KGG KG G
Sbjct: 159 QGGQKGGGGQGGQKGGG 175
Score = 33.5 bits (75), Expect = 0.20
Identities = 24/76 (31%), Positives = 28/76 (36%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG G+ GG G GG G GG G+G + G G GG +
Sbjct: 154 GGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGRGQGGMKGGGGGGQGGHKG 213
Query: 118 NVGKGGVNVHAGKGRG 133
G GG H G G G
Sbjct: 214 GGGGGGQGGHKGGGGG 229
Score = 33.5 bits (75), Expect = 0.20
Identities = 30/88 (34%), Positives = 37/88 (41%), Gaps = 11/88 (12%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G+GG+ G GG + G GG G GG G+GG G G+ GGG
Sbjct: 196 GQGGMKGGGG----GGQGGHKGGGGGGGQGGHKGGGGGG---GQGGHKGGGGGGQGGGGH 248
Query: 116 RVNVGKGGVNVHAGKGRGKPVHVSVGNK 143
+ G GG H G G G HV G +
Sbjct: 249 K--GGGGGQGGHKGGGGGG--HVGGGGR 272
Score = 32.7 bits (73), Expect = 0.34
Identities = 27/77 (35%), Positives = 30/77 (38%), Gaps = 4/77 (5%)
Query: 57 KGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTR 116
KGG G K GG G+ G G K GG KGG G K GGG
Sbjct: 88 KGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGG- 146
Query: 117 VNVGKGGVNVHAGKGRG 133
G+GG G G+G
Sbjct: 147 ---GQGGQKGGGGGGQG 160
Score = 32.3 bits (72), Expect = 0.44
Identities = 32/92 (34%), Positives = 39/92 (41%), Gaps = 8/92 (8%)
Query: 43 LDWTEEKSTSVAVGKGGVNVDSGKTKPGGTSVNVG-KGGVHVDAGKTKPGGTAVNVGKGG 101
LD EE+ + +GG+ G K GG G KGG G K GG KGG
Sbjct: 65 LDGEEEEESKN--NQGGIGRGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGQGGQKGG 122
Query: 102 VHVNAGKGKPGGGTRVNVGKGGVNVHAGKGRG 133
G+ K GGG G+GG G G+G
Sbjct: 123 GGGQGGQ-KGGGGG----GQGGQKGGGGGGQG 149
Score = 30.8 bits (68), Expect = 1.3
Identities = 28/83 (33%), Positives = 34/83 (40%), Gaps = 9/83 (10%)
Query: 57 KGGVNVDSGKTKPGGTSVNVGK--GGVHVDAGKTKPGGTAVNVGKGGVHVNAG----KGK 110
KGG G K GG G+ GG G+ GG G GG G G+
Sbjct: 130 KGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGQGGQKGGGGQGGQKGGGGQGGQ 189
Query: 111 PGGGTRVNVGKGGVNVHAGKGRG 133
GGG R G+GG+ G G+G
Sbjct: 190 KGGGGR---GQGGMKGGGGGGQG 209
Score = 30.4 bits (67), Expect = 1.7
Identities = 27/77 (35%), Positives = 28/77 (36%), Gaps = 1/77 (1%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVG-KGGVHVNAGKGKPGGG 114
G+ G G K GG KGG G K GG G KGG G K GGG
Sbjct: 108 GQKGGGGGQGGQKGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGGGGQGGQKGGGG 167
Query: 115 TRVNVGKGGVNVHAGKG 131
G GG G G
Sbjct: 168 QGGQKGGGGQGGQKGGG 184
Score = 30.0 bits (66), Expect = 2.2
Identities = 27/78 (34%), Positives = 33/78 (41%), Gaps = 8/78 (10%)
Query: 56 GKGGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGT 115
G+GG G + GG G G G K GG +VG GG G G+ GGG+
Sbjct: 231 GQGGHKGGGGGGQGGGGHKGGGGG-----QGGHKGGGGGGHVGGGG-RGGGGGGRGGGGS 284
Query: 116 RVNVGKGGVNVHAGKGRG 133
G GG + G G G
Sbjct: 285 --GGGGGGGSGRGGGGGG 300
>At1g27710 unknown protein
Length = 212
Score = 35.8 bits (81), Expect = 0.040
Identities = 27/77 (35%), Positives = 32/77 (41%), Gaps = 9/77 (11%)
Query: 58 GGVNVDSGKTKPGGTSVNVGKGGVHVDAGKTKPGGTAVNVGKGGVHVNAGKGKPGGGTRV 117
GG+ D G GG G GG + G GG + G GG + G GGG
Sbjct: 144 GGLGWDGGN---GGGGPGYGSGGGGIGGGGGIGGGVIIGGGGGGCGGSCSGGGGGGG--- 197
Query: 118 NVGKGGVNVHAGKGRGK 134
G GGV+ KG GK
Sbjct: 198 GYGHGGVST---KGSGK 211
Score = 33.1 bits (74), Expect = 0.26
Identities = 31/94 (32%), Positives = 38/94 (39%), Gaps = 5/94 (5%)
Query: 42 HLDWTEEKSTSVAVGKGGVNVDSGKTKPGGTS--VNVGKGGVHVDAGKTKPGGTAVNVGK 99
H+ + + T+ G GG G PGG V +G GG AG GG + G
Sbjct: 94 HVTGFKGELTAGGYGGGGPGYGGGGYGPGGGGGGVVIG-GGFGGGAGYGSGGGLGWDGGN 152
Query: 100 GGVHVNAGKGKPGGGTRVNVGKGGVNVHAGKGRG 133
GG G G GGG G GG + G G G
Sbjct: 153 GG--GGPGYGSGGGGIGGGGGIGGGVIIGGGGGG 184
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.131 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,151,201
Number of Sequences: 26719
Number of extensions: 437349
Number of successful extensions: 1969
Number of sequences better than 10.0: 106
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 71
Number of HSP's that attempted gapping in prelim test: 1163
Number of HSP's gapped (non-prelim): 463
length of query: 380
length of database: 11,318,596
effective HSP length: 101
effective length of query: 279
effective length of database: 8,619,977
effective search space: 2404973583
effective search space used: 2404973583
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148532.8


