

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.4 - phase: 0
(340 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
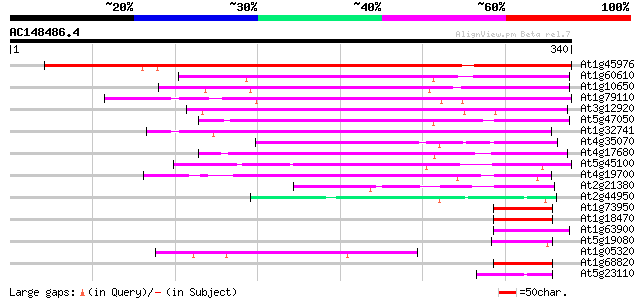
Score E
Sequences producing significant alignments: (bits) Value
At1g45976 S-ribonuclease binding like protein (SBP1) 433 e-122
At1g60610 unknown protein 161 5e-40
At1g10650 similar to putative inhibitor of apoptosis gi|3924605;... 154 5e-38
At1g79110 unknown protein 124 6e-29
At3g12920 unknown protein 119 2e-27
At5g47050 unknown protein 118 5e-27
At1g32741 unknown protein 115 4e-26
At4g35070 unknown protein 114 6e-26
At4g17680 unknown protein 110 1e-24
At5g45100 unknown protein 94 8e-20
At4g19700 unknown protein 91 9e-19
At2g21380 putative kinesin heavy chain 54 9e-08
At2g44950 unknown protein 49 5e-06
At1g73950 unknown protein (At1g73950) 48 9e-06
At1g18470 unknown protein 47 1e-05
At1g63900 putative RING zinc finger protein 45 6e-05
At5g19080 unknown protein 44 2e-04
At1g05320 unknown protein 44 2e-04
At1g68820 unknown protein (At1g68820) 43 3e-04
At5g23110 putative protein 42 6e-04
>At1g45976 S-ribonuclease binding like protein (SBP1)
Length = 325
Score = 433 bits (1114), Expect = e-122
Identities = 212/323 (65%), Positives = 261/323 (80%), Gaps = 11/323 (3%)
Query: 22 TKSFRNLQTIEGQMSQQMAFYNPTDLQDQSQHPPYIPPFHVVGFAPGPVIPADGSDGG-- 79
+K+FR+ I+GQ+S ++ F +L DQSQHPPYIPPFHV GFAPGPV+ DGSDGG
Sbjct: 10 SKNFRDFCGIDGQISPELGFNRSENLHDQSQHPPYIPPFHVAGFAPGPVVQIDGSDGGNG 69
Query: 80 VDLHWNFGL--EPERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDS 137
D WN+GL EP R+R+KEQDFLENNSQISS+DFLQ RSVSTGLGLSLDN R+AS+ S
Sbjct: 70 ADFEWNYGLGLEPRRERIKEQDFLENNSQISSIDFLQARSVSTGLGLSLDNARVASSDGS 129
Query: 138 ALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLR 197
ALLSL+GDDIDRELQ+QD ++DRFLK+QG+QLR IL+K++ Q ++VS++E+KV+QKLR
Sbjct: 130 ALLSLVGDDIDRELQRQDADIDRFLKIQGDQLRHAILDKIKRGQQKTVSLMEEKVVQKLR 189
Query: 198 EKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKE 257
EK+ E+E IN++N ELE +MEQL++EA AWQQRA+YNENMIAAL +NL +A + RDS E
Sbjct: 190 EKDEELERINRKNKELEVRMEQLTMEAEAWQQRAKYNENMIAALNYNLDRAQGRPRDSIE 249
Query: 258 GCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKD 317
GCGDSEVDDTASC NGR N N+N K +M C+ C V E+ M+LLPC H+CLCK+
Sbjct: 250 GCGDSEVDDTASCFNGR-------DNSNNNTKTMMMCRFCGVREMCMLLLPCNHMCLCKE 302
Query: 318 CESKLSFCPLCQSSKFIGMEVYM 340
CE KLS CPLCQSSKF+GMEVYM
Sbjct: 303 CERKLSSCPLCQSSKFLGMEVYM 325
>At1g60610 unknown protein
Length = 340
Score = 161 bits (407), Expect = 5e-40
Identities = 93/257 (36%), Positives = 147/257 (57%), Gaps = 28/257 (10%)
Query: 103 NNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSL-------IGDDIDRELQQQD 155
NN+ + + + VSTGL LS D+ S+ SA S+ +GD+I +L +Q+
Sbjct: 90 NNNTVVQDEVPKQNLVSTGLRLSYDDDERNSSVTSANGSITTPVYQSLGDNIRLDLNRQN 149
Query: 156 LEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELED 215
E+D+F+K + +Q+ + + + Q V+ +E V +KL+EK+ E+E++NK+N EL D
Sbjct: 150 DELDQFIKFRADQMAKGVRDIKQRHVTSFVTALEKDVSKKLQEKDHEIESMNKKNRELVD 209
Query: 216 QMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDS-------------KEGCGDS 262
+++Q++VEA W +A+YNE+++ ALK NLQQ G D+ KEG GDS
Sbjct: 210 KIKQVAVEAQNWHYKAKYNESVVNALKVNLQQVMSHGNDNNAVGGGVADHHQMKEGFGDS 269
Query: 263 EVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKL 322
E+DD A+ N N M+CK C V V+++L+PC+HL LCKDC+
Sbjct: 270 EIDDEAASYN--------YLNIPGMPSTGMRCKLCNVKNVSVLLVPCRHLSLCKDCDVFT 321
Query: 323 SFCPLCQSSKFIGMEVY 339
CP+CQS K ++V+
Sbjct: 322 GVCPVCQSLKTSSVQVF 338
>At1g10650 similar to putative inhibitor of apoptosis gi|3924605;
similar to ESTs gb|AI994578.1, gb|T04171, gb|AA728525
Length = 339
Score = 154 bits (390), Expect = 5e-38
Identities = 93/267 (34%), Positives = 152/267 (56%), Gaps = 22/267 (8%)
Query: 91 ERKRLKEQDFLENNSQISSVDFLQPRS--VSTGLGLSLDNTRLASTGDSALLSLIG---- 144
+R++ K Q L N +SV P+ VSTGL LS D+ S+ SA S++
Sbjct: 75 QRQQQKLQMSLNYNYNNTSVREEVPKENLVSTGLRLSYDDDEHNSSVTSASGSILAASPI 134
Query: 145 -----DDIDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREK 199
D + +L +Q E D+F+K+Q Q+ + + + Q ++ +E V +KL+EK
Sbjct: 135 FQSLDDSLRIDLHRQKDEFDQFIKIQAAQMAKGVRDMKQRHIASFLTTLEKGVSKKLQEK 194
Query: 200 ETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGR------ 253
+ E+ ++NK+N EL ++++Q+++EA W RA+YNE+++ LK NLQQA
Sbjct: 195 DHEINDMNKKNKELVERIKQVAMEAQNWHYRAKYNESVVNVLKANLQQAMSHNNSVIAAA 254
Query: 254 -DSKEGCGDSEVDDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHL 312
KEG GDSE+DD AS +D + +N N + M+CK C V EV+++++PC+HL
Sbjct: 255 DQGKEGFGDSEIDDAAS----SYIDPNNNNNNNMGIHQRMRCKMCNVKEVSVLIVPCRHL 310
Query: 313 CLCKDCESKLSFCPLCQSSKFIGMEVY 339
LCK+C+ CP+C+S K ++V+
Sbjct: 311 SLCKECDVFTKICPVCKSLKSSCVQVF 337
>At1g79110 unknown protein
Length = 355
Score = 124 bits (312), Expect = 6e-29
Identities = 91/301 (30%), Positives = 136/301 (44%), Gaps = 29/301 (9%)
Query: 58 PPFHVVGFAPG-PVIPADGSDGGVDLHWNFGLEPERKRLKEQDFLENNSQISSVDFLQPR 116
P HV G + P +DG +L F P RKR ++ + S + PR
Sbjct: 64 PLLHVYGGSDTIPTTAGYYADGATNLDCEFFPLPTRKRSRDS----SRSNYHHLLLQNPR 119
Query: 117 SVSTGLGLSLDNTRLASTGDSALLSLIGDDID--RELQQQDLEMDRFLKLQGEQLRQTIL 174
S S +T + L S +G DID + QQ E+DRF+ L E+++ I
Sbjct: 120 SSSCV-------NAATTTTTTTLFSFLGQDIDISSHMNQQQHEIDRFVSLHMERVKYEIE 172
Query: 175 EKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYN 234
EK + + IE ++++LR KE E E I K N LE++++ LS+E W+ A+ N
Sbjct: 173 EKRKRQARTIMEAIEQGLVKRLRVKEEERERIGKVNHALEERVKSLSIENQIWRDLAQTN 232
Query: 235 ENMIAALKFNLQQAYLQGRDSKEGCG----DSEVDDTASCCNG-----------RSLDFH 279
E L+ NL+ Q +D G G +E DD SCC R +
Sbjct: 233 EATANHLRTNLEHVLAQVKDVSRGAGLEKNMNEEDDAESCCGSSCGGGGEETVRRRVGLE 292
Query: 280 LLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
+ + + + C+ C E ++LLPC+HLCLC C S + CP+C S K + V
Sbjct: 293 REAQDKAERRRRRMCRNCGEEESCVLLLPCRHLCLCGVCGSSVHTCPICTSPKNASVHVN 352
Query: 340 M 340
M
Sbjct: 353 M 353
>At3g12920 unknown protein
Length = 335
Score = 119 bits (298), Expect = 2e-27
Identities = 72/246 (29%), Positives = 123/246 (49%), Gaps = 15/246 (6%)
Query: 108 SSVDFLQP-----RSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFL 162
++VDFL+P RS + L+ + L +G D+ +QQ ++DR +
Sbjct: 86 NNVDFLRPVSSRKRSREESVVLNPSAYMQIQKNPTDPLMFLGQDLSSNVQQHHFDIDRLI 145
Query: 163 KLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSV 222
E++R I EK + + V +E +++ LR K+ E+ +I K N+ LE++++ L V
Sbjct: 146 SNHVERMRMEIEEKRKTQGRRIVEAVEQGLMKTLRAKDDEINHIGKLNLFLEEKVKSLCV 205
Query: 223 EAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGR-----SLD 277
E W+ A+ NE + AL+ NLQQ ++ + DD SCC +
Sbjct: 206 ENQIWRDVAQSNEATVNALRSNLQQVLAAVERNRWEEPPTVADDAQSCCGSNDEGDSEEE 265
Query: 278 FHLLSNENSNMKDLMK-----CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSK 332
L+ E + K + + C++C E +++LLPC+H+CLC C S L+ CP+C+S K
Sbjct: 266 RWKLAGEAQDTKKMCRVGMSMCRSCGKGEASVLLLPCRHMCLCSVCGSSLNTCPICKSPK 325
Query: 333 FIGMEV 338
+ V
Sbjct: 326 TASLHV 331
>At5g47050 unknown protein
Length = 300
Score = 118 bits (295), Expect = 5e-27
Identities = 73/232 (31%), Positives = 117/232 (49%), Gaps = 16/232 (6%)
Query: 115 PRSVSTGLGLSLDNTRLASTGDSALLSL-IGDDIDRELQQQDLEMDRFLKLQGEQLRQTI 173
P VSTGL LS + ++ + LS I D+ E++ Q E++RFL++QGEQL++ +
Sbjct: 76 PNVVSTGLRLSREQSQ---NQEQRFLSFPITGDVAGEIKSQTDELNRFLQIQGEQLKRML 132
Query: 174 LEKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARY 233
E + + + E+ V ++LREKE E+E +R++ELE + Q+ EA AWQ RA
Sbjct: 133 AENSERNYRELLRTTEESVRRRLREKEAEIEKATRRHVELEARATQIETEARAWQMRAAA 192
Query: 234 NENMIAALKFNLQQAYLQGRDS------KEGCGDSEVDDTASCCNGRSLDFHLLSNENSN 287
E +L+ L QA + + G + D A +D +
Sbjct: 193 REAEATSLQAQLHQAVVVAHGGGVITTVEPQSGSVDGVDEAEDAESAYVDPDRVEMIGPG 252
Query: 288 MKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
C+ CR T++ LPC+HL +C +C+ + CPLC S+K +EV+
Sbjct: 253 ------CRICRRRSATVLALPCRHLVMCTECDGSVRICPLCLSTKNSSVEVF 298
>At1g32741 unknown protein
Length = 312
Score = 115 bits (287), Expect = 4e-26
Identities = 82/258 (31%), Positives = 125/258 (47%), Gaps = 17/258 (6%)
Query: 84 WNFGLEPERKRLKEQDFLENNSQISSVDFLQ-PRSVSTGL---------GLSLDNTRLAS 133
+N G +RKR +E NN Q+ + Q P+ + L + RL S
Sbjct: 46 FNNGSSNQRKRRRET----NNHQLLPMQSHQFPQVIDLSLLHNYNHPPSNMVHTGLRLFS 101
Query: 134 TGDSAL-LSLIGDDI-DRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDK 191
D A +S + +D+ + +Q E+D FL Q E+LR+T+ EK + + +E+
Sbjct: 102 GEDQAQKISHLSEDVFAAHINRQSEELDEFLHAQAEELRRTLAEKRKMHYKALLGAVEES 161
Query: 192 VLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ 251
+++KLREKE E+E +R+ EL + QL E WQ+RA+ +E+ A+L+ LQQA Q
Sbjct: 162 LVRKLREKEVEIERATRRHNELVARDSQLRAEVQVWQERAKAHEDAAASLQSQLQQAVNQ 221
Query: 252 GRDSKEGCGDSEVDDTASCCNGRS-LDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCK 310
DS + C S +D + CKACR E T+V+LPC+
Sbjct: 222 CAGGCVSAQDSRAAEEGLLCTTISGVDDAESVYVDPERVKRPNCKACREREATVVVLPCR 281
Query: 311 HLCLCKDCESKLSFCPLC 328
HL +C C+ CPLC
Sbjct: 282 HLSICPGCDRTALACPLC 299
>At4g35070 unknown protein
Length = 265
Score = 114 bits (286), Expect = 6e-26
Identities = 66/187 (35%), Positives = 101/187 (53%), Gaps = 11/187 (5%)
Query: 150 ELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENINKR 209
++++Q E+D+F+K+Q E+LR + E+ + + +E K L + +KE E+ +
Sbjct: 77 QMEKQKQEIDQFIKIQNERLRYVLQEQRKREMEMILRKMESKALLLMSQKEEEMSKALNK 136
Query: 210 NMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGC---GDSEVDD 266
NMELED + ++ +E WQ+ AR NE ++ L L+Q R+ C G++EV+D
Sbjct: 137 NMELEDLLRKMEMENQTWQRMARENEAIVQTLNTTLEQV----RERAATCYDAGEAEVED 192
Query: 267 TASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVL-LPCKHLCLCKDCESKLSFC 325
S C G D + L + M C C N VT VL LPC+HLC C DCE L C
Sbjct: 193 EGSFCGGEG-DGNSLPAKKMKMSSC--CCNCGSNGVTRVLFLPCRHLCCCMDCEEGLLLC 249
Query: 326 PLCQSSK 332
P+C + K
Sbjct: 250 PICNTPK 256
>At4g17680 unknown protein
Length = 314
Score = 110 bits (275), Expect = 1e-24
Identities = 73/232 (31%), Positives = 116/232 (49%), Gaps = 21/232 (9%)
Query: 115 PRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMDRFLKLQGEQLRQTIL 174
P VSTGL L D S S + D+ ++++Q E+DRF++ QGE+LR+T+
Sbjct: 94 PNVVSTGLRLFHDQ----SQNQQQFFSSLPGDVTGKIKRQRDELDRFIQTQGEELRRTLA 149
Query: 175 EKVQATQLQSVSIIEDKVLQKLREKETEVENINKRNMELEDQMEQLSVEAGAWQQRARYN 234
+ + ++ + E+ V +KLR+KE E+E +R+ ELE ++ + EA WQ RA
Sbjct: 150 DNRERRYVELLCAAEEIVGRKLRKKEAELEKATRRHAELEARVAHIVEEARNWQLRAATR 209
Query: 235 ENMIAALKFNLQQAYLQGRDSK-------EGCGDSEVDDTASCCNGRSLDFHLLSNENSN 287
E +++L +LQQA D+ E GD+E + A L+
Sbjct: 210 EAEVSSLHAHLQQAIANRLDTAAKQSTFGEDGGDAEEAEDAESVYVDPERIELIG----- 264
Query: 288 MKDLMKCKACRVNEVTMVLLPCKHLCLCKDCE-SKLSFCPLCQSSKFIGMEV 338
C+ CR T++ LPC+HL LC C+ + CP+C + K G+EV
Sbjct: 265 ----PSCRICRRKSATVMALPCQHLILCNGCDVGAVRVCPICLAVKTSGVEV 312
>At5g45100 unknown protein
Length = 294
Score = 94.4 bits (233), Expect = 8e-20
Identities = 73/258 (28%), Positives = 118/258 (45%), Gaps = 39/258 (15%)
Query: 100 FLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALLSLIGDDIDRELQQQDLEMD 159
F ++ S+++++ Q S L R A A SLI ++ ++QQQ+ E+D
Sbjct: 57 FNKSESELTAISKRQRDSTFDSDALIASQKRRAIAFSPA--SLIDAELVSQIQQQNSEID 114
Query: 160 RFLKLQGEQLRQTILEKVQATQLQSV-SIIEDKVLQKLREKETEVENINKRNMELEDQME 218
RF+ Q E LR LE Q TQ + + S +++ +L+KL+ K+ E+ + K N L+++++
Sbjct: 115 RFVAQQTETLR-IELEARQRTQTRMLASAVQNAILKKLKAKDEEIIRMGKLNWVLQERVK 173
Query: 219 QLSVEAGAWQQRARYNENMIAALKFNLQQAYLQ--------------GRDSKEGCGDSEV 264
L VE W+ A+ NE L+ NL+Q Q D++ CG +
Sbjct: 174 NLYVENQIWRDLAQTNEATANNLRSNLEQVLAQVDDLDAFRRPLVEEADDAESSCGSCDG 233
Query: 265 DDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESK--L 322
D + NG CK C ++++LPC+HLCLC C S L
Sbjct: 234 GDVTAVVNG-------------------GCKRCGELTASVLVLPCRHLCLCTVCGSSALL 274
Query: 323 SFCPLCQSSKFIGMEVYM 340
CP+C + V M
Sbjct: 275 RTCPVCDMVMTASVHVNM 292
>At4g19700 unknown protein
Length = 304
Score = 90.9 bits (224), Expect = 9e-19
Identities = 73/253 (28%), Positives = 117/253 (45%), Gaps = 34/253 (13%)
Query: 82 LHWNFGLEP-ERKRLKEQDFLENNSQISSVDFLQPRSVSTGLGLSLDNTRLASTGDSALL 140
L +NF + P KR ++ F ++N+ + RSV+ DS+
Sbjct: 66 LSYNFTVPPLSTKRQRDFQFSDSNAPVKR------RSVAF---------------DSSSP 104
Query: 141 SLIGDDIDRELQ-QQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREK 199
SLI ++ ++Q QQ E+DRF+ Q E+LR I + Q S +++ + +KL+EK
Sbjct: 105 SLINVELVSQIQNQQQSEIDRFVAQQTEKLRIEIEARQQTQTRMLASAVQNVIAKKLKEK 164
Query: 200 ETEVENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGC 259
+ E+ I N L+++++ L VE W+ A+ NE L+ NL Q Q
Sbjct: 165 DDEIVRIRNLNWVLQERVKSLYVENQIWRDIAQTNEANANTLRTNLDQVLAQLETFPTAS 224
Query: 260 GDSEVDDTASC--CNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKD 317
E D +SC C G + + CK C E ++++LPC+HLCLC
Sbjct: 225 AVVEDDAESSCGSCCGDGGGEAVTAVGGG-------CKRCGEREASVLVLPCRHLCLCTV 277
Query: 318 C--ESKLSFCPLC 328
C + L CP+C
Sbjct: 278 CGGSALLRTCPVC 290
>At2g21380 putative kinesin heavy chain
Length = 1058
Score = 54.3 bits (129), Expect = 9e-08
Identities = 39/164 (23%), Positives = 73/164 (43%), Gaps = 33/164 (20%)
Query: 173 ILEKVQATQLQSVSIIEDKVLQKLREKE-TEVENINKRNMELEDQM-----EQLSVEAGA 226
++ K++ ++SI + + +E E TE++N N++N L+++ E++ V A
Sbjct: 911 LVAKLKKANSGALSIQKSDEAEPAKEDEVTELDNKNEQNAILKERQLVNGHEEVIV---A 967
Query: 227 WQQRARYNENMIAALKFNLQQAYLQGRDSKEGCGDSEVDDTASCCNGRSLDFHLLSNENS 286
+ E ++A LK +Q+ + E+ A+ + H+
Sbjct: 968 KAEETPKEEPLVARLKARMQEMK-----------EKEMKSQAAAAANADANSHI------ 1010
Query: 287 NMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQS 330
CK C + +LLPC+H CLCK C S CP+C++
Sbjct: 1011 -------CKVCFESPTATILLPCRHFCLCKSCSLACSECPICRT 1047
>At2g44950 unknown protein
Length = 518
Score = 48.5 bits (114), Expect = 5e-06
Identities = 44/192 (22%), Positives = 76/192 (38%), Gaps = 14/192 (7%)
Query: 147 IDRELQQQDLEMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETEVENI 206
ID+ + +D++ + I ++++ Q + EDK +K +EN+
Sbjct: 323 IDKYIMDKDIQQGSAYASFLSKKSSRIEDQLRFCTDQFQKLAEDKY-----QKSVSLENL 377
Query: 207 NKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQAYLQGRDSKEGC--GDSEV 264
K+ ++ + +EQ + + AL+ L+ R +E +V
Sbjct: 378 QKKRADIGNGLEQARSRLEESHSKVEQSRLDYGALELELEIERFNRRRIEEEMEIAKKKV 437
Query: 265 DDTASCCNGRSLDFHLLSNENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLS- 323
S G S L E S K+++KCKAC +V+ C HL C C KL+
Sbjct: 438 SRLRSLIEGSSA-IQKLRQELSEFKEILKCKACNDRPKEVVITKCYHL-FCNPCVQKLTG 495
Query: 324 ----FCPLCQSS 331
CP C +S
Sbjct: 496 TRQKKCPTCSAS 507
>At1g73950 unknown protein (At1g73950)
Length = 466
Score = 47.8 bits (112), Expect = 9e-06
Identities = 16/36 (44%), Positives = 24/36 (66%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C+ C E+++VLLPC+H LC++C K CP C+
Sbjct: 419 CRVCFEREISVVLLPCRHRVLCRNCSDKCKKCPFCR 454
>At1g18470 unknown protein
Length = 467
Score = 47.4 bits (111), Expect = 1e-05
Identities = 15/36 (41%), Positives = 25/36 (68%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C+ C ++++VLLPC+H LC+ C K + CP+C+
Sbjct: 420 CRVCFEKDISLVLLPCRHRVLCRTCADKCTTCPICR 455
>At1g63900 putative RING zinc finger protein
Length = 115
Score = 45.1 bits (105), Expect = 6e-05
Identities = 17/46 (36%), Positives = 24/46 (51%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQSSKFIGMEVY 339
C C E V +PC H+C C C S L+ CPLC+ + ++ Y
Sbjct: 68 CVICLEQEYNAVFVPCGHMCCCTACSSHLTSCPLCRRRIDLAVKTY 113
>At5g19080 unknown protein
Length = 378
Score = 43.5 bits (101), Expect = 2e-04
Identities = 16/41 (39%), Positives = 23/41 (56%), Gaps = 4/41 (9%)
Query: 293 KCKACRVNEVTMVLLPCKHLCLCKDCESKLSF----CPLCQ 329
+C C ++PC+HLCLC DC +L F CP+C+
Sbjct: 320 ECVICLTEPKDTAVMPCRHLCLCSDCAEELRFQTNKCPICR 360
>At1g05320 unknown protein
Length = 841
Score = 43.5 bits (101), Expect = 2e-04
Identities = 43/173 (24%), Positives = 81/173 (45%), Gaps = 14/173 (8%)
Query: 89 EPERKRLKEQDFLENNSQISSV---DFLQPRSVSTGLGLSLDNTR--LASTGDSALLSLI 143
+ + K K+ LE+ Q+ V + + + GLGL L+N+R + D +S +
Sbjct: 65 DDDEKAEKQLKSLEDALQLHDVKHKELTEVKEAFDGLGLELENSRKKMIELEDRIRISAL 124
Query: 144 GDDIDRELQQQDL-EMDRFLKLQGEQLRQTILEKVQATQLQSVSIIEDKVLQKLREKETE 202
+ ELQ+Q E++ LK+ E+ +T QA SV + K L++L EK +E
Sbjct: 125 EAEKLEELQKQSASELEEKLKISDERYSKTDALLSQALSQNSVLEQKLKSLEELSEKVSE 184
Query: 203 V--------ENINKRNMELEDQMEQLSVEAGAWQQRARYNENMIAALKFNLQQ 247
+ E K ++++++ E++S + Q + N + L+ LQ+
Sbjct: 185 LKSALIVAEEEGKKSSIQMQEYQEKVSKLESSLNQSSARNSELEEDLRIALQK 237
>At1g68820 unknown protein (At1g68820)
Length = 468
Score = 42.7 bits (99), Expect = 3e-04
Identities = 15/36 (41%), Positives = 22/36 (60%)
Query: 294 CKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
C+ C + + +VLLPC+H LC C K CP+C+
Sbjct: 421 CRVCFEDPINVVLLPCRHHVLCSTCCEKCKKCPICR 456
>At5g23110 putative protein
Length = 4706
Score = 41.6 bits (96), Expect = 6e-04
Identities = 16/46 (34%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Query: 284 ENSNMKDLMKCKACRVNEVTMVLLPCKHLCLCKDCESKLSFCPLCQ 329
E K C+ C+ EV + ++PC H+ LC+ C + +S CP C+
Sbjct: 4650 EAETAKSQWLCQICQTKEVEVTIVPCGHV-LCRHCSTSVSRCPFCR 4694
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,598,402
Number of Sequences: 26719
Number of extensions: 339637
Number of successful extensions: 1732
Number of sequences better than 10.0: 179
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 108
Number of HSP's that attempted gapping in prelim test: 1506
Number of HSP's gapped (non-prelim): 291
length of query: 340
length of database: 11,318,596
effective HSP length: 100
effective length of query: 240
effective length of database: 8,646,696
effective search space: 2075207040
effective search space used: 2075207040
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC148486.4


