

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148528.7 - phase: 0 /pseudo
(1391 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
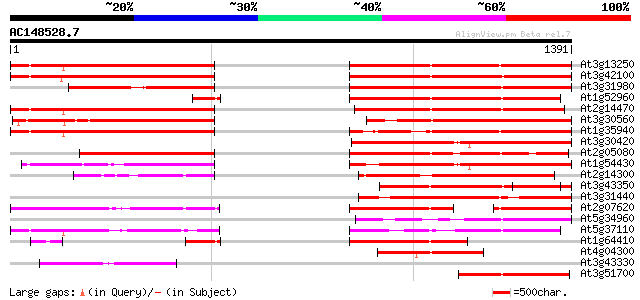
At3g13250 hypothetical protein 569 e-162
At3g42100 putative protein 567 e-161
At3g31980 hypothetical protein 551 e-156
At1g52960 hypothetical protein 538 e-152
At2g14470 pseudogene 510 e-144
At3g30560 hypothetical protein 493 e-139
At1g35940 hypothetical protein 480 e-135
At3g30420 hypothetical protein 471 e-132
At2g05080 putative helicase 458 e-128
At1g54430 hypothetical protein 455 e-128
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 442 e-124
At3g43350 putative protein 430 e-120
At3g31440 hypothetical protein 426 e-119
At2g07620 putative helicase 369 e-102
At5g34960 putative protein 368 e-101
At5g37110 putative helicase 350 4e-96
At1g64410 unknown protein 293 6e-79
At4g04300 hypothetical protein 254 3e-67
At3g43330 putative protein 250 3e-66
At3g51700 unknown protein 237 3e-62
>At3g13250 hypothetical protein
Length = 1419
Score = 569 bits (1466), Expect = e-162
Identities = 298/553 (53%), Positives = 405/553 (72%), Gaps = 9/553 (1%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDA-LK 901
LT+ QI+N TL IEK M N +L++ + FP P+ ++ NRLI DEL Y+ + LK
Sbjct: 872 LTEAQIQNYTLQEIEKIMLFNGATLEDIEHFPKPSREGIDN-SNRLIIDELRYNNQSDLK 930
Query: 902 LEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKI 961
+ +T EQR IY +I +AV GGVFF++G+GGTGKTF+W TLAA++RS+G+I
Sbjct: 931 KKHSDWIQKLTPEQRGIYDQITNAVFNDLGGVFFVYGFGGTGKTFIWKTLAAAVRSKGQI 990
Query: 962 VLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
L VA+SGIASLLL GGRTAHS+F IP+ E S C I SDLA L+K +LIIWDEAP
Sbjct: 991 CLNVASSGIASLLLEGGRTAHSRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDEAP 1050
Query: 1022 MAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSIN 1081
M KFCFEALD++ DI+ +K+FGGKV+VFGGDFRQ+LPV+ A R++I+ SS+N
Sbjct: 1051 MMSKFCFEALDKSFSDIIKRV--DNKVFGGKVMVFGGDFRQVLPVINGAGRAEIVMSSLN 1108
Query: 1082 SSYIWDECIVLTLTKNMRLRFNVGSSD-ADELKTFSEWILKVGEGKISEPNDGIVDFEIP 1140
+SY+WD C VL LTKNMRL N S D A E++ FS+W+L VG+G+++EPNDG V +IP
Sbjct: 1109 ASYLWDHCKVLRLTKNMRLLNNDLSVDEAKEIQEFSDWLLAVGDGRVNEPNDGEVIIDIP 1168
Query: 1141 DDLLIKEFDDPIEAIMKSTY--PNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIP 1198
++LLI+E D+PIEAI + Y P L+ ++P + Q+RAILA + V+ IN Y+L +
Sbjct: 1169 EELLIQEADNPIEAISREIYGDPTKLHEISDPKFFQRRAILAPKNEDVNTINQYMLEHLD 1228
Query: 1199 GEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLD 1258
EE+ Y S+DSID S+ +D ++ + TP+FL++++ SG+P+H ++LKVG P+MLLRNLD
Sbjct: 1229 SEERIYLSADSIDPSD-SDSLKN-PVITPDFLNSIKVSGMPHHSLRLKVGAPVMLLRNLD 1286
Query: 1259 QVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPI 1318
GLCNGTRL +T++ +H++EAK+I+G +G + YIP ++++PS + PFK+ RRQFP+
Sbjct: 1287 PKGGLCNGTRLQITQLCSHIVEAKVITGDRIGQIVYIPLINITPSDTKLPFKMRRRQFPL 1346
Query: 1319 IVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSN 1378
V++ MTINKSQGQ+L+ VGLYLP+ VFSHGQLYVA SRV +K GLKILI D E K
Sbjct: 1347 SVAFVMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKTGLKILILDKEGKIQKQ 1406
Query: 1379 TTNVVYKEVFDNL 1391
TTNVV+KEVF N+
Sbjct: 1407 TTNVVFKEVFQNI 1419
Score = 502 bits (1292), Expect = e-142
Identities = 252/519 (48%), Positives = 348/519 (66%), Gaps = 15/519 (2%)
Query: 1 MWYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRL 60
MW+ ER K S NP+F +CCG G +++PFL P L+ L+ N + S+ + Q IR+
Sbjct: 1 MWFNERINKKSNSENPKFSLCCGQGSVKLPFLKESPELIKKLLKGNDAL-SRHYRQFIRI 59
Query: 61 YNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDN 120
YN +FA +S G KVDK + GRGP R+QG + H++GS+ P G K++QLYI DT+N
Sbjct: 60 YNMIFAMTSLGGKVDKSMPKGRGPAMFRLQGGNYHQIGSLKPKDGDYAKYSQLYIVDTEN 119
Query: 121 ELQNRVQGIRN----------PNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVEN 170
E++NR I NLN Q + + +ML+ N + + FR AR+R+ N E
Sbjct: 120 EVENRANVIGKGNNGSSTKGKKNLNKQLIDAIIKMLNQVNPYVEKFRSARERIDSTNDEP 179
Query: 171 LKLKLISDRT-TDGRIYNQPTVSEVAALIVGD-VDSAARRDIIMERQS-GRLERIDEFHP 227
++++SDR TDGR+YN PT EVAALI GD V RDII+E++S GRL+RI + H
Sbjct: 180 FHMRIVSDRKGTDGRLYNMPTAGEVAALIPGDFVSQMPVRDIILEKKSTGRLKRISQIHI 239
Query: 228 AYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILC 287
+YLA QYPL+F YGEDGY + + +G K +++R+W FR+Q R+ E+ T+L
Sbjct: 240 SYLALQYPLIFCYGEDGYTPGI-EKCYKSGYTKKKKCISMRQWYAFRIQEREDESHTLLQ 298
Query: 288 ARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLP 347
++RLFQQFL D +T +E++RL +++ NQSKLR + S+ + + ++G +V++P
Sbjct: 299 SKRLFQQFLCDAYTTIESNRLAYIKFNQSKLRCENFNSIKESASSGSTTMSEEGNQVLIP 358
Query: 348 SSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDII 407
SS+ GG RYM Q Y+D MAI GFPDLFITFTCNP WPEI R+ +GL DRPDI+
Sbjct: 359 SSFTGGPRYMLQTYYDAMAICKHFGFPDLFITFTCNPKWPEITRYCEKRGLTADDRPDIV 418
Query: 408 ARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHIN 467
AR+FKIK D L+ DLT++H+LGK VA MYT+EFQKRGLPHAHIL+F+ +SK PT D I+
Sbjct: 419 ARIFKIKLDSLMKDLTERHLLGKTVASMYTVEFQKRGLPHAHILLFMAANSKLPTADDID 478
Query: 468 QIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCM 506
+IISAEIP+ + + ELY+++ MMHGPCG A SSPCM
Sbjct: 479 KIISAEIPNKDKEPELYEVIKNSMMHGPCGSANTSSPCM 517
>At3g42100 putative protein
Length = 1752
Score = 567 bits (1462), Expect = e-161
Identities = 295/554 (53%), Positives = 404/554 (72%), Gaps = 9/554 (1%)
Query: 842 RLTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDA-L 900
+LT +I+N TL IEK M +N +LKE + FP P+ ++ NRL+ DEL Y+ D+ L
Sbjct: 1203 KLTLAEIRNYTLQEIEKIMLRNGATLKEIQDFPQPSREGIDN-SNRLVVDELRYNIDSNL 1261
Query: 901 KLEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGK 960
K + F + EQR IY +I AV GGVFF++G+GGTGKTF+W TLAA++RS+G+
Sbjct: 1262 KEKHDEWFQMLNTEQRGIYDEITGAVFNDLGGVFFIYGFGGTGKTFIWKTLAAAVRSRGQ 1321
Query: 961 IVLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEA 1020
IVL VA+SGIASLLL GGRTAHS+F IP+ E S C I SDLA L+K +LIIWDEA
Sbjct: 1322 IVLNVASSGIASLLLEGGRTAHSRFAIPLNPDEFSVCKITPKSDLANLIKEASLIIWDEA 1381
Query: 1021 PMAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSI 1080
PM KFCFE+LD++ DI+++ +K+FGGKV+VFGGDFRQ+LPV+ A R +I+ SS+
Sbjct: 1382 PMMSKFCFESLDKSFYDILNNK--DNKVFGGKVVVFGGDFRQVLPVINGAGRVEIVMSSL 1439
Query: 1081 NSSYIWDECIVLTLTKNMRLRFN-VGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEI 1139
N+SY+WD C VL LTKNMRL + S +A E++ FS+W+L VG+G+I+EPNDG +I
Sbjct: 1440 NASYLWDHCKVLKLTKNMRLLSGGLSSEEAKEIQQFSDWLLAVGDGRINEPNDGEALIDI 1499
Query: 1140 PDDLLIKEFDDPIEAIMKSTY--PNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSII 1197
P++LLIKE +PIEAI K Y P+ L+M N+P + Q+RAILA T + V+ IN Y+L +
Sbjct: 1500 PEELLIKEAGNPIEAISKEIYGDPSELHMINDPKFFQRRAILAPTNEDVNTINQYMLEHL 1559
Query: 1198 PGEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNL 1257
EE+ Y S+DSID ++ + + TP+FL++++ +G+P+H ++LKVG P+MLLRNL
Sbjct: 1560 KSEERIYLSADSIDPTDSDSLANP--VITPDFLNSIQLTGMPHHALRLKVGAPVMLLRNL 1617
Query: 1258 DQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFP 1317
D GLCNGTRL +T++A V++AK+I+ +G++ IP ++++PS + PFK+ RRQFP
Sbjct: 1618 DPKGGLCNGTRLQITQLAKQVVQAKVITRDRIGDIVLIPLINLTPSDTKLPFKMRRRQFP 1677
Query: 1318 IIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLS 1377
+ V++AMTINKSQGQ+L+ VGLYLP+ VFSHGQLYVA SRV +K GLKILI D +
Sbjct: 1678 LSVAFAMTINKSQGQSLEQVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGNMQK 1737
Query: 1378 NTTNVVYKEVFDNL 1391
TTNVV+KEVF N+
Sbjct: 1738 QTTNVVFKEVFQNI 1751
Score = 457 bits (1177), Expect = e-128
Identities = 236/519 (45%), Positives = 331/519 (63%), Gaps = 14/519 (2%)
Query: 1 MWYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRL 60
MW+ ER K +++ +P F +CCG G +++P L P L+N+L+ + + S+ F +NIR+
Sbjct: 332 MWFAERINKKQQNKSPTFTLCCGKGNVKLPLLKDSPALINNLLTGDDAL-SRNFRENIRI 390
Query: 61 YNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDN 120
YN +FA +S G +VD + G+GP R+QG + H +GS+ P G K++QLYI DT+N
Sbjct: 391 YNMIFAMTSLGGRVDNSMPKGKGPNMFRLQGGNYHLIGSLKPNPGDYAKYSQLYIVDTEN 450
Query: 121 ELQNRV------QGIRNP----NLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVEN 170
E+ NR +G RN L + + L +ML+ N + FR+AR+R++ N E
Sbjct: 451 EVDNRATVINKGKGRRNTPAKQKLKKEVIEALIEMLNKVNPYVDKFRQARERIQDDNDEP 510
Query: 171 LKLKLISDRT-TDGRIYNQPTVSEVAALIVGDVD-SAARRDIIMERQS-GRLERIDEFHP 227
+++++DR D R Y+ PT SEVAALI G S RDI++E ++ G L RI + H
Sbjct: 511 FHMRIVADRKGVDRRTYSMPTSSEVAALIPGGFQPSMFDRDIVLEEKTTGHLTRISQIHI 570
Query: 228 AYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILC 287
+YLA QYPL+ YGEDGY + + K+ K +++R+W FR+Q R E T+
Sbjct: 571 SYLALQYPLILCYGEDGYTPGIEKCLPNSAKKKKKKCISMRQWFAFRIQERPNECKTLTR 630
Query: 288 ARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLP 347
++RLFQQFL D +T +E++RL +++ QSKLR Y SL ++G +V++P
Sbjct: 631 SKRLFQQFLCDAYTTIESNRLSYIKFKQSKLRCENYNSLKKASEAGTTSMNEEGNQVLIP 690
Query: 348 SSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDII 407
SS GG RYM Q Y+D MAI GFPDLFITFTCNP WPEI R A+GL DRPDI+
Sbjct: 691 SSLTGGPRYMVQNYYDAMAICKHYGFPDLFITFTCNPKWPEITRHCQARGLSVDDRPDIV 750
Query: 408 ARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHIN 467
AR+FKIK D L+ DLT +LGK VA M+T+EFQKRGLPHAHIL+F+ SK PT D I+
Sbjct: 751 ARIFKIKLDSLMKDLTDGKMLGKTVASMHTVEFQKRGLPHAHILLFMDAKSKLPTADDID 810
Query: 468 QIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCM 506
+IISAEIP + + ELY+++ M+HGPCG A +SPCM
Sbjct: 811 KIISAEIPDKDKEPELYEVIKNSMIHGPCGAANMNSPCM 849
>At3g31980 hypothetical protein
Length = 1099
Score = 551 bits (1419), Expect = e-156
Identities = 286/552 (51%), Positives = 395/552 (70%), Gaps = 8/552 (1%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKL 902
LTD + +N L IE + N +L++FK P PT + NR I +E NY+ + L+
Sbjct: 551 LTDAERRNYALLEIEDMLLCNGSTLQDFKDMPKPTKEGTDH-SNRFITEEKNYNIEKLRE 609
Query: 903 EFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIV 962
+ F+ +T EQ+ IY +I+ AV +GG+FF++G+GGT KTFMW TL+A++R +G I
Sbjct: 610 DHDDWFNKMTSEQKGIYDEIIKAVLENSGGIFFVYGFGGTSKTFMWKTLSAAVRMRGLIS 669
Query: 963 LTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPM 1022
+ VA+SGIASLLL GGRTAHS+F IP+ + +TC+I +SDLA +LK +LIIWDEAPM
Sbjct: 670 VNVASSGIASLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDEAPM 729
Query: 1023 AHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINS 1082
++CFE+LDR+L D++ + DG K FGGKV+VFGGDFRQ+LPV+ A R++I+ +++NS
Sbjct: 730 MSRYCFESLDRSLNDVIGNI-DG-KPFGGKVVVFGGDFRQVLPVIHGAGRAEIVLAALNS 787
Query: 1083 SYIWDECIVLTLTKNMRLRFN-VGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPD 1141
SY+W+ C VLTLTKNMRL N + +A+E+K FS W+L VG+G++SEPNDG V +IP+
Sbjct: 788 SYLWEHCKVLTLTKNMRLMSNDLDKDEAEEIKEFSNWLLAVGDGRVSEPNDGEVLIDIPE 847
Query: 1142 DLLIKEFDDPIEAIMKSTYPNF--LNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPG 1199
+LLIK+ +DPIEAI K+ Y + L N+P + QQRAIL V+ IND +L + G
Sbjct: 848 ELLIKDANDPIEAITKAVYGDLDLLQPNNDPKFFQQRAILCPRNTDVNTINDIMLDKLNG 907
Query: 1200 EEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQ 1259
E Y S+DSID + + + TP+FL++++ SGLPNH + LK+GTP+MLLRN+D
Sbjct: 908 ELVTYLSADSIDPQDAASL--NNPVLTPDFLNSIKLSGLPNHNLTLKIGTPVMLLRNIDP 965
Query: 1260 VEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPII 1319
GLCNGTRL VT+M NH++EA++I+G VG+ I + +SPS + PF++ RRQFPI
Sbjct: 966 KGGLCNGTRLQVTQMGNHILEARVITGDRVGDKVIIIKSQISPSDTKLPFRMRRRQFPIA 1025
Query: 1320 VSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNT 1379
V++AMTINKSQGQ+L VG+YLP+ VFSHGQLYVA SRV +K GLK+LI D E S T
Sbjct: 1026 VAFAMTINKSQGQSLKEVGIYLPKPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQT 1085
Query: 1380 TNVVYKEVFDNL 1391
NVV+KE+F N+
Sbjct: 1086 MNVVFKEIFQNI 1097
Score = 316 bits (810), Expect = 5e-86
Identities = 168/367 (45%), Positives = 228/367 (61%), Gaps = 36/367 (9%)
Query: 145 MLDDTNCHAKSFRKARDRLRQGNVENLKLKLISDRT-TDGRIYNQPTVSEVAALIVGDV- 202
MLD +N + + FR A+D+L N E ++++SDR DG+ Y PT SEV ALI GD
Sbjct: 1 MLDASNPYVEKFRLAKDKLDSNNGEPFYMQIVSDRVGKDGKTYCNPTTSEVTALIPGDFR 60
Query: 203 -DSAARRDIIMERQSGRLERIDEFHPAYLAYQYPLLFPYGEDGYRDDVLHRGIVA--GKQ 259
+ R I+ ++++G L RI E HP+Y+ QYPL+F YGEDG+R + +G GKQ
Sbjct: 61 PEMPTRDIIVQDKKTGHLSRISEVHPSYVPMQYPLIFNYGEDGFRPGI-QKGYTGRTGKQ 119
Query: 260 SKMDRLTIREWLTFRLQSRKTEAMTILCARRLFQQFLVDCFTMMEADRLKWLRKNQSKLR 319
+ +++R+W FR+Q R EA T+L ++RLFQQFLVD +T
Sbjct: 120 ANKC-ISMRQWYAFRIQERSDEAQTLLRSKRLFQQFLVDGYT------------------ 160
Query: 320 VGKYRSLSNNQGNVDGQTKKQGKRVVLPSSYVGGRRYMDQLYFDGMAISSDVGFPDLFIT 379
GN + +QGK + +P S+ GG RYM Y+D MAI+ GFP+LFIT
Sbjct: 161 ---------TGGNTN--MSEQGKSIRIPQSFTGGPRYMLNSYYDVMAITKMYGFPNLFIT 209
Query: 380 FTCNPSWPEIQRFVGAKGLKPHDRPDIIARVFKIKFDELLNDLTKKHILGKVVAYMYTIE 439
FTCNP WPEI R+ + L DRPD + +FK+K D L+ DLT++H+LGK V+ MYT+E
Sbjct: 210 FTCNPKWPEITRYCKERKLNAEDRPDGVCWIFKMKLDSLMKDLTEEHLLGKTVSAMYTVE 269
Query: 440 FQKRGLPHAHILIFLHPSSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLA 499
FQKRGLPHAHIL+F+ S K+PT D I+ IISAEIP D +LY++V M+HGPCG A
Sbjct: 270 FQKRGLPHAHILLFMDKSCKFPTSDDIDNIISAEIPDKSKDPKLYEVVKDCMIHGPCGAA 329
Query: 500 KPSSPCM 506
SPC+
Sbjct: 330 NKESPCI 336
>At1g52960 hypothetical protein
Length = 924
Score = 538 bits (1385), Expect = e-152
Identities = 276/524 (52%), Positives = 378/524 (71%), Gaps = 5/524 (0%)
Query: 842 RLTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALK 901
+L+D++ + L I + + KN SL ++K P +D +E+ N I DE YD L
Sbjct: 404 QLSDEERQQYCLQEIARLLTKNGVSLSKWKQMPQISDEHVEKC-NHFILDERKYDRAYLI 462
Query: 902 LEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKI 961
+ + + VT EQ+ IY +IM AV GGVFF++G+GGTGKTF+W L+A++RS+G I
Sbjct: 463 EKHEEWLTMVTSEQKKIYDEIMDAVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKGDI 522
Query: 962 VLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
L VA+SGIA+LLL GGRT HS+F IP+ E+STCNI SDL +L+K NLIIWDE P
Sbjct: 523 SLNVASSGIAALLLDGGRTTHSRFGIPINPNESSTCNISRGSDLGELVKEANLIIWDETP 582
Query: 1022 MAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSIN 1081
M K CFE+LDRTL+DIM++ GDK FGGK IVFGGDFRQ+LPV+ A R +I+ +++N
Sbjct: 583 MMSKHCFESLDRTLRDIMNNP--GDKPFGGKGIVFGGDFRQVLPVINGAGREEIVFAALN 640
Query: 1082 SSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPD 1141
SSYIW+ C VL LTKNMRL N+ + +++ FS+WIL VG+GKIS+PNDGI +IP+
Sbjct: 641 SSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGIALIDIPE 700
Query: 1142 DLLIKEFDDPIEAIMKSTYPNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPGEE 1201
+ LI +DP+E+I+++ Y N +P + Q RAIL T + V+ IN++++S++ GEE
Sbjct: 701 EFLINGDNDPVESIIEAVYGNTFMEEKDPKFFQGRAILCPTNEDVNSINEHMMSMLDGEE 760
Query: 1202 KEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVE 1261
+ Y SSDSID ++ + + ++ +FL+++R SGLPNH ++LKVG P+MLLRN+D +
Sbjct: 761 RIYLSSDSIDPADTSSA--NNDAYSADFLNSVRVSGLPNHCLRLKVGCPVMLLRNMDPNK 818
Query: 1262 GLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVS 1321
GLCNGTRL VT+MA+ VI+A+ I+G VG + IPRM ++PS + PFK+ RRQFP+ V+
Sbjct: 819 GLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTRLPFKMRRRQFPLSVA 878
Query: 1322 YAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLK 1365
+AMTINKSQGQTL+SVGLYLPR VFSHGQLYVA SRV +K G K
Sbjct: 879 FAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 922
Score = 58.5 bits (140), Expect = 3e-08
Identities = 28/68 (41%), Positives = 43/68 (63%), Gaps = 1/68 (1%)
Query: 454 LHPSSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMKKKR*MF 513
+HP+SK T + ++II+AEIP + LY +V M+HGPCG+ P+SPCM+ +
Sbjct: 1 MHPTSKLSTAEDTDKIITAEIPDKKKKPGLYAVVKDCMIHGPCGVGHPNSPCMENGK-CK 59
Query: 514 QVFPEKVS 521
+ FP+ S
Sbjct: 60 KYFPKSYS 67
>At2g14470 pseudogene
Length = 1265
Score = 510 bits (1313), Expect = e-144
Identities = 264/527 (50%), Positives = 373/527 (70%), Gaps = 12/527 (2%)
Query: 855 LIEKHMEKNRRSLK---EFKGFPYPTDYVMEELGNRLIYDELNYDEDA-LKLEFKRLFST 910
L E ++K R K + FP PT ++ NRLI +EL Y+ ++ LK + +
Sbjct: 743 LAEDILKKKRDEFKNPEDIDEFPKPTIDGIDN-SNRLIVEELRYNRESNLKEKHEEWKQM 801
Query: 911 VTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGI 970
+T EQR +Y++I AV GGVFF++G+GGTGKTF+W TL+A++R + +IVL VA+SGI
Sbjct: 802 LTPEQRGVYNEITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSATIRYRDQIVLNVASSGI 861
Query: 971 ASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEA 1030
ASLLL GGRTAHS+F IP+ E S C I SDLA L+K +L+IWDEAPM +FCFEA
Sbjct: 862 ASLLLEGGRTAHSRFGIPLNPDEFSVCKIKPKSDLANLVKKASLVIWDEAPMMSRFCFEA 921
Query: 1031 LDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSYIWDECI 1090
LD++ DI+ +T + +FGGKV+VFGGDFRQ+ PV+ A R++I+ SS+N+SY+WD C
Sbjct: 922 LDKSFSDIIKNT--DNTVFGGKVVVFGGDFRQVFPVINGAGRAEIVMSSLNASYLWDNCK 979
Query: 1091 VLTLTKNMRLRFN-VGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFD 1149
VL LTKN RL N + ++A E++ FS+W+L VG+G+I+E NDG+ +IP+DLLI D
Sbjct: 980 VLKLTKNTRLLANNLSETEAKEIQEFSDWLLAVGDGRINESNDGVAIIDIPEDLLITNAD 1039
Query: 1150 DPIEAIMKSTY--PNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSS 1207
PIE+I Y P L+ +P + Q RAILAS + V+ IN+Y+L + EE+ Y S+
Sbjct: 1040 KPIESITNEIYGDPKILHEITDPKFFQGRAILASKNEDVNTINEYLLDQLHAEERIYLSA 1099
Query: 1208 DSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGT 1267
DSID ++ + S + TP+FL++++ GLPNH ++LKVG P++LLRNLD GLCNGT
Sbjct: 1100 DSIDPTDSDSL--SNPVITPDFLNSIKLPGLPNHSLRLKVGAPVLLLRNLDPKGGLCNGT 1157
Query: 1268 RLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTIN 1327
RL +T++ ++EAK+I+G +G++ IP ++++P+ + PFK+ RRQFP+ V++ MTIN
Sbjct: 1158 RLQITQLCTQIVEAKVITGDRIGHIILIPTVNLTPTNTKLPFKMRRRQFPLSVAFVMTIN 1217
Query: 1328 KSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKK 1374
KS+GQ+L+ VGLYLP+ VFSHGQLYVA SRV +K GLKILI D + K
Sbjct: 1218 KSEGQSLEHVGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGK 1264
Score = 473 bits (1216), Expect = e-133
Identities = 242/519 (46%), Positives = 334/519 (63%), Gaps = 14/519 (2%)
Query: 1 MWYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRL 60
MWY ER RK F +CCG G +++PFL P LL +L+ N+ SK + N R
Sbjct: 1 MWYDERIRKKETKKESGFTLCCGEGSVKLPFLKESPDLLKNLLSGNHPL-SKHYRDNART 59
Query: 61 YNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDN 120
+N +FA +S G KVDK + GRGP R+QG + H +GS+ P G K++QLYI DT+N
Sbjct: 60 FNMVFAVTSLGGKVDKSMPKGRGPAMFRLQGGNYHLIGSLKPTPGDYAKYSQLYIVDTEN 119
Query: 121 ELQNRVQGIRN----------PNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVEN 170
E++NR I PNL+ + + +ML+ N + + FR AR+R++ + E
Sbjct: 120 EVENRAIVIGKGKIAKPASGKPNLDKNLIEVIIKMLNRCNPYVRKFRTARERIQTNDEEP 179
Query: 171 LKLKLISDRT-TDGRIYNQPTVSEVAALIVGDV-DSAARRDIIMERQS-GRLERIDEFHP 227
+++I+DR DGR Y+ T SEVAALI GD RDI++E++S G L+RI++ H
Sbjct: 180 FHMRIIADRQGVDGRTYSMHTTSEVAALIPGDFRHGMPDRDIVIEKKSNGHLKRINQIHI 239
Query: 228 AYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILC 287
+YLA QYPL+F YGEDG+R + K+ +++R+W FR+Q R+ E T+L
Sbjct: 240 SYLALQYPLIFCYGEDGFRPGIEKCFKSKSKKKNKKCISMRQWFAFRIQEREVECQTLLR 299
Query: 288 ARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLP 347
++RLFQQFL D +T +E++RL +++ NQSKLR Y S+ +++G ++++P
Sbjct: 300 SKRLFQQFLCDAYTTIESNRLNYIKFNQSKLRCENYTSVKEAAAAGATTMEEEGNQLLIP 359
Query: 348 SSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDII 407
+S+ GG RYM Q Y+D MAI GFPDLFITFTCNP WP I R+ +GL P DR DII
Sbjct: 360 ASFNGGPRYMVQSYYDAMAICKLYGFPDLFITFTCNPKWPHITRYCDKRGLNPKDRLDII 419
Query: 408 ARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHIN 467
AR+FKIK D L+NDLT K +LGK VA MYT+EFQKRGLPHAHIL+F+H SK PT D I+
Sbjct: 420 ARIFKIKLDSLMNDLTVKKMLGKTVASMYTVEFQKRGLPHAHILLFMHAKSKLPTSDDID 479
Query: 468 QIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCM 506
++ISAEIP E + ELY+++ M+HGPCG A SPCM
Sbjct: 480 KLISAEIPDKEKEPELYEVIKNSMIHGPCGSANVKSPCM 518
>At3g30560 hypothetical protein
Length = 1473
Score = 493 bits (1270), Expect = e-139
Identities = 259/506 (51%), Positives = 344/506 (67%), Gaps = 31/506 (6%)
Query: 886 NRLIYDELNYDEDALKLEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKT 945
N+LI DE NY+ + LK +T EQ+ +Y KIM AV GG
Sbjct: 999 NQLILDERNYNRETLKTIHDDWLKMLTTEQKKVYDKIMDAVLNNKGG------------- 1045
Query: 946 FMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDL 1005
I L VA+SGIASLLL GGRTAHS+F IP+ E STCN++ SDL
Sbjct: 1046 --------------DICLNVASSGIASLLLEGGRTAHSRFGIPLTPHETSTCNMERGSDL 1091
Query: 1006 AKLLKVTNLIIWDEAPMAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILP 1065
A+L+ LIIWDEAPM K+CFE+LD++LKDI+S D FGGK+I+FGGDFRQILP
Sbjct: 1092 AELVTAAKLIIWDEAPMMSKYCFESLDKSLKDILSTPEDMP--FGGKLIIFGGDFRQILP 1149
Query: 1066 VVPRADRSDIINSSINSSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEG 1125
V+ A R I+ SS+NSS++W C V LTKNMRL ++ ++A E++ FS+WIL VGEG
Sbjct: 1150 VILAAGRELIVKSSLNSSHLWQYCKVFKLTKNMRLLQDIDINEAREIEDFSKWILAVGEG 1209
Query: 1126 KISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTYPNFLNMYNNPDYLQQRAILASTIDV 1185
K+++PNDG+ +I DD+LI E D+PIE+I+K+ Y + +P + Q RAIL T D
Sbjct: 1210 KLNQPNDGVTQIQIRDDILIPEGDNPIESIIKAVYGTSFDEERDPKFFQDRAILCPTNDD 1269
Query: 1186 VDKINDYVLSIIPGEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKL 1245
V+ IND++LS + GEEK Y SSDSID S+ + ++TP+FL+ ++ SGLPNH + L
Sbjct: 1270 VNSINDHMLSKLTGEEKIYRSSDSIDPSDT--RADKNPVYTPDFLNKIKISGLPNHLLWL 1327
Query: 1246 KVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQS 1305
KVG P+MLLRNLD GL NGTRL + R+ + +++ +I++G VG L IPRM ++PS
Sbjct: 1328 KVGCPVMLLRNLDSHGGLMNGTRLQIVRLGDKLVQGRILTGTRVGKLVIIPRMPLTPSDR 1387
Query: 1306 PWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLK 1365
PFK+ RRQFP+ V++AMTINKSQGQ+L +VG+YLP+ VFSHGQLYVA SRV++K GLK
Sbjct: 1388 RLPFKMKRRQFPLSVAFAMTINKSQGQSLGNVGIYLPKPVFSHGQLYVAMSRVKSKGGLK 1447
Query: 1366 ILIHDLEKKPLSNTTNVVYKEVFDNL 1391
+LI D + K + TTNVV+KE+F NL
Sbjct: 1448 VLITDSKGKQKNETTNVVFKEIFRNL 1473
Score = 444 bits (1142), Expect = e-124
Identities = 236/527 (44%), Positives = 331/527 (62%), Gaps = 30/527 (5%)
Query: 6 RRRKSRE----SSNPEFGM-----CCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQ 56
RR ++E +SN E G CC G+I +P L P + L F + ++K F
Sbjct: 104 RRNLNKEFAAAASNKEHGSSTTSRCCMQGQIVLPMLKESPEYMWWL-FTSDHPDAKNFRA 162
Query: 57 NIRLYNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIF 116
N+R YN +F+F+S G KVD+ V GRGP +QG++ H + ++ P G KF QLY+
Sbjct: 163 NVRPYNMLFSFTSLGGKVDRSVKKGRGPSMFALQGENYHLIDALKPKPGDYAKFQQLYVM 222
Query: 117 DTDNELQNRVQGIRNPN----------LNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQG 166
DTDNE+ NR++ + N +TVS L +MLD+ N H +FR ARDR
Sbjct: 223 DTDNEVDNRIEVMSKGNNGKTEGVKQKFKKETVSALLKMLDEINPHVANFRIARDRF--- 279
Query: 167 NVE----NLKLKLISDRTTDGRIYNQPTVSEVAALIVGDVD-SAARRDIIMERQSGRLER 221
N+E N +++IS+R TDGR+YN P+V EVA LI GD D + +RDI+++ +S +L R
Sbjct: 280 NIEKEDANFHMRIISNRDTDGRVYNLPSVGEVA-LIPGDFDDNLDKRDIVLQIKSEKLRR 338
Query: 222 IDEFHPAYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDR-LTIREWLTFRLQSRKT 280
I E H +YL+ QYPLLFP GEDGYR + K+ K + +++R+W +RLQ RK
Sbjct: 339 IHECHVSYLSLQYPLLFPKGEDGYRLGIKKTETNTSKRKKKQKDVSMRQWFDYRLQERKD 398
Query: 281 EAMTILCARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQ 340
E +L ++RL QQF+VD FTM+E++RL++++KNQ+KLR +++ + D +
Sbjct: 399 EKHILLRSKRLLQQFIVDAFTMIESNRLRFIKKNQTKLRSTNKQAVQDASDAGDNDLSNK 458
Query: 341 GKRVVLPSSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKP 400
GK V++P S+ GG YM Q Y D M GFPDLFITFTCN WP+I RFV + L
Sbjct: 459 GKSVIIPPSFTGGPAYMQQNYLDAMGTCKHFGFPDLFITFTCNSKWPKITRFVKERKLNA 518
Query: 401 HDRPDIIARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKY 460
D+PDII R++K+K + L++DLTK HI GK YTIEFQKRGLPHAHI++++ P K+
Sbjct: 519 EDKPDIICRIYKMKLENLMDDLTKNHIFGKTKLATYTIEFQKRGLPHAHIIVWMDPRYKF 578
Query: 461 PTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMK 507
P DH+++II AEIP END +LY+ V M+HGPC L P+SPCM+
Sbjct: 579 PIADHVDKIIFAEIPDKENDPKLYQAVSECMIHGPCRLVNPNSPCME 625
>At1g35940 hypothetical protein
Length = 1678
Score = 480 bits (1236), Expect = e-135
Identities = 260/552 (47%), Positives = 363/552 (65%), Gaps = 66/552 (11%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKL 902
LT+ +IKN TL IEK M N +L++ FP PT +
Sbjct: 1189 LTETEIKNYTLQEIEKIMLSNGATLEDIDEFPKPT-----------------------RD 1225
Query: 903 EFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIV 962
E+K++ +T EQR +Y+ I AV GGVFF++G+GGTGKTF+W TL+A++R +G+IV
Sbjct: 1226 EWKQM---LTPEQRGVYNAITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSAAIRCRGQIV 1282
Query: 963 LTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPM 1022
L VA+SGIASLLL GGRTAHS+F IP+ E S
Sbjct: 1283 LNVASSGIASLLLEGGRTAHSRFGIPLNHDEFSV-------------------------- 1316
Query: 1023 AHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINS 1082
+LD++ DI+ +T +K+FGGKV+VFGGDFRQ+LPV+ A R++I+ SS+N+
Sbjct: 1317 -------SLDKSFSDIIKNT--NNKVFGGKVVVFGGDFRQVLPVINGAGRAEIVMSSLNA 1367
Query: 1083 SYIWDECIVLTLTKNMRLRFN-VGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPD 1141
SY+WD C VL LTKNMRL N + +++A E++ FS+W+L V +G+I+EPNDG+ +IP+
Sbjct: 1368 SYLWDHCKVLKLTKNMRLLANNLSATEAKEIQEFSDWLLAVSDGRINEPNDGVATIDIPE 1427
Query: 1142 DLLIKEFDDPIEAIMKSTY--PNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPG 1199
DLLI D PIE I Y P L+ +P + Q RAILA + V+ IN+Y+L +
Sbjct: 1428 DLLITNADKPIETITNEIYGDPKILHEITDPKFFQGRAILAPKNEDVNTINEYLLEQLDA 1487
Query: 1200 EEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQ 1259
EE+ Y S+DSID ++ +D + + TP+FL++++ GLPNH + LKVG P+MLLRNLD
Sbjct: 1488 EERIYLSADSIDPTD-SDSLNN-PVITPDFLNSIKLPGLPNHSLCLKVGAPVMLLRNLDP 1545
Query: 1260 VEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPII 1319
GLCNGTRL +T++ ++EAK+I+G +GN+ IP ++++P+ + PFK+ RRQFP+
Sbjct: 1546 KGGLCNGTRLQITQLCTQIVEAKVITGDRIGNIVLIPTVNLTPTDTKLPFKMRRRQFPLS 1605
Query: 1320 VSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNT 1379
V++AMTINKSQGQ+L+ +GLYLP+ VFSHGQLYVA SRV +K GLKILI D + K T
Sbjct: 1606 VAFAMTINKSQGQSLEHIGLYLPKPVFSHGQLYVALSRVTSKKGLKILILDKDGKLQKQT 1665
Query: 1380 TNVVYKEVFDNL 1391
TNVV+KEVF N+
Sbjct: 1666 TNVVFKEVFQNI 1677
Score = 475 bits (1222), Expect = e-134
Identities = 239/519 (46%), Positives = 337/519 (64%), Gaps = 14/519 (2%)
Query: 1 MWYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRL 60
MWY ER RK + F +CCG G +++PFL P LL +L+ N+ SK + N R+
Sbjct: 342 MWYDERIRKKETNKESVFTLCCGEGSVKLPFLKESPHLLKNLLSGNHPL-SKHYRDNARI 400
Query: 61 YNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDN 120
+N +FA +S G KVDK + GRGP R+QG + H +GS+ G K++QLYI DT+N
Sbjct: 401 FNMVFAMTSFGGKVDKSMPKGRGPAMFRLQGGNYHLIGSLKLTPGDYAKYSQLYIIDTEN 460
Query: 121 ELQNRVQGIRN----------PNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVEN 170
E++NR I PNL+ + + +ML+ N + + FR AR+R++ + E
Sbjct: 461 EVENRATVINKGKNAKPASGKPNLDKNLIEAIIKMLNRCNPYVRRFRTARERIQTNDEEP 520
Query: 171 LKLKLISDRT-TDGRIYNQPTVSEVAALIVGDV-DSAARRDIIMERQS-GRLERIDEFHP 227
+++I+DR +GR Y+ PT SEVAALI GD RDI++ ++S G L+RI++ H
Sbjct: 521 FHMRIIADRQGVEGRTYSMPTTSEVAALIPGDFRHGMPDRDIVIGKKSNGHLKRINQIHI 580
Query: 228 AYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILC 287
+YLA QYPL+F YGEDG+R + K+ +++R+W FR+Q R+ E T+L
Sbjct: 581 SYLALQYPLIFCYGEDGFRPGIEKCSKSKSKKKNKKCISMRQWFAFRIQEREVECQTLLR 640
Query: 288 ARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLP 347
++RLFQQ L D +T +E++RL +++ NQSKLR Y S+ +++G ++++P
Sbjct: 641 SKRLFQQCLCDAYTTIESNRLNYIKFNQSKLRCENYTSVKEAAAAGATTMEEEGNQLLIP 700
Query: 348 SSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDII 407
+S+ GG RYM Q Y+D MAI GFPDLFITFTCNP WPEI R+ +GL P DRP+II
Sbjct: 701 ASFTGGPRYMVQSYYDAMAICKHYGFPDLFITFTCNPKWPEITRYYDKRGLNPEDRPNII 760
Query: 408 ARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHIN 467
AR+FKIK D L+NDLT K +LGK VA MYT+EFQKRGLPHAHIL+F+H SK PT D I+
Sbjct: 761 ARIFKIKLDSLMNDLTVKKMLGKTVASMYTVEFQKRGLPHAHILLFMHAKSKLPTADDID 820
Query: 468 QIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCM 506
++ISAEIP E + +LY+++ M+HGPCG A +SPCM
Sbjct: 821 KLISAEIPDKEKEPDLYEVIKNSMIHGPCGSANVNSPCM 859
>At3g30420 hypothetical protein
Length = 837
Score = 471 bits (1212), Expect = e-132
Identities = 255/555 (45%), Positives = 367/555 (65%), Gaps = 20/555 (3%)
Query: 847 QIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKLEFKR 906
+++ TL IE + ++ +SL ++ P P ++EE+ N L+ E + D +
Sbjct: 285 ELEKYTLIEIETLLRQHEKSLSDYPEMPQPEKSILEEVNNSLLRQEFQINIDKERETHAN 344
Query: 907 LFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVA 966
LFS + ++QR IY ++ +V + G +FFL+G GGTGKTF++ T+ ++LRS GK V+ VA
Sbjct: 345 LFSKLNEQQRIIYDDVLKSVTNKEGKLFFLYGDGGTGKTFLYKTIISALRSNGKNVMPVA 404
Query: 967 TSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKF 1026
+S IA+LLLPGGRTAHS FKIP+ E+ C+I S LA +L +LIIWDEAPMAH+
Sbjct: 405 SSAIAALLLPGGRTAHSWFKIPINVHEDFICDIKIGSMLANVLSKVDLIIWDEAPMAHRH 464
Query: 1027 CFEALDRTLKDIMS--DTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSY 1084
FEA+DRTL+DI+S D K GGK ++ GGDFRQILPV+P+ R + ++++IN SY
Sbjct: 465 TFEAVDRTLRDILSVGDEKALTKTLGGKTVLLGGDFRQILPVIPQRTRQETVSAAINRSY 524
Query: 1085 IWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFE------ 1138
+W+ C L++NMR++ +E+K F+EWIL++G+G+ GI D +
Sbjct: 525 LWESCHKYLLSQNMRVQ-------PEEIK-FAEWILQIGDGEAPRKTHGIDDDQEEDNII 576
Query: 1139 IPDDLLIKEFDDPIEAIMKSTYPNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIP 1198
I +LL+ E ++P+E + +S P+F N + + + L+ A+L + VD+INDY+LS +P
Sbjct: 577 IDKNLLLPETENPLEVLCQSVSPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLLSKVP 636
Query: 1199 GEEKEYFSSDSIDRSEVNDQCQSFQLFTP-EFLSTLRTSGLPNHKIKLKVGTPIMLLRNL 1257
G KEYFS+DSID+ E + + F++ P E+L++L GLP H++ LKVG PIMLLRNL
Sbjct: 637 GLAKEYFSADSIDQDEALTE-EGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIMLLRNL 695
Query: 1258 DQVEGLCNGTRLIVTRMANHVIEAKIISG-KNVGNLTYIPRMSMSPSQSPWPFKLIRRQF 1316
+Q EGLCNGTRL VT + + V++A+I+S IPR+ +SP S PF L RRQF
Sbjct: 696 NQKEGLCNGTRLTVTHLGDKVLKAEILSDTTKKRKKVLIPRIILSPQDSKHPFTLRRRQF 755
Query: 1317 PIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPL 1376
P+ + YAMT+NKSQGQTL+ V LYLP+ VFSHGQLYVA SRV + GL +L +KK
Sbjct: 756 PVRMCYAMTVNKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVL-DTSKKKEG 814
Query: 1377 SNTTNVVYKEVFDNL 1391
TN+VY+EVF+ L
Sbjct: 815 KYVTNIVYREVFNGL 829
>At2g05080 putative helicase
Length = 1219
Score = 458 bits (1178), Expect = e-128
Identities = 254/547 (46%), Positives = 353/547 (64%), Gaps = 38/547 (6%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPY-PTDYVMEELGNRLIYDELNYDEDALK 901
L+D++ K L I+ + +N SL +K P P D + N LI DE YD ++
Sbjct: 680 LSDEEKKVYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDT--NVLILDEKGYDRESET 737
Query: 902 LEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKI 961
+ +T EQ+ +Y I+ AVN GGVFF++G+GGTGKTF+W TL+A+LRS+G I
Sbjct: 738 KKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFGGTGKTFLWKTLSAALRSKGDI 797
Query: 962 VLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
VL VA+SGIASLLL GGRTAHS+ IP+ E +TCN+ SD A L+K +LIIWDEAP
Sbjct: 798 VLNVASSGIASLLLEGGRTAHSRSGIPLNPNEFTTCNMKAGSDRANLVKEASLIIWDEAP 857
Query: 1022 MAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSIN 1081
M + CFE+LDR+L DI + +K FGGKV+VFGGDFRQ+LPV+P AD +DI+ +++N
Sbjct: 858 MMSRHCFESLDRSLSDICGNC--DNKPFGGKVVVFGGDFRQVLPVIPGADTADIVMAALN 915
Query: 1082 SSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFS-EWILKVGEGKISEPNDGIVDFEIP 1140
SSY+W C VLTLTKNM L FS EWIL VG+G+I EPNDG +IP
Sbjct: 916 SSYLWSHCKVLTLTKNMCL--------------FSEEWILAVGDGRIGEPNDGEALIDIP 961
Query: 1141 DDLLIKEFDDPIEAIMKSTYPNFLNMY--NNPDYLQQRAILASTIDVVDKINDYVLSIIP 1198
+ LI + DPI+AI Y + ++ +P + Q+RAIL T + V++IN+ +L +
Sbjct: 962 SEFLITKAKDPIQAICTEIYGDITKIHEQKDPVFFQERAILCPTNEDVNQINETMLDNLQ 1021
Query: 1199 GEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLD 1258
GEE + SSDS+D +++ ++ + TPEFL+ ++ GL NHK++LK+G+P+MLLRN+D
Sbjct: 1022 GEELTFLSSDSLDTADIGS--RNNPVLTPEFLNNVKVLGLSNHKLRLKIGSPVMLLRNID 1079
Query: 1259 QVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPI 1318
+ GL NGTRL + +M+ +++A I++G + + PF++ R Q P+
Sbjct: 1080 PIGGLMNGTRLQIMQMSPFILQAMILTGDR--------------ADTKLPFRMRRTQLPL 1125
Query: 1319 IVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSN 1378
V +AMTINKSQGQ+L VG++LPR FSH QLYVA SRV +K+GLKILI + E KP
Sbjct: 1126 AVCFAMTINKSQGQSLKRVGIFLPRPCFSHSQLYVAISRVTSKSGLKILIVNDEGKPQKQ 1185
Query: 1379 TTNVVYK 1385
T K
Sbjct: 1186 TKKFTKK 1192
Score = 342 bits (877), Expect = 9e-94
Identities = 168/336 (50%), Positives = 221/336 (65%), Gaps = 1/336 (0%)
Query: 173 LKLISDRTTDGRIYNQPTVSEVAALIVGDVD-SAARRDIIMERQSGRLERIDEFHPAYLA 231
++++S R TDGR+YN PT SEVA LI GD RDII+E +SG+L+RI E P YL
Sbjct: 1 MRIVSKRETDGRVYNVPTTSEVAMLIPGDFTIDIPCRDIIVEEKSGKLQRISEILPCYLP 60
Query: 232 YQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILCARRL 291
QYPLLFPYGEDG+R + AGK K ++IR+W FR+ RK E +L ++RL
Sbjct: 61 LQYPLLFPYGEDGFRTGIEKHQTGAGKDKKNKFISIRQWFAFRIHERKHEKHILLRSKRL 120
Query: 292 FQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLPSSYV 351
+QQFLVD + +E++RL +++ NQS LR Y S+ K QG LP+++
Sbjct: 121 WQQFLVDSYIAIESNRLGYIKLNQSSLRADNYNSVQKASEEGKCDLKYQGLACYLPATFT 180
Query: 352 GGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDIIARVF 411
GG RYM +Y D MA+ GFPD FITFTCNP WPE+ RF G + L+ DRP+II ++F
Sbjct: 181 GGPRYMRNMYLDAMAVCKHFGFPDYFITFTCNPKWPELIRFCGERNLRVDDRPEIICKIF 240
Query: 412 KIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHINQIIS 471
K+K D L+ DLTK++ILGK MYTIEFQKRGLPHAHILI+L K +HI++ IS
Sbjct: 241 KMKLDSLMLDLTKRNILGKTSTSMYTIEFQKRGLPHAHILIWLDSKCKLTRAEHIDKAIS 300
Query: 472 AEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMK 507
AEIP D EL++++ M+HGPCG+ P PCM+
Sbjct: 301 AEIPDKLKDPELFEVIKEMMVHGPCGVVNPKCPCME 336
>At1g54430 hypothetical protein
Length = 1639
Score = 455 bits (1171), Expect = e-128
Identities = 254/559 (45%), Positives = 360/559 (63%), Gaps = 51/559 (9%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKL 902
L ++++ TL IE + ++ +SL ++ P P + + E+
Sbjct: 1114 LKAEELEKYTLIEIETLLRQHEKSLSDYPEMPQPENKLNEQ------------------- 1154
Query: 903 EFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIV 962
QR IY ++ +V + G +FFL+G GGTGKTF++ T+ ++LRS GK V
Sbjct: 1155 ------------QRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGKNV 1202
Query: 963 LTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPM 1022
+ VA+S IA+LLLPGGRTAHS+FKIP+ E+S C+I S LA +L +LIIWDEAPM
Sbjct: 1203 MPVASSAIAALLLPGGRTAHSRFKIPINVHEDSICDIKIGSMLANVLSKVDLIIWDEAPM 1262
Query: 1023 AHKFCFEALDRTLKDIMS--DTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSI 1080
AH+ FEA+DRTL+DI+S D K FGGK ++ GGDFRQILPV+P+ R + ++++I
Sbjct: 1263 AHRHTFEAVDRTLRDILSVGDEKALTKTFGGKTVLLGGDFRQILPVIPQGTRQETVSAAI 1322
Query: 1081 NSSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFE-- 1138
N SY+W+ C L++NMR++ +E+K F+EWIL+VG+G+ GI D +
Sbjct: 1323 NRSYLWESCHKYLLSQNMRVQ-------PEEIK-FAEWILQVGDGEAPRKTHGIDDDQEE 1374
Query: 1139 ----IPDDLLIKEFDDPIEAIMKSTYPNFLNMYNNPDYLQQRAILASTIDVVDKINDYVL 1194
I +LL+ E ++P+E + +S +P+F N + + + L+ A+L + VD+INDY+L
Sbjct: 1375 DNIIIDKNLLLPETENPLEVLCRSVFPDFTNTFQDLENLKGTAVLTPRNETVDEINDYLL 1434
Query: 1195 SIIPGEEKEYFSSDSIDRSEVNDQCQSFQLFTP-EFLSTLRTSGLPNHKIKLKVGTPIML 1253
S +PG KEYFS+DSIDR E + + F++ P E+L++L GLP H++ LKVG PIML
Sbjct: 1435 SKVPGLAKEYFSADSIDRDEALTE-EGFEMSYPMEYLNSLEFPGLPAHRLCLKVGVPIML 1493
Query: 1254 LRNLDQVEGLCNGTRLIVTRMANHVIEAKIISG-KNVGNLTYIPRMSMSPSQSPWPFKLI 1312
LRNL+Q EGLCNGTRLIVT + + V++A+I+S IPR+ +SP S PF L
Sbjct: 1494 LRNLNQKEGLCNGTRLIVTHLGDKVLKAEILSDTTKERKKVLIPRIILSPQDSKHPFTLR 1553
Query: 1313 RRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLE 1372
RRQFP+ + YAMTINKSQGQTL+ V LYLP+ VFSHGQLYVA SRV + GL +L +
Sbjct: 1554 RRQFPVRMCYAMTINKSQGQTLNRVALYLPKPVFSHGQLYVALSRVTSPKGLTVL-DTSK 1612
Query: 1373 KKPLSNTTNVVYKEVFDNL 1391
KK TN+VY+EVF+ L
Sbjct: 1613 KKEGKYVTNIVYREVFNGL 1631
Score = 396 bits (1017), Expect = e-110
Identities = 219/486 (45%), Positives = 287/486 (58%), Gaps = 39/486 (8%)
Query: 29 IPFLPTPPPLLNDLMFNNYSTESKQFNQNIRLYNSMFAFSSPGFKVDKGVIPGRGPPTIR 88
+P P PP +L L+ TES + +NIR YNS+ AF+S G ++DK V+ GP T R
Sbjct: 320 LPPEPQPPQMLKKLL-----TESPHYQRNIRTYNSILAFTSMGAQIDKNVMHKGGPFTFR 374
Query: 89 IQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDNELQNRVQGIRNPN----LNMQTVSKLQQ 144
I GQ+ H++GS++P GK PK QLYIFDT NE+ NR+ ++ LN + V L
Sbjct: 375 IHGQNHHKLGSLVPEEGKPPKILQLYIFDTANEVHNRISAVKRTTKVGELNEKIVKDLIT 434
Query: 145 MLDDTNCHAKSFRKARDRLRQGNVENLKLKLISDRTTDGRIYNQPTVSEVAALIVGDVD- 203
+D NC AK FRKARDR G+ +KLI + G+ Y+ PT E+A LIVGD
Sbjct: 435 TVDTFNCLAKVFRKARDRYEAGDCPEFSIKLIGQKKK-GKQYDMPTTDEIAGLIVGDFSK 493
Query: 204 SAARRDIIMERQSGRLERIDEFHPAYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMD 263
+ RD+I+ +S L++I + HP ++ QYPLLFPYGE G+ H GI ++
Sbjct: 494 NIGERDVIVHHKSSGLQQISDLHPLFMTLQYPLLFPYGEIGF-----HEGIPVVEKG--- 545
Query: 264 RLTIREWLTFRLQSRKTEAMTILCARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKY 323
MTI+ ++RL Q++VD +T +E +RL+W R NQ KLR +Y
Sbjct: 546 -------------------MTIVRSKRLLHQYIVDAYTSIEQERLRWYRLNQKKLRADQY 586
Query: 324 RSLSNNQGNVDGQTKKQGKRVVLPSSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCN 383
++ + D K GKRV+LP+SY G RYM + Y D MAI G PDLFIT T N
Sbjct: 587 NNVKDAVARGDTDAKSIGKRVILPASYTGSPRYMVEKYHDAMAICRWYGNPDLFITMTTN 646
Query: 384 PSWPEIQRFVGAKGLKPHD-RPDIIARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQK 442
P W EI + G + RPDI RVFKIK DELL D K K +A +YTIEFQK
Sbjct: 647 PKWEEISEHLKTYGNDAANVRPDIECRVFKIKLDELLADFNKGLFFPKPIAIVYTIEFQK 706
Query: 443 RGLPHAHILIFLHPSSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPS 502
RGLPHAHIL++L K PTP+ I++ ISAEIP + D E + LV HMMHGPCG +PS
Sbjct: 707 RGLPHAHILLWLQGDLKKPTPNDIDKYISAEIPVKDKDPEGHTLVEQHMMHGPCGKDRPS 766
Query: 503 SPCMKK 508
SPCM+K
Sbjct: 767 SPCMEK 772
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 442 bits (1136), Expect = e-124
Identities = 234/487 (48%), Positives = 319/487 (65%), Gaps = 32/487 (6%)
Query: 864 RRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKLEFKRLFSTVTDEQRDIYH-KI 922
R L +K P D E N LI DE NY+ ++LK + T+T E + +YH +I
Sbjct: 775 RPDLWRYKMLLEPGD---EPAFNPLIIDERNYNRESLKKKHDNWLKTLTPEHKKVYHDEI 831
Query: 923 MSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAH 982
M V GGVFFL+ +GGTGKTF+W L+A++R +G L VA+S IASLLL GGRTAH
Sbjct: 832 MDDVLNDKGGVFFLYAFGGTGKTFLWKVLSAAIRCKGDTCLNVASSSIASLLLEGGRTAH 891
Query: 983 SKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEALDRTLKDIMSDT 1042
S+F IP+ E STCN++ SDLA+L+ LIIWDE
Sbjct: 892 SRFGIPLTPHETSTCNMERGSDLAELVTAAKLIIWDE----------------------- 928
Query: 1043 ADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSYIWDECIVLTLTKNMRLRF 1102
D FG KVI+FGGDFRQIL V+P A R I+ SS+NSSY+W C VL LTKNMRL
Sbjct: 929 ---DMPFGRKVILFGGDFRQILHVIPAAGRELIVKSSLNSSYLWQHCKVLKLTKNMRLLQ 985
Query: 1103 NVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTYPN 1162
++ ++A E++ F +WIL VGEGK++EP+DG+ +IPDD+LI E D+PIE+I+K+ Y
Sbjct: 986 DIDINEAREIEDFLKWILTVGEGKLNEPSDGVTHIQIPDDILIPEGDNPIESIIKAVYGT 1045
Query: 1163 FLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSIDRSEVNDQCQSF 1222
+P + Q +AIL T D V+ IND++LS + GEE+ Y SS+SID S+ +
Sbjct: 1046 TFAQKRDPKFFQHKAILCPTNDDVNSINDHMLSKLTGEERIYRSSNSIDPSDT--RADKN 1103
Query: 1223 QLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAK 1282
++TP+FL+ ++ SGL NH ++LKVG P+MLLRN GL NGTRL + R+ + +++ +
Sbjct: 1104 PIYTPDFLNKIKISGLANHLLRLKVGCPVMLLRNFYPHGGLMNGTRLQIVRLGDKLVQGR 1163
Query: 1283 IISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLP 1342
I++G VG L IPRMS++PS PFK+ RR FP+ V++AMTINKSQGQ+L +VG+YLP
Sbjct: 1164 ILTGTRVGKLVIIPRMSLTPSDRRLPFKMKRRHFPLSVAFAMTINKSQGQSLGNVGMYLP 1223
Query: 1343 RSVFSHG 1349
++VFSHG
Sbjct: 1224 KAVFSHG 1230
Score = 292 bits (748), Expect = 8e-79
Identities = 157/351 (44%), Positives = 209/351 (58%), Gaps = 37/351 (10%)
Query: 159 ARDRLRQGNVE-NLKLKLISDRTTDGRIYNQPTVSEVAALIVGDVD-SAARRDIIMERQS 216
A DR E N +++IS R TDGR+YN P+V+EVAALI GD D + ++DI+++ +S
Sbjct: 162 AHDRFNIEKEEANFHMRIISKRETDGRVYNLPSVAEVAALIPGDFDDNLDKKDIVLQMKS 221
Query: 217 GRLERIDEFHPAYLAYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQ 276
G+L RI E H YPLLFP GEDGYR + K K ++R+W +RLQ
Sbjct: 222 GKLRRIHECH-------YPLLFPKGEDGYRLGIKKTPTKTSKGKK----SMRQWFDYRLQ 270
Query: 277 SRKTEAMTILCARRLFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQ 336
RK E +L ++RL QQF+ +KLR +++ + D
Sbjct: 271 ERKDEKHILLRSKRLLQQFM-------------------TKLRSTNKQAVQDASDAGDND 311
Query: 337 TKKQGKRVVLPSSYVGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAK 396
+GK ++P S+ GG YM Q Y D + GFPDLFITFTCNP WPEI RFV +
Sbjct: 312 LSNKGKSYIIPPSFTGGPAYMQQNYLDAIGTCKHFGFPDLFITFTCNPKWPEITRFVKER 371
Query: 397 GLKPHDRPDIIARVFKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHP 456
L DRPDII R++K+K D L++DLTK HI MYT+EFQKRGLPHAHI++++ P
Sbjct: 372 KLNAEDRPDIICRIYKMKLDNLMDDLTKNHIFA-----MYTVEFQKRGLPHAHIIVWMDP 426
Query: 457 SSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMK 507
K+ T DH+++II AEIP E ELY+ V M+HGPC L P+SPCM+
Sbjct: 427 RYKFHTADHVDKIIFAEIPDKEKHPELYQAVSECMIHGPCRLVNPNSPCME 477
Score = 36.6 bits (83), Expect = 0.10
Identities = 14/35 (40%), Positives = 21/35 (60%)
Query: 2 WYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPP 36
WY ER + ++S+NP + CC G+I +P L P
Sbjct: 76 WYGERLNRRKKSANPVYTGCCMQGQIVLPMLKESP 110
>At3g43350 putative protein
Length = 830
Score = 430 bits (1105), Expect = e-120
Identities = 235/484 (48%), Positives = 325/484 (66%), Gaps = 47/484 (9%)
Query: 916 RDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLL 975
+ IY +IM V GGVFF++G+GGTGKTF+W L+A++RS+G I L VA+SGIA+L L
Sbjct: 181 KKIYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAVRSKGDISLNVASSGIAALRL 240
Query: 976 PGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEALDRTL 1035
GGRTAHS+F IP+ E+STCNI SDL +L+K LIIWDEAPM K CFE+LDRTL
Sbjct: 241 DGGRTAHSRFDIPINPNESSTCNISRGSDLGELVKEAKLIIWDEAPMMSKHCFESLDRTL 300
Query: 1036 KDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSYIWDECIVLTLT 1095
KDI+++ GDK GGKVIVFGGDFRQ+LPV+ A R +I+ +++NSSYIW+ VL LT
Sbjct: 301 KDIVNN--PGDKPLGGKVIVFGGDFRQVLPVINGAGREEIVFAALNSSYIWEHSKVLELT 358
Query: 1096 KNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAI 1155
KNMRL ++ + +++ FS+WIL VG+GKIS+PNDGI +IP++ LI +DP+E+I
Sbjct: 359 KNMRLLADISEHEKRDIEDFSKWILDVGDGKISQPNDGIALIDIPEEFLINGDNDPVESI 418
Query: 1156 MKSTYPNFLNMYNNP---DY--LQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSI 1210
+++ Y N +P DY Q RAIL T + V+ IN++++ ++ GEE+ Y SSDSI
Sbjct: 419 IEAVYGNTFMEEKDPKKTDYPQYQGRAILCPTNEDVNSINEHMMRMLDGEERIYLSSDSI 478
Query: 1211 DRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLI 1270
D ++++ + + +FL+ +R GLPNH ++LKVG P+MLLRN+D +GLCNGTRL
Sbjct: 479 DPADISSANNA--AYLADFLNNVRVYGLPNHCLRLKVGCPVMLLRNMDPNKGLCNGTRLQ 536
Query: 1271 VTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQ 1330
VT+M + +I+A+ I+ ++AMTINKSQ
Sbjct: 537 VTQMTDTIIQARFIT-----------------------------------AFAMTINKSQ 561
Query: 1331 GQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKA-GLKI-LIHDLE-KKPLSNTTNVVYKEV 1387
GQTL+SVGLYLPR VFSHGQLYVA SRV +K G + L+ +++ K L N T + ++
Sbjct: 562 GQTLESVGLYLPRPVFSHGQLYVAISRVTSKTIGCPVMLLRNMDPNKGLCNGTRLQVTQM 621
Query: 1388 FDNL 1391
D +
Sbjct: 622 ADTV 625
Score = 155 bits (391), Expect = 2e-37
Identities = 76/119 (63%), Positives = 95/119 (78%)
Query: 1247 VGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSP 1306
VG P+MLLRN+D +GLCNGTRL VT+MA+ VI+A+ I+G VG + IPRM ++P +
Sbjct: 710 VGCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPLDTR 769
Query: 1307 WPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLK 1365
PFK+ R+QF + V++AMTINKSQGQTL+SVGLYLPR VFSHGQLYVA SRV +K G K
Sbjct: 770 LPFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTGTK 828
Score = 154 bits (388), Expect = 4e-37
Identities = 81/148 (54%), Positives = 110/148 (73%), Gaps = 3/148 (2%)
Query: 1247 VGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSP 1306
+G P+MLLRN+D +GLCNGTRL VT+MA+ VI+A+ I+G VG + IPRM ++PS +
Sbjct: 594 IGCPVMLLRNMDPNKGLCNGTRLQVTQMADTVIQARFITGNRVGKIVLIPRMLITPSDTR 653
Query: 1307 WPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTK-AGLK 1365
PFK+ R+QF + V++AMTINKSQGQTL+SVGLYLPR VFSHGQLYVA SRV +K G
Sbjct: 654 LPFKMRRKQFALSVAFAMTINKSQGQTLESVGLYLPRPVFSHGQLYVAISRVTSKTVGCP 713
Query: 1366 I-LIHDLE-KKPLSNTTNVVYKEVFDNL 1391
+ L+ +++ K L N T + ++ D +
Sbjct: 714 VMLLRNMDPNKGLCNGTRLQVTQMADTV 741
>At3g31440 hypothetical protein
Length = 536
Score = 426 bits (1096), Expect = e-119
Identities = 239/530 (45%), Positives = 338/530 (63%), Gaps = 57/530 (10%)
Query: 866 SLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDA-LKLEFKRLFSTVTDEQRDIYHKIMS 924
+L++ FP PT ++ NRLI +EL Y+ ++ LK + + +T EQR
Sbjct: 59 TLEDIDEFPKPTRDGIDN-SNRLIVEELRYNRESNLKEKHEEWKQMLTSEQR-------- 109
Query: 925 AVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLLPGGRTAHSK 984
GT KT +W TL A++R + +IVL +A+SGIASLLL GGRTAHS+
Sbjct: 110 ----------------GTWKTIIWKTLFAAIRRRDQIVLNMASSGIASLLLEGGRTAHSR 153
Query: 985 FKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEALDRTLKDIMSDTAD 1044
F I + E S C I SDLA L+K +L+I D+APM +FCFEALD++ DI+ +T +
Sbjct: 154 FGIRLNPDEFSVCKIKPKSDLANLVKEASLVICDKAPMMSRFCFEALDKSFSDIIKNTYN 213
Query: 1045 GDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSYIWDECIVLTLTKNMRLRFN- 1103
K+FGGKV+VF GDFRQ+LPV+ A R++I+ SS+N+SY+WD C VL LTKNMRL N
Sbjct: 214 --KVFGGKVVVFSGDFRQVLPVINGAGRAEIVMSSLNASYLWDHCKVLKLTKNMRLLANN 271
Query: 1104 VGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTY--P 1161
+ ++A E+ FS+W+L VG+G+I+EPND + +IP DLLI D PIE I Y P
Sbjct: 272 LSETEAKEIHEFSDWLLAVGDGRINEPNDDVAIIDIPKDLLITNADKPIEWITNEIYGDP 331
Query: 1162 NFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSIDRSEVNDQCQS 1221
L+ +P + Q RAILA + V+ IN+Y+L + EE+ Y S+DSID ++ +D +
Sbjct: 332 KILHEITDPKFFQGRAILAPKNEDVNTINEYLLEQLHAEERIYLSADSIDPTD-SDSLNN 390
Query: 1222 FQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEA 1281
+ TP+FL++++ G GLCNG RL +T++ ++EA
Sbjct: 391 -PVITPDFLNSIKLPG------------------------GLCNGARLQITQLFTEIVEA 425
Query: 1282 KIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYL 1341
K+I+G +G++ IP ++++P+ + PFK+ RRQFP+ V++AMTINKSQGQ+L+ VGLYL
Sbjct: 426 KVITGDRIGHIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTINKSQGQSLEHVGLYL 485
Query: 1342 PRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
P+ VFSHGQLYVA SRV +K GLKILI D K TTN+V+KEVF N+
Sbjct: 486 PKPVFSHGQLYVALSRVTSKKGLKILILDKNGKLQKQTTNIVFKEVFQNI 535
>At2g07620 putative helicase
Length = 1241
Score = 369 bits (946), Expect = e-102
Identities = 207/526 (39%), Positives = 303/526 (57%), Gaps = 47/526 (8%)
Query: 2 WYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRLY 61
WY ER K R+S P+F +CC G +++PFL P L+ +L+ + + + F +NIR Y
Sbjct: 3 WYDERVNKRRKSKKPKFSLCCLQGSVKLPFLTESPELIRELLSCDDALR-RHFRENIRAY 61
Query: 62 NSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDNE 121
N +F+ +S G KVD+ G+GP ++ G + H +GSM+P G KF+QLYI DT NE
Sbjct: 62 NMLFSMTSLGGKVDRSNPQGKGPNQFQLHGANYHLIGSMLPGEGDYAKFSQLYIVDTGNE 121
Query: 122 LQNRVQGIRNPNLNMQT----VSKLQQMLDDTNCHAKSFRKARDRLRQGNVENLKLKLIS 177
++NR + Q + KL +MLD N + ++FR A+ +L N E ++++S
Sbjct: 122 VENRSTNPKKATFKNQVRKDLIEKLIRMLDACNPYIENFRLAKYKLDSNNGEPFYMQIVS 181
Query: 178 DRT-TDGRIYNQPTVSEVAALIVGDVDSAAR-RDIIME-RQSGRLERIDEFHPAYLAYQY 234
DR DGR Y P SEVAALI GD RDII++ +++G+L RI + HP+Y+ QY
Sbjct: 182 DRVGKDGRTYCNPRTSEVAALIPGDFRPKMHTRDIIVQDKKTGQLSRISKVHPSYVPMQY 241
Query: 235 PLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILCARRLFQQ 294
PL+F YGED +R GI G + + Q+ K
Sbjct: 242 PLIFNYGEDDFRP-----GIQKGYTGRTGK-----------QANKC-------------- 271
Query: 295 FLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLPSSYVGGR 354
++ +T +E++RL +++ NQS LR Y ++ + + +QGK + +P S+ GG
Sbjct: 272 --INGYTTIESNRLAYIKFNQSNLRCKNYDTVKAAREAGNTNMSEQGKSIRIPQSFTGGP 329
Query: 355 RYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDIIARVFKIK 414
R+M Y+D MAI GFP+LFITFTCNP PEI + + L DRPD++ +FK+K
Sbjct: 330 RHMLNSYYDAMAICKMYGFPNLFITFTCNPKLPEITSYCKERKLNAEDRPDVVCWIFKMK 389
Query: 415 FDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHINQIISAEI 474
D L+ DLT+ H+LGK T+ FQKRGLPHAHIL+F+ S K+PT D I+ I+SAEI
Sbjct: 390 LDSLMKDLTEDHLLGK------TVMFQKRGLPHAHILLFMDKSCKFPTSDDIDNILSAEI 443
Query: 475 PHPENDRELYKLVGTHMMHGPCGLAKPSSPCMKKKR*MFQVFPEKV 520
P D +LY++V M+HGPCG A SPC+ + + FP+K+
Sbjct: 444 PDKAKDPKLYEVVNDCMIHGPCGAANKESPCIVDGK-CSKFFPKKL 488
Score = 253 bits (646), Expect = 5e-67
Identities = 132/258 (51%), Positives = 185/258 (71%), Gaps = 3/258 (1%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALKL 902
L+D + N L IE + N +L++FK P PT + NR I +E NY+ + LK
Sbjct: 780 LSDAEKINYALLEIEDMLLCNGSTLEDFKHMPKPTKEGTDH-SNRFITEEKNYNVEKLKE 838
Query: 903 EFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIV 962
+ F+ +T EQ++IY +IM AV +GG+FF++G+GGTGKTFMW TL+A++R +G I
Sbjct: 839 DHDDWFNKMTSEQKEIYDEIMKAVLENSGGIFFVYGFGGTGKTFMWKTLSAAVRMKGLIS 898
Query: 963 LTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPM 1022
+ VA+SGIA LLL GGRTAHS+F IP+ + +TC+I +SDLA +LK +LIIWDEAPM
Sbjct: 899 VNVASSGIAFLLLQGGRTAHSRFGIPINPDDFTTCHIVPNSDLANMLKEASLIIWDEAPM 958
Query: 1023 AHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINS 1082
++CFE+LDR+L D++ + DG K FGGKV+VFGGDFRQ+L V+ A R++I+ +++NS
Sbjct: 959 MSRYCFESLDRSLNDVIGN-VDG-KPFGGKVVVFGGDFRQVLHVIHGAGRAEIVLAALNS 1016
Query: 1083 SYIWDECIVLTLTKNMRL 1100
SY+W+ C VLTLTKNM L
Sbjct: 1017 SYLWEHCNVLTLTKNMSL 1034
Score = 202 bits (515), Expect = 8e-52
Identities = 103/192 (53%), Positives = 136/192 (70%), Gaps = 2/192 (1%)
Query: 1200 EEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQ 1259
E Y S+DSID + + +FTP FL++++ SGL NH + LK+GTP+MLL+N+D
Sbjct: 1050 ESITYLSADSIDPQD--PASLNNPVFTPYFLNSIKLSGLSNHNLTLKIGTPVMLLKNIDP 1107
Query: 1260 VEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPII 1319
GLCNGTRL VT+M NH++EA++I+G V + I + +SPS + PF++ RRQFPI
Sbjct: 1108 KGGLCNGTRLQVTQMGNHILEARVITGDRVRDKVIIIKAQISPSDTKLPFRMRRRQFPIA 1167
Query: 1320 VSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNT 1379
V++AM I KSQGQ+L V +YLPR VFSHGQLYVA SRV +K GLK+LI D E S T
Sbjct: 1168 VAFAMRIKKSQGQSLKEVEIYLPRPVFSHGQLYVALSRVTSKKGLKVLIVDKEGNTQSQT 1227
Query: 1380 TNVVYKEVFDNL 1391
NVV+KE+F NL
Sbjct: 1228 MNVVFKEIFQNL 1239
>At5g34960 putative protein
Length = 1033
Score = 368 bits (944), Expect = e-101
Identities = 220/538 (40%), Positives = 307/538 (56%), Gaps = 103/538 (19%)
Query: 857 EKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDA-LKLEFKRLFSTVTDEQ 915
EK M N +L++ FP PT ++ NRLI +EL Y+ ++ LK + + +T EQ
Sbjct: 595 EKIMLSNGATLEDIDEFPKPTRDGIDN-SNRLIVEELRYNRESNLKEKHEEWKQMLTPEQ 653
Query: 916 RDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSGIASLLL 975
R +Y++I AV + +IVL VA+SGIASLLL
Sbjct: 654 RGVYNEITEAV----------------------------FNNLDQIVLNVASSGIASLLL 685
Query: 976 PGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAPMAHKFCFEALDRTL 1035
GGRTAHS+F I + E S C I SDLA L+K +L+IWDEAPM
Sbjct: 686 EGGRTAHSRFGISLNPDEFSVCKIKPKSDLANLVKEASLVIWDEAPMMS----------- 734
Query: 1036 KDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSINSSYIWDECIVLTLT 1095
RQ+L V+ A R++I+ SS+N+SY+WD C VL
Sbjct: 735 -------------------------RQVLLVINGAGRAEIVMSSLNASYLWDHCKVL--- 766
Query: 1096 KNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAI 1155
KI+EPNDG+ +IP+DLLI D PIE+I
Sbjct: 767 ------------------------------KINEPNDGVAIIDIPEDLLITNTDKPIESI 796
Query: 1156 MKSTY--PNFLNMYNNPDYLQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSIDRS 1213
Y P L+ +P + Q RAILA T + V+ IN+Y+L + EE+ Y S+DSID +
Sbjct: 797 TNEIYGDPKILHEITDPKFFQGRAILAPTNEDVNTINEYLLEQLHAEERIYLSADSIDPT 856
Query: 1214 EVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLIVTR 1273
+ N + + TP+FL++++ +GLPNH ++LKV P+MLLRNLD GLCNGTRL +T+
Sbjct: 857 DSNSL--NNPVITPDFLNSIKLAGLPNHSLRLKVSAPVMLLRNLDPKGGLCNGTRLQITQ 914
Query: 1274 MANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQT 1333
+ ++EAK+I+G +G++ IP ++++P+ + PFK+ RRQFP+ V++AMTIN SQGQ+
Sbjct: 915 LCTQIVEAKVITGDIIGHIVLIPTVNLTPTDTKLPFKMRRRQFPLSVAFAMTINTSQGQS 974
Query: 1334 LDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKILIHDLEKKPLSNTTNVVYKEVFDNL 1391
L+ VGLYLP++VFSHGQLYVA SRV +K GLK LI D + K TTNVV+KEVF N+
Sbjct: 975 LEHVGLYLPKAVFSHGQLYVALSRVTSKKGLKFLILDKDGKLQKQTTNVVFKEVFQNI 1032
>At5g37110 putative helicase
Length = 1307
Score = 350 bits (897), Expect = 4e-96
Identities = 210/528 (39%), Positives = 297/528 (55%), Gaps = 89/528 (16%)
Query: 843 LTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPY-PTDYVMEELGNRLIYDELNYDEDALK 901
L+D++ K L I+ + +N SL +K P P D + N LI DE YD D L
Sbjct: 850 LSDEEKKLYALQEIDHILRRNGTSLTYYKTMPQVPRDPRFDT--NVLILDEKGYDRDNLT 907
Query: 902 LEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKI 961
+ + +T EQ+ IY I+ AVN G V F++G+GGT
Sbjct: 908 EKHAKWIKMLTPEQKSIYDDIIGAVNENVGVVVFVYGFGGT------------------- 948
Query: 962 VLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
GGRTAHS+F IP+ E +TCN+ SD A L+K +LIIWDEAP
Sbjct: 949 --------------EGGRTAHSRFGIPLNPNEFTTCNMKVGSDRANLVKEASLIIWDEAP 994
Query: 1022 MAHKFCFEALDRTLKDIMSDTADGD-KIFGGKVIVFGGDFRQILPVVPRADRSDIINSSI 1080
M ++CFE+LDR+L DI + GD K FGGKV+VFGG
Sbjct: 995 MMSRYCFESLDRSLSDICGN---GDNKPFGGKVVVFGG---------------------- 1029
Query: 1081 NSSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGIVDFEIP 1140
+ S+A ++K FSEWIL VG+G+I EPNDG IP
Sbjct: 1030 -----------------------LSVSEAKDIKEFSEWILAVGDGRIVEPNDGEALIVIP 1066
Query: 1141 DDLLIKEFDDPIEAIMKSTYPNFLNMY--NNPDYLQQRAILASTIDVVDKINDYVLSIIP 1198
+ LI + DPIEAI Y + ++ N+P + Q++AIL T + V++IN+ +L +
Sbjct: 1067 SEFLITKAKDPIEAICTEIYGDITKIHEQNDPIFFQEKAILCPTNEDVNQINETMLDNLQ 1126
Query: 1199 GEEKEYFSSDSIDRSEVNDQCQSFQLFTPEFLSTLRTSGLPNHKIKLKVGTPIMLLRNLD 1258
GEE + SSDS+D +++ + + TP+FL++++ S LPNHK++LK+G P+MLLRN+D
Sbjct: 1127 GEEFTFLSSDSLDPADIGGK--NNPALTPDFLNSVKVSRLPNHKLRLKIGCPVMLLRNID 1184
Query: 1259 QVEGLCNGTRLIVTRMANHVIEAKIISGKNVGNLTYIPRMSMSPSQSPWPFKLIRRQFPI 1318
+ GL NGTRL +T+M +++A I++G G+L IPR+ ++PS + PF++ R Q P+
Sbjct: 1185 PIGGLMNGTRLRITQMGPFILQAMILTGDRAGHLVLIPRLKLAPSDTKLPFRMRRTQLPL 1244
Query: 1319 IVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQLYVAFSRVRTKAGLKI 1366
V +AMTINKSQGQ+L VG++L R FSHGQLYVA SRV +K LKI
Sbjct: 1245 AVCFAMTINKSQGQSLKRVGIFLLRPCFSHGQLYVAISRVTSKTRLKI 1292
Score = 338 bits (866), Expect = 2e-92
Identities = 194/529 (36%), Positives = 277/529 (51%), Gaps = 70/529 (13%)
Query: 1 MWYQERRRKSRESSNPEFGMCCGNGKIQIPFLPTPPPLLNDLMFNNYSTESKQFNQNIRL 60
MW+ ER K+ ++ +F CC G++++PFL P LL L N+ S+ F +NIR
Sbjct: 112 MWHGERIEKTVKNKKSKFTSCCLQGQVKLPFLKNSPELLYALPTNDDEI-SRHFRENIRA 170
Query: 61 YNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDN 120
YN +F F+S G + V GP +I ++ HR+GS+ P PKF QLYI DT+N
Sbjct: 171 YNMIFYFTSLGGDTENSVRASGGPQMFQIHRENYHRIGSLKPDNDIPPKFMQLYIVDTEN 230
Query: 121 ELQNRVQGIRN---------PNLNMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVENL 171
E+ NR + + N L + + + ++L+ N + R ARDR E L
Sbjct: 231 EVDNRSEALSNGVNADKYKKKELRKEIIDAVIKLLNSINPYVSHLRTARDRFNANPEETL 290
Query: 172 KLKLISDRTTDGRIYNQPTVSEVAALIVGDVD-SAARRDIIMERQSGRLERIDEFHPAYL 230
++++S R DGR+YN PT SEVA LI GD RDI+++ +SG+L+RI E P YL
Sbjct: 291 HMRIVSKREKDGRVYNVPTTSEVAMLIPGDFTIDMPSRDIVVQEKSGKLQRISEILPCYL 350
Query: 231 AYQYPLLFPYGEDGYRDDVLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILCARR 290
QYPLLFPYGEDG+R ++ +T
Sbjct: 351 PLQYPLLFPYGEDGFR--------------------------IGIEKHQT---------G 375
Query: 291 LFQQFLVDCFTMMEADRLKWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLPSSY 350
L+QQFLVD +T +E++ L +++ NQ+ LR Y S+ K QG LP+++
Sbjct: 376 LWQQFLVDSYTAIESNWLGYIKLNQTSLRAYNYNSVQKASEEGKSDLKDQGLACYLPATF 435
Query: 351 VGGRRYMDQLYFDGMAISSDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDIIARV 410
GG RYM +Y D MA+ GFPD FITFTCNP WPE+ RF G + L+ DRP+II ++
Sbjct: 436 TGGPRYMRNMYLDAMALCKHFGFPDYFITFTCNPKWPELIRFCGERNLRADDRPEIICKI 495
Query: 411 FKIKFDELLNDLTKKHILGKVVAYMYTIEFQKRGLPHAHILIFLHPSSKYPTPDHINQII 470
K+K D L+ DLTKK K + I+ K +HI+++I
Sbjct: 496 LKMKLDSLMLDLTKK----KPIREDKYIKLTK--------------------AEHIDKVI 531
Query: 471 SAEIPHPENDRELYKLVGTHMMHGPCGLAKPSSPCMKKKR*MFQVFPEK 519
SAEIP D EL++++ M+HGPCG+ P PCM+ + F EK
Sbjct: 532 SAEIPDKLKDPELFEVIKESMVHGPCGVVNPKCPCMENGKRRTDDFVEK 580
>At1g64410 unknown protein
Length = 753
Score = 293 bits (749), Expect = 6e-79
Identities = 153/293 (52%), Positives = 205/293 (69%), Gaps = 3/293 (1%)
Query: 842 RLTDDQIKNLTLTLIEKHMEKNRRSLKEFKGFPYPTDYVMEELGNRLIYDELNYDEDALK 901
+L+D++ + L I + + KN SL ++K P +D +E+ N I DE YD L
Sbjct: 454 QLSDEERQQYCLQEIARLLTKNGVSLSKWKQMPQISDEHVEKC-NHFILDERKYDRAYLT 512
Query: 902 LEFKRLFSTVTDEQRDIYHKIMSAVNGQNGGVFFLHGYGGTGKTFMWTTLAASLRSQGKI 961
+ + + VT EQ+ IY +IM V GGVFF++G+GGTGKTF+W L+A++RS+G I
Sbjct: 513 EKHEEWLTMVTLEQKKIYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAIRSKGDI 572
Query: 962 VLTVATSGIASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLAKLLKVTNLIIWDEAP 1021
L VA+SGIA+LLL GGRTAHS+F IP+ E+STCNI D +L+K NLIIWDEA
Sbjct: 573 SLNVASSGIAALLLDGGRTAHSRFGIPINPNESSTCNISRGLDFGELVKEANLIIWDEAH 632
Query: 1022 MAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSDIINSSIN 1081
M K CFE+LDRTL+DIM++ GDK FGGKVIVFGGDFRQ+L V+ A R +I+ +++N
Sbjct: 633 MMSKHCFESLDRTLRDIMNN--PGDKPFGGKVIVFGGDFRQVLSVINGAGREEIVFAALN 690
Query: 1082 SSYIWDECIVLTLTKNMRLRFNVGSSDADELKTFSEWILKVGEGKISEPNDGI 1134
SSYIW+ C VL LTKNMRL N+ + +++ FS+WIL VG+GKIS+PNDGI
Sbjct: 691 SSYIWEHCKVLELTKNMRLLANISEHEKRDIEYFSKWILDVGDGKISQPNDGI 743
Score = 99.4 bits (246), Expect = 1e-20
Identities = 46/87 (52%), Positives = 63/87 (71%), Gaps = 1/87 (1%)
Query: 435 MYTIEFQKRGLPHAHILIFLHPSSKYPTPDHINQIISAEIPHPENDRELYKLVGTHMMHG 494
MYTIEFQKRGLPHAHIL+F+HP+SK T + +++I+AEIP + ELY +V M+HG
Sbjct: 179 MYTIEFQKRGLPHAHILLFMHPTSKLSTAEDTDKVITAEIPDKKKKPELYAVVKDCMIHG 238
Query: 495 PCGLAKPSSPCMKKKR*MFQVFPEKVS 521
PCG+ P+SPCM+ + + FP+ S
Sbjct: 239 PCGVGHPNSPCMENGK-CKKYFPKSYS 264
Score = 63.2 bits (152), Expect = 1e-09
Identities = 35/80 (43%), Positives = 47/80 (58%), Gaps = 7/80 (8%)
Query: 51 SKQFNQNIRLYNSMFAFSSPGFKVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKF 110
+K F +NIR YN +F+F+S G KVD + GRGP IQ G++ P + KF
Sbjct: 72 AKHFRENIRAYNMLFSFTSIGGKVDHCLPKGRGPNMFAIQ-------GALKPKSVAKAKF 124
Query: 111 AQLYIFDTDNELQNRVQGIR 130
QLYI DT+NE+ NR +R
Sbjct: 125 QQLYIVDTENEVNNRYNIMR 144
>At4g04300 hypothetical protein
Length = 286
Score = 254 bits (648), Expect = 3e-67
Identities = 139/284 (48%), Positives = 189/284 (65%), Gaps = 21/284 (7%)
Query: 911 VTDEQRDIYHKIMSAVNGQNGGVFF-LHGYGGTGKTFMWTTLAASLRSQGKIVLTVATSG 969
+T EQR IY +I +AV GGVFF ++G GG GKTF+W TLAA RS+G+ L +A+SG
Sbjct: 2 LTPEQRGIYDQITNAVFNDMGGVFFFVYGSGGIGKTFIWKTLAAVGRSKGQTCLNIASSG 61
Query: 970 IASLLLPGGRTAHSKFKIPVPSFENSTCNIDGDSDLA---------------KLLKVTNL 1014
IASLLL GGR AH +F IP+ E S C I SDLA L+K +L
Sbjct: 62 IASLLLEGGRIAHYRFSIPLNPDEFSVCKIKPKSDLADLIKEASLIIWDKLVDLIKKASL 121
Query: 1015 IIWDEAPMAHKFCFEALDRTLKDIMSDTADGDKIFGGKVIVFGGDFRQILPVVPRADRSD 1074
IIWD+APM KFCFEALD++ DI+ +K+F GKV+VFGGDFRQ+LPV+ A R++
Sbjct: 122 IIWDKAPMKSKFCFEALDKSFSDIIKRV--DNKVFCGKVMVFGGDFRQVLPVINGAGRAE 179
Query: 1075 IINSSINSSYIWDECIVLTLTKNMRLRFN-VGSSDADELKTFSEWILKVGEGKISEPNDG 1133
+ SS+N+ YIWD C VL LTKNMRL N + +A E++ F +W+L VG+G+++EPNDG
Sbjct: 180 TVMSSLNAVYIWDHCKVLKLTKNMRLLNNDLSVDEAKEIQEFFDWLLVVGDGRVNEPNDG 239
Query: 1134 IVDFEIPDDLLIKEFDDPIEAIMKSTYPNFLNMY--NNPDYLQQ 1175
+IP++LLI+E D PIEAI + Y + ++ N+P + ++
Sbjct: 240 EALIDIPEELLIQEADIPIEAISREIYGDATKLHEINDPKFFRR 283
>At3g43330 putative protein
Length = 489
Score = 250 bits (639), Expect = 3e-66
Identities = 141/344 (40%), Positives = 202/344 (57%), Gaps = 22/344 (6%)
Query: 73 KVDKGVIPGRGPPTIRIQGQSCHRMGSMIPAAGKTPKFAQLYIFDTDNELQNRVQGIRNP 132
KVD + RGP IQG++ + MG++ P +G KF QLYI DT+NE++NR + N
Sbjct: 164 KVDHCLPKDRGPNMFAIQGENYYLMGALKPKSGAKAKFQQLYIVDTENEVKNRYNIMSNE 223
Query: 133 NL--NMQTVSKLQQMLDDTNCHAKSFRKARDRLRQGNVENLKLKLISDRTTDGRIYNQPT 190
N N Q K+ ++L+ N H K+FR ARDR E+ +++I+ R TD RIYN PT
Sbjct: 224 NESENGQKKKKIMKLLNRVNPHVKAFRYARDRFNTDAQESFHMRIIAARKTDPRIYNLPT 283
Query: 191 VSEVAALIVGDV-DSAARRDIIM-ERQSGRLERIDEFHPAYLAYQYPLLFPYGEDGYRDD 248
SEVAALI GD + + DI++ E SG+L+RI+ + + +G+ D
Sbjct: 284 ASEVAALIPGDFHEDMDKLDIVLQETTSGKLKRINGYRLGIKKAR--------TNGHGD- 334
Query: 249 VLHRGIVAGKQSKMDRLTIREWLTFRLQSRKTEAMTILCARRLFQQFLVDCFTMMEADRL 308
GK+ + D +++R+W +RLQ RK E + ++RL QQF+VD +TM+E++RL
Sbjct: 335 --------GKKEQKD-VSMRQWFAYRLQERKAEKHILFRSKRLLQQFIVDAYTMIESNRL 385
Query: 309 KWLRKNQSKLRVGKYRSLSNNQGNVDGQTKKQGKRVVLPSSYVGGRRYMDQLYFDGMAIS 368
++++KNQ K R + N + + + QGK V +P+S+ GG RYM Y D MAI
Sbjct: 386 RYIKKNQPKFRSSTREPIQNASNSGNNDLENQGKEVKIPASFTGGPRYMTNNYLDAMAIC 445
Query: 369 SDVGFPDLFITFTCNPSWPEIQRFVGAKGLKPHDRPDIIARVFK 412
GFP LFITFTCNP WPEI RF + LK DR DII R+ +
Sbjct: 446 KHFGFPSLFITFTCNPKWPEIVRFCKERNLKSEDRQDIICRILR 489
>At3g51700 unknown protein
Length = 344
Score = 237 bits (605), Expect = 3e-62
Identities = 126/279 (45%), Positives = 182/279 (65%), Gaps = 7/279 (2%)
Query: 1113 KTFSEWILKVGEGKISEPNDGIVDFEIPDDLLIKEFDDPIEAIMKSTYPNFLNMYNNPDY 1172
+ F++WI +G I++PNDG +I +DLLI E DPI+ I+ Y NPD+
Sbjct: 58 EAFTKWITNIGGENINKPNDGETKIDIHEDLLITECKDPIKTIVDEVYGESFTESYNPDF 117
Query: 1173 LQQRAILASTIDVVDKINDYVLSIIPGEEKEYFSSDSI--DRSEVNDQCQSFQLFTPEFL 1230
Q+RAIL T DV D+INDY+LS + GEE + + +D+I + ND+ L+ EFL
Sbjct: 118 YQERAILCHTNDVADEINDYMLSQLQGEETKCYGADTIYPTHASPNDK----MLYPLEFL 173
Query: 1231 STLRTSGLPNHKIKLKVGTPIMLLRNLDQVEGLCNGTRLIVTRMANHVIEAKIISGKNVG 1290
++++ G P+ K++LKVG P+MLLR+L L GTRL +TR+ V+EA II+G N G
Sbjct: 174 NSIKIPGFPDFKLRLKVGAPVMLLRDLAPYGWLRKGTRLQITRVETFVLEAMIITGNNHG 233
Query: 1291 NLTYIPRMSMSPSQSPWPFKLIRRQFPIIVSYAMTINKSQGQTLDSVGLYLPRSVFSHGQ 1350
IPR+ ++ +P K+ RRQFP+ +++AMTI++SQ QTL VG+YLPR + HGQ
Sbjct: 234 EKVLIPRIPSDLREAKFPIKMRRRQFPVKLAFAMTIDESQRQTLSKVGIYLPRQLLFHGQ 293
Query: 1351 LYVAFSRVRTKAGLKILIHDLEKKP-LSNTTNVVYKEVF 1388
YVA S+V+++AGLK+LI D + KP T NVV+KE+F
Sbjct: 294 RYVAISKVKSRAGLKVLITDKDGKPDQEETKNVVFKELF 332
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.336 0.147 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 30,229,147
Number of Sequences: 26719
Number of extensions: 1316160
Number of successful extensions: 5026
Number of sequences better than 10.0: 39
Number of HSP's better than 10.0 without gapping: 34
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 4776
Number of HSP's gapped (non-prelim): 104
length of query: 1391
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1279
effective length of database: 8,326,068
effective search space: 10649040972
effective search space used: 10649040972
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC148528.7


