

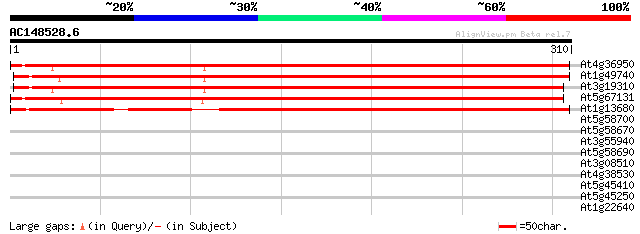
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148528.6 + phase: 0 /partial
(310 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g36950 MAP3K-like protein kinase 358 2e-99
At1g49740 unknown protein 346 1e-95
At3g19310 unknown protein 325 1e-89
At5g67131 unknown protein 317 7e-87
At1g13680 hypothetical protein 271 4e-73
At5g58700 phosphoinositide-specific phospholipase C4 (PLC4) 33 0.26
At5g58670 phosphoinositide specific phospholipase C (AtPLC1) 33 0.26
At3g55940 phosphoinositide-specific phospholipase C - like protein 33 0.26
At5g58690 phosphoinositide-specific phospholipase C-line (MZN1.13) 32 0.44
At3g08510 phosphoinositide specific phospholipase C (AtPLC2) 32 0.44
At4g38530 phosphoinositide-specific phospholipase C 30 2.2
At5g45410 unknown protein 28 6.3
At5g45250 disease resistance protein RPS4 28 6.3
At1g22640 transcription factor like protein (MYB3) 28 6.3
>At4g36950 MAP3K-like protein kinase
Length = 799
Score = 358 bits (919), Expect = 2e-99
Identities = 177/316 (56%), Positives = 222/316 (70%), Gaps = 8/316 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNS 56
++GETCS ++C GL C C N+ S CTR++ +P SKV LPFN YSWLTTHNS
Sbjct: 338 EMGETCS-STSECDAGLSCQSCPANGNTGSTCTRIQPLNPTSKVNGLPFNKYSWLTTHNS 396
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFT 113
YA AN + S + S NQEDSIT+QL+NGVRGIMLD +D+ DIWLC G C FT
Sbjct: 397 YAITGANSATGSFLVSPKNQEDSITNQLKNGVRGIMLDTYDFQNDIWLCHSTGGTCFNFT 456
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AFQPAIN L+EIN FL + +EIVT+ ++D V S G+ VFN +GL KF P+ +MPK+
Sbjct: 457 AFQPAINALKEINDFLESNLSEIVTIILEDYVKSQMGLTNVFNASGLSKFLLPISRMPKD 516
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G+DW TV M++ N RL+VFTS KEASE +AY+WNY+VEN+YGNDGM C R+ES
Sbjct: 517 GTDWPTVDDMVKQNQRLVVFTSKKDKEASEGLAYQWNYMVENQYGNDGMKDGSCSSRSES 576
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
++T ++SLV NY+ NS +AC DNSSPLI M TC++ AG RWPN+IAVDFY+R
Sbjct: 577 SSLDTMSRSLVFQNYFETSPNSTQACADNSSPLIEMMRTCHEAAGKRWPNFIAVDFYQRS 636
Query: 294 DGGGAPEALDVANRNL 309
D GGA EA+D AN L
Sbjct: 637 DSGGAAEAVDEANGRL 652
>At1g49740 unknown protein
Length = 359
Score = 346 bits (887), Expect = 1e-95
Identities = 171/314 (54%), Positives = 220/314 (69%), Gaps = 8/314 (2%)
Query: 3 GETCSRDVNDCGTGLQCLECNSQS----RCTRVRTSSPISKVMELPFNHYSWLTTHNSYA 58
G+TC + N C GL C C + + RC+R + +PI+K LPFN YSWLTTHNS+A
Sbjct: 30 GKTCITNSN-CDAGLHCETCIANTDFRPRCSRTQPINPITKAKGLPFNKYSWLTTHNSFA 88
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTAF 115
S I + NQ+DSIT QL NGVRG MLDM+D+ DIWLC G C FTAF
Sbjct: 89 RLGEVSRTGSAILAPTNQQDSITSQLNNGVRGFMLDMYDFQNDIWLCHSFDGTCFNFTAF 148
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
QPAIN+LRE FL +++ E+VT+ I+D V SP G+ KVF+ AGLRKF FPV +MPKNG
Sbjct: 149 QPAINILREFQVFLEKNKEEVVTIIIEDYVKSPKGLTKVFDAAGLRKFMFPVSRMPKNGG 208
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW + M+R N RL+VFTS++ KEA+E IAY+W Y+VEN+YGN G+ C +RA+S P
Sbjct: 209 DWPRLDDMVRKNQRLLVFTSDSHKEATEGIAYQWKYMVENQYGNGGLKVGVCPNRAQSAP 268
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
M+ +KSLVL+N++ + + ACK NS+ L+ + TCY+ AG RWPN+IAVDFYKR DG
Sbjct: 269 MSDKSKSLVLVNHFPDAADVIVACKQNSASLLESIKTCYQAAGQRWPNFIAVDFYKRSDG 328
Query: 296 GGAPEALDVANRNL 309
GGAP+A+DVAN NL
Sbjct: 329 GGAPQAVDVANGNL 342
>At3g19310 unknown protein
Length = 413
Score = 325 bits (834), Expect = 1e-89
Identities = 157/311 (50%), Positives = 212/311 (67%), Gaps = 8/311 (2%)
Query: 3 GETCSRDVNDCGTGLQCLEC----NSQSRCTRVRTSSPISKVMELPFNHYSWLTTHNSYA 58
GETC N C GL C C + + RC+R++ +P +KV LP+N YSWLTTHNS+A
Sbjct: 30 GETCIVSKN-CDRGLHCESCLASDSFRPRCSRMQPINPTTKVKGLPYNKYSWLTTHNSFA 88
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCR---GPCTIFTAF 115
A S I + NQ+DSIT QL NGVRG MLD++D+ DIWLC G C +TAF
Sbjct: 89 RMGAKSGTGSMILAPSNQQDSITSQLLNGVRGFMLDLYDFQNDIWLCHSYGGNCFNYTAF 148
Query: 116 QPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGS 175
QPA+N+L+E FL +++ +VT+ ++D V SPNG+ +VF+ +GLR F FPV +MPKNG
Sbjct: 149 QPAVNILKEFQVFLDKNKDVVVTLILEDYVKSPNGLTRVFDASGLRNFMFPVSRMPKNGE 208
Query: 176 DWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYP 235
DW T+ M+ N RL+VFTSN KEASE IA+ W Y++EN+YG+ GM C +R ES
Sbjct: 209 DWPTLDDMICQNQRLLVFTSNPQKEASEGIAFMWRYMIENQYGDGGMKAGVCTNRPESVA 268
Query: 236 MNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDG 295
M ++SL+L+NY+ + + +CK NS+PL+ + C + +G RWPN+IAVDFYKR DG
Sbjct: 269 MGDRSRSLILVNYFPDTADVIGSCKQNSAPLLDTVKNCQEASGKRWPNFIAVDFYKRSDG 328
Query: 296 GGAPEALDVAN 306
GGAP+A+DVAN
Sbjct: 329 GGAPKAVDVAN 339
>At5g67131 unknown protein
Length = 426
Score = 317 bits (811), Expect = 7e-87
Identities = 157/313 (50%), Positives = 213/313 (67%), Gaps = 8/313 (2%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQSR----CTRVRTSSPISKVMELPFNHYSWLTTHNS 56
Q+ ++CS DC +GL C +C + R CTR + +SP S + LPFN Y+WL THN+
Sbjct: 35 QLLDSCS-SATDCVSGLYCGDCPAVGRSKPVCTRGQATSPTSIINGLPFNKYTWLMTHNA 93
Query: 57 YASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLC---RGPCTIFT 113
+++ A L + + NQED+IT+QL+NGVRG+MLDM+D+ DIWLC RG C FT
Sbjct: 94 FSNANAPLLPGVERITFYNQEDTITNQLQNGVRGLMLDMYDFNNDIWLCHSLRGQCFNFT 153
Query: 114 AFQPAINVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKN 173
AFQPAIN+LRE+ FL+++ TEIVT+ I+D V P G++ +F AGL K+WFPV KMP+
Sbjct: 154 AFQPAINILREVEAFLSQNPTEIVTIIIEDYVHRPKGLSTLFANAGLDKYWFPVSKMPRK 213
Query: 174 GSDWLTVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAES 233
G DW TV M++ NHRL+VFTS A KE E +AY+W Y+VEN+ G+ G+ R C +R ES
Sbjct: 214 GEDWPTVTDMVQENHRLLVFTSVAAKEDEEGVAYQWRYMVENESGDPGVKRGSCPNRKES 273
Query: 234 YPMNTTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRG 293
P+N+ + SL LMNY+ +ACK++S+PL + TC K GNR PN++AV+FY R
Sbjct: 274 QPLNSKSSSLFLMNYFPTYPVEKDACKEHSAPLAEMVGTCLKSGGNRMPNFLAVNFYMRS 333
Query: 294 DGGGAPEALDVAN 306
DGGG E LD N
Sbjct: 334 DGGGVFEILDRMN 346
>At1g13680 hypothetical protein
Length = 300
Score = 271 bits (692), Expect = 4e-73
Identities = 143/311 (45%), Positives = 193/311 (61%), Gaps = 24/311 (7%)
Query: 1 QIGETCSRDVNDCGTGLQCLECNSQ-SRCTRVRTSSPISKVME-LPFNHYSWLTTHNSYA 58
Q+G+ CS D DC GL C +C +RC R + S V +PFN Y++LTTHNSYA
Sbjct: 2 QLGDQCSSD-EDCNVGLGCFKCGIDVARCVRSNITDQFSIVNNSMPFNKYAFLTTHNSYA 60
Query: 59 SRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDMHDYYGDIWLCRGPCTIFTAFQPA 118
I+ K V QED+I QL +GVR +MLD +DY GD A
Sbjct: 61 -------IEGKALHVATQEDTIVQQLNSGVRALMLDTYDYEGD--------------NRA 99
Query: 119 INVLREINTFLTRHRTEIVTVFIKDRVTSPNGVNKVFNKAGLRKFWFPVYKMPKNGSDWL 178
I+ +EI FLT + +EIVT+ ++D V S NG+ KVF +GL+KFWFPV MP G DW
Sbjct: 100 IDTFKEIFAFLTANPSEIVTLILEDYVKSQNGLTKVFTDSGLKKFWFPVQNMPIGGQDWP 159
Query: 179 TVKKMLRMNHRLIVFTSNATKEASERIAYEWNYVVENKYGNDGMGRDHCLHRAESYPMNT 238
VK M+ NHRLIVFTS +K+ +E IAY+WNY+VEN+YG+DG+ D C +RA+S +
Sbjct: 160 LVKDMVANNHRLIVFTSAKSKQETEGIAYQWNYMVENQYGDDGVKPDECSNRADSALLTD 219
Query: 239 TTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDAGNRWPNYIAVDFYKRGDGGGA 298
TK+LV +N+++ V C++NS L+ + TCY AGNRW N++AV+FYKR +GGG
Sbjct: 220 KTKALVSVNHFKTVPVKILTCEENSEQLLDMIKTCYVAAGNRWANFVAVNFYKRSNGGGT 279
Query: 299 PEALDVANRNL 309
+A+D N L
Sbjct: 280 FQAIDKLNGEL 290
>At5g58700 phosphoinositide-specific phospholipase C4 (PLC4)
Length = 597
Score = 32.7 bits (73), Expect = 0.26
Identities = 21/55 (38%), Positives = 30/55 (54%), Gaps = 8/55 (14%)
Query: 41 MELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDM 95
M+ P +HY T HNSY + +++SS + E I D LR GVR + LD+
Sbjct: 116 MDAPLSHYFIFTGHNSYLT-------GNQLSSNCS-ELPIADALRRGVRVVELDL 162
>At5g58670 phosphoinositide specific phospholipase C (AtPLC1)
Length = 561
Score = 32.7 bits (73), Expect = 0.26
Identities = 22/55 (40%), Positives = 28/55 (50%), Gaps = 8/55 (14%)
Query: 41 MELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDM 95
M P +HY T HNSY + L+ +S I + I LRNGVR I LD+
Sbjct: 107 MNQPLSHYFLYTGHNSYLT-GNQLNSNSSI-------EPIVKALRNGVRVIELDL 153
>At3g55940 phosphoinositide-specific phospholipase C - like protein
Length = 584
Score = 32.7 bits (73), Expect = 0.26
Identities = 25/68 (36%), Positives = 34/68 (49%), Gaps = 13/68 (19%)
Query: 33 TSSPISKV-----MELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNG 87
T+SP+S + M+ P +HY T HNSY + LS D E I + L+ G
Sbjct: 92 TNSPLSSLEVHQDMDAPLSHYFIYTGHNSYLT-GNQLSSDC-------SELPIIEALKKG 143
Query: 88 VRGIMLDM 95
VR I LD+
Sbjct: 144 VRVIELDI 151
>At5g58690 phosphoinositide-specific phospholipase C-line (MZN1.13)
Length = 578
Score = 32.0 bits (71), Expect = 0.44
Identities = 21/61 (34%), Positives = 31/61 (50%), Gaps = 8/61 (13%)
Query: 35 SPISKVMELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLD 94
S + + M P +HY T+HNSY + I+S+ S V + L+ GVR + LD
Sbjct: 108 SKVHQDMASPLSHYFIYTSHNSYLT---GNQINSECSDV-----PLIKALKRGVRALELD 159
Query: 95 M 95
M
Sbjct: 160 M 160
>At3g08510 phosphoinositide specific phospholipase C (AtPLC2)
Length = 581
Score = 32.0 bits (71), Expect = 0.44
Identities = 22/55 (40%), Positives = 27/55 (49%), Gaps = 8/55 (14%)
Query: 41 MELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIMLDM 95
M+ P +HY T HNSY + LS D E I D L+ GVR I LD+
Sbjct: 105 MDAPISHYFIFTGHNSYLT-GNQLSSDC-------SEVPIIDALKKGVRVIELDI 151
>At4g38530 phosphoinositide-specific phospholipase C
Length = 526
Score = 29.6 bits (65), Expect = 2.2
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 8/62 (12%)
Query: 34 SSPISKVMELPFNHYSWLTTHNSYASRAANLSIDSKISSVMNQEDSITDQLRNGVRGIML 93
S + M+ P +HY T HNSY + ++S+ SSV + I LR GV+ I L
Sbjct: 63 SGQVHHDMKAPLSHYFVYTGHNSYLT---GNQVNSR-SSV----EPIVQALRKGVKVIEL 114
Query: 94 DM 95
D+
Sbjct: 115 DL 116
>At5g45410 unknown protein
Length = 342
Score = 28.1 bits (61), Expect = 6.3
Identities = 14/35 (40%), Positives = 19/35 (54%)
Query: 96 HDYYGDIWLCRGPCTIFTAFQPAINVLREINTFLT 130
H+ GD WL G C I +F+ A V+ I+ LT
Sbjct: 138 HEAVGDEWLKTGNCPIAKSFRAASKVMPLISKALT 172
>At5g45250 disease resistance protein RPS4
Length = 1217
Score = 28.1 bits (61), Expect = 6.3
Identities = 16/53 (30%), Positives = 30/53 (56%), Gaps = 4/53 (7%)
Query: 17 LQCLECNSQSRCTRVRT-SSPISKVMELPFNHYSWLTTHNSYASRAANLSIDS 68
LQCL+ + C+ ++T S P++++M NH +++ T+ +AA I S
Sbjct: 885 LQCLDAHG---CSSLKTVSKPLARIMPTEQNHSTFIFTNCENLEQAAKEEITS 934
>At1g22640 transcription factor like protein (MYB3)
Length = 257
Score = 28.1 bits (61), Expect = 6.3
Identities = 25/77 (32%), Positives = 33/77 (42%), Gaps = 12/77 (15%)
Query: 222 MGRDHCLHRAESYPMN----TTTKSLVLMNYYRNVLNSNEACKDNSSPLIRKMHTCYKDA 277
MGR C +A MN T + +L++Y R E C S P + C K
Sbjct: 1 MGRSPCCEKAH---MNKGAWTKEEDQLLVDYIRK---HGEGCW-RSLPRAAGLQRCGKSC 53
Query: 278 GNRWPNYIAVDFYKRGD 294
RW NY+ D KRG+
Sbjct: 54 RLRWMNYLRPDL-KRGN 69
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,288,023
Number of Sequences: 26719
Number of extensions: 301900
Number of successful extensions: 700
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 684
Number of HSP's gapped (non-prelim): 14
length of query: 310
length of database: 11,318,596
effective HSP length: 99
effective length of query: 211
effective length of database: 8,673,415
effective search space: 1830090565
effective search space used: 1830090565
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148528.6


