

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148486.10 - phase: 0
(659 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
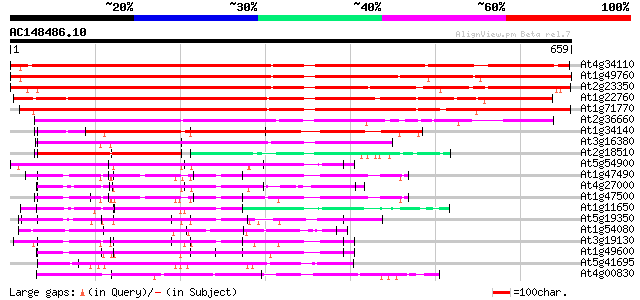
Score E
Sequences producing significant alignments: (bits) Value
At4g34110 poly(A)-binding protein 862 0.0
At1g49760 Putative Poly-A Binding Protein (F14J22.3) 839 0.0
At2g23350 putative poly(A) binding protein 782 0.0
At1g22760 putative polyA-binding protein, PAB3 607 e-174
At1g71770 polyadenylate-binding protein 5 597 e-171
At2g36660 putative poly(A) binding protein 375 e-104
At1g34140 putative protein 305 5e-83
At3g16380 putative poly(A) binding protein 298 8e-81
At2g18510 putative spliceosome associated protein 115 6e-26
At5g54900 unknown protein 103 4e-22
At1g47490 unknown protein 102 9e-22
At4g27000 putative DNA binding protein 101 2e-21
At1g47500 unknown protein 100 2e-21
At1g11650 putative DNA binding protein 100 3e-21
At5g19350 DNA binding protein ACBF - like 96 6e-20
At1g54080 unknown protein 95 1e-19
At3g19130 nuclear acid binding protein, putative 94 2e-19
At1g49600 DNA binding protein ACBF, putative 92 7e-19
At5g41695 putative protein 89 6e-18
At4g00830 unknown protein 89 8e-18
>At4g34110 poly(A)-binding protein
Length = 629
Score = 862 bits (2226), Expect = 0.0
Identities = 446/665 (67%), Positives = 523/665 (78%), Gaps = 44/665 (6%)
Query: 1 MAQIQVQHQTP-------APVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFN 53
MAQ+Q+Q QTP PA S G A QF TSLYVGDL+ NV DSQL+D F
Sbjct: 1 MAQVQLQGQTPNGSTAAVTSAPATSGGAT---ATQFGNTSLYVGDLDFNVTDSQLFDAFG 57
Query: 54 QVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPS 113
Q+G VV+VRVCRDL TRRSLGYGYVNFTNPQDAARA+ LN+ P+ K IRVMYSHRDPS
Sbjct: 58 QMGTVVTVRVCRDLVTRRSLGYGYVNFTNPQDAARAIQELNYIPLYGKPIRVMYSHRDPS 117
Query: 114 SRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSA 173
R+SG NIFIKNLD++IDHKALHDTFSSFG I+SCK+A D SGQSKGYGFVQ+ E+SA
Sbjct: 118 VRRSGAGNIFIKNLDESIDHKALHDTFSSFGNIVSCKVAVDSSGQSKGYGFVQYANEESA 177
Query: 174 QNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA 233
Q AI+KLNGML+NDKQV+VG FLR+Q+RD+ +KTKF NVYVKNL+ES T+DDLKN FG
Sbjct: 178 QKAIEKLNGMLLNDKQVYVGPFLRRQERDSTANKTKFTNVYVKNLAESTTDDDLKNAFGE 237
Query: 234 YGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSERE 293
YG ITSAV+M+D +G+SK FGFVNFENA+DAA+AVE+LNG K DD+EWYVG+AQKKSERE
Sbjct: 238 YGKITSAVVMKDGEGKSKGFGFVNFENADDAARAVESLNGHKFDDKEWYVGRAQKKSERE 297
Query: 294 QELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFT 353
EL+ R+EQ +KE+ DKFQ NLY+KNLD SI+DEKLKE+FS FGT+TS KV
Sbjct: 298 TELRVRYEQNLKEA-ADKFQSSNLYVKNLDPSISDEKLKEIFSPFGTVTSSKV------- 349
Query: 354 DIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQ 413
MRDPNG S+GSGFVAF+TPEEA+ A+ +++GKMI SKPLYVA+AQRKEDRR RLQ
Sbjct: 350 -----MRDPNGTSKGSGFVAFATPEEATEAMSQLSGKMIESKPLYVAIAQRKEDRRVRLQ 404
Query: 414 AQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMR 473
AQFSQ+RPVA+ PSV PRMP+YPPG PG+GQQ YGQ PPAM+PPQ G+GYQQQLVPGMR
Sbjct: 405 AQFSQVRPVAMQPSVGPRMPVYPPGGPGIGQQMFYGQAPPAMIPPQPGYGYQQQLVPGMR 464
Query: 474 PGGGPMPSYFVPMVQQGQQGQRP-GGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRS 532
PGGGP+PS+F+PMVQ Q QRP GGRR G Q QQ PMMQQQM PR R++RYP GR
Sbjct: 465 PGGGPVPSFFMPMVQ--PQQQRPGGGRRPGGIQHSQQQNPMMQQQMHPRGRMFRYPQGRG 522
Query: 533 NIQDAPVQNIGGGMMSYDMGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLV 592
D P ++G M P+ I ALA+ L+NA PEQQRTMLGE LYPLV
Sbjct: 523 GSGDVPPYDMGNNM---------------PLTIGALASNLSNATPEQQRTMLGEVLYPLV 567
Query: 593 DQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGNSPADQLA 652
+Q+E +SAAKVTGMLLEMDQ EVLHL+ESP+ALKAKVAEAM+VLR+V+ +QLA
Sbjct: 568 EQVEAESAAKVTGMLLEMDQTEVLHLLESPEALKAKVAEAMDVLRSVA---AGGATEQLA 624
Query: 653 SLSLN 657
SL+L+
Sbjct: 625 SLNLS 629
>At1g49760 Putative Poly-A Binding Protein (F14J22.3)
Length = 671
Score = 839 bits (2168), Expect = 0.0
Identities = 446/684 (65%), Positives = 523/684 (76%), Gaps = 41/684 (5%)
Query: 1 MAQIQVQHQTP-----------APVPAPSNGVVPNVAN--QFVTTSLYVGDLEVNVNDSQ 47
MAQIQ Q Q A A + G A Q TTSLYVGDL+ V DSQ
Sbjct: 1 MAQIQHQGQNANGGVAVPGAAAAEAAAAAAGAAAAAAGAAQQGTTSLYVGDLDATVTDSQ 60
Query: 48 LYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMY 107
L++ F Q GQVVSVRVCRD+ TRRSLGYGYVN+ PQDA+RAL+ LNF +N ++IRVMY
Sbjct: 61 LFEAFTQAGQVVSVRVCRDMTTRRSLGYGYVNYATPQDASRALNELNFMALNGRAIRVMY 120
Query: 108 SHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQF 167
S RDPS RKSG NIFIKNLDK+IDHKALH+TFS+FG I+SCK+A D SGQSKGYGFVQ+
Sbjct: 121 SVRDPSLRKSGVGNIFIKNLDKSIDHKALHETFSAFGPILSCKVAVDPSGQSKGYGFVQY 180
Query: 168 EAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDL 227
+ +++AQ AIDKLNGML+NDKQV+VG F+ K RD K KF NVYVKNLSES ++++L
Sbjct: 181 DTDEAAQGAIDKLNGMLLNDKQVYVGPFVHKLQRDPSGEKVKFTNVYVKNLSESLSDEEL 240
Query: 228 KNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQ 287
FG +G TS V+MRD +G+SK FGFVNFEN++DAA+AV+ALNGK DD+EW+VGKAQ
Sbjct: 241 NKVFGEFGVTTSCVIMRDGEGKSKGFGFVNFENSDDAARAVDALNGKTFDDKEWFVGKAQ 300
Query: 288 KKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVC 347
KKSERE ELK +FEQ++KE+ DK QG NLY+KNLD+S+TD+KL+E F+ FGTITS KV
Sbjct: 301 KKSERETELKQKFEQSLKEA-ADKSQGSNLYVKNLDESVTDDKLREHFAPFGTITSCKV- 358
Query: 348 TMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKED 407
MRDP+GVSRGSGFVAFSTPEEA+RA+ EMNGKMIV+KPLYVA+AQRKED
Sbjct: 359 -----------MRDPSGVSRGSGFVAFSTPEEATRAITEMNGKMIVTKPLYVALAQRKED 407
Query: 408 RRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQ 467
R+ARLQAQFSQMRPV + P+V PRM +YPPG P +GQQ YGQGPPAM+ PQ GFGYQQQ
Sbjct: 408 RKARLQAQFSQMRPVNMPPAVGPRMQMYPPGGPPMGQQLFYGQGPPAMI-PQPGFGYQQQ 466
Query: 468 LVPGMRPGGGPMPSYFVPMVQQG-------QQGQRPGGRRGGPGQQPPQQVPMMQQQMLP 520
LVPGMRPGG PMP++F+PM+QQG QQ QRPGG R G QP Q PMMQQQM P
Sbjct: 467 LVPGMRPGGSPMPNFFMPMMQQGQQQQQQQQQQQRPGGGRRGALPQPQQPSPMMQQQMHP 526
Query: 521 RQRVYRYPPGRSNIQDAPVQNIGGGMMSYDM---GGLPLRD--VVPPMPIHALATALANA 575
R R+YRYP N P QN+ + YD+ GG+ RD P+PI ALAT LANA
Sbjct: 527 RGRMYRYPQRDVNTMPGPTQNMLS--VPYDVSSGGGVHHRDSPTSQPVPIVALATRLANA 584
Query: 576 PPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEV 635
PEQQRTMLGE LYPLV+QLE +SAAKVTGMLLEMDQ EVLHL+ESP+ALKAKV EAM+V
Sbjct: 585 APEQQRTMLGENLYPLVEQLEPESAAKVTGMLLEMDQTEVLHLLESPEALKAKVTEAMDV 644
Query: 636 LRNVSQQQGNSPADQLASLSLNDS 659
LR+V+QQQ ADQLASLSL D+
Sbjct: 645 LRSVAQQQAGGAADQLASLSLGDN 668
>At2g23350 putative poly(A) binding protein
Length = 662
Score = 782 bits (2019), Expect = 0.0
Identities = 418/687 (60%), Positives = 502/687 (72%), Gaps = 56/687 (8%)
Query: 1 MAQIQVQHQTPAPVPAPSN----------GVVPNVANQFVT----TSLYVGDLEVNVNDS 46
MAQ+Q P PA N G+ VT SLYVGDL+ NV DS
Sbjct: 1 MAQVQAPSSHSPPPPAVVNDGAATASATPGIGVGGGGDGVTHGALCSLYVGDLDFNVTDS 60
Query: 47 QLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVM 106
QLYD F +V QVVSVRVCRD AT SLGYGYVN++N DA +A+ LN++ +N K IR+
Sbjct: 61 QLYDYFTEVCQVVSVRVCRDAATNTSLGYGYVNYSNTDDAEKAMQKLNYSYLNGKMIRIT 120
Query: 107 YSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQ 166
YS RD S+R+SG N+F+KNLDK++D+K LH+ FS G I+SCK+ATD GQS+GYGFVQ
Sbjct: 121 YSSRDSSARRSGVGNLFVKNLDKSVDNKTLHEAFSGCGTIVSCKVATDHMGQSRGYGFVQ 180
Query: 167 FEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDD 226
F+ EDSA+NAI+KLNG ++NDKQ+FVG FLRK++R++ K KF NVYVKNLSE+ T+D+
Sbjct: 181 FDTEDSAKNAIEKLNGKVLNDKQIFVGPFLRKEERESAADKMKFTNVYVKNLSEATTDDE 240
Query: 227 LKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKA 286
LK FG YG+I+SAV+MRD DG+S+CFGFVNFEN EDAA+AVEALNGKK DD+EWYVGKA
Sbjct: 241 LKTTFGQYGSISSAVVMRDGDGKSRCFGFVNFENPEDAARAVEALNGKKFDDKEWYVGKA 300
Query: 287 QKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKV 346
QKKSERE EL R+EQ + +KF GLNLY+KNLDD++TDEKL+E+F+EFGTITS KV
Sbjct: 301 QKKSERELELSRRYEQGSSDG-GNKFDGLNLYVKNLDDTVTDEKLRELFAEFGTITSCKV 359
Query: 347 CTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKE 406
MRDP+G S+GSGFVAFS EASR L EMNGKM+ KPLYVA+AQRKE
Sbjct: 360 ------------MRDPSGTSKGSGFVAFSAASEASRVLNEMNGKMVGGKPLYVALAQRKE 407
Query: 407 DRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQ 466
+RRA+LQAQFSQMRP A P V PRMP++ G PGLGQQ YGQGPP ++P Q GFGYQ
Sbjct: 408 ERRAKLQAQFSQMRP-AFIPGVGPRMPIFTGGAPGLGQQIFYGQGPPPIIPHQPGFGYQP 466
Query: 467 QLVPGMRPG--GGPMPSYFVPMVQQGQQGQRPGGRRGGPG---QQPPQQVPMMQQQMLPR 521
QLVPGMRP GG PM+Q GQQG RPGGRR G G Q Q +P MQ QM+PR
Sbjct: 467 QLVPGMRPAFFGG-------PMMQPGQQGPRPGGRRSGDGPMRHQHQQPMPYMQPQMMPR 519
Query: 522 QRVYRYPPGRSNIQDAPVQNIGGGM--MSYDMGGLPLRDVVPPMPIHALATALANAPPEQ 579
R YRYP G N+ D P+ GGM ++YDM +P PM LAT+LANA P Q
Sbjct: 520 GRGYRYPSGGRNMPDGPMP---GGMVPVAYDMNVMPYSQ---PMSAGQLATSLANATPAQ 573
Query: 580 QRTMLGEALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNV 639
QRT+LGE+LYPLVDQ+E + AAKVTGMLLEMDQ EVLHL+ESP+AL AKV+EA++VLRNV
Sbjct: 574 QRTLLGESLYPLVDQIESEHAAKVTGMLLEMDQTEVLHLLESPEALNAKVSEALDVLRNV 633
Query: 640 SQ-----QQGN---SPADQLASLSLND 658
+Q +GN SP+D LASLS+ND
Sbjct: 634 NQPSSQGSEGNKSGSPSDLLASLSIND 660
>At1g22760 putative polyA-binding protein, PAB3
Length = 660
Score = 607 bits (1565), Expect = e-174
Identities = 346/647 (53%), Positives = 438/647 (67%), Gaps = 40/647 (6%)
Query: 5 QVQHQTPAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVC 64
Q Q P +P PS VV + + +SLY GDL+ V ++ L+DLF V VVSVRVC
Sbjct: 24 QTSVQVPVSIPLPSPVVVADQTHP--NSSLYAGDLDPKVTEAHLFDLFKHVANVVSVRVC 81
Query: 65 RDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFI 124
RD RRSLGY Y+NF+NP DA RA++ LN+TP+ ++ IR+M S+RDPS+R SG NIFI
Sbjct: 82 RD-QNRRSLGYAYINFSNPNDAYRAMEALNYTPLFDRPIRIMLSNRDPSTRLSGKGNIFI 140
Query: 125 KNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGML 184
KNLD +ID+KAL +TFSSFG I+SCK+A D +G+SKGYGFVQFE E+SAQ AIDKLNGML
Sbjct: 141 KNLDASIDNKALFETFSSFGTILSCKVAMDVTGRSKGYGFVQFEKEESAQAAIDKLNGML 200
Query: 185 INDKQVFVGHFLRKQD--RDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVL 242
+NDKQVFVGHF+R+Q+ RD +F NVYVKNL + ED+L+ FG +G I+SAV+
Sbjct: 201 MNDKQVFVGHFIRRQERARDENTPTPRFTNVYVKNLPKEIGEDELRKTFGKFGVISSAVV 260
Query: 243 MRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQ 302
MRD G S+CFGFVNFE E AA AVE +NG + D+ YVG+AQKKSERE+EL+ +FEQ
Sbjct: 261 MRDQSGNSRCFGFVNFECTEAAASAVEKMNGISLGDDVLYVGRAQKKSEREEELRRKFEQ 320
Query: 303 TVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDP 362
+ + +K QG NLYLKNLDDS+ DEKLKEMFSE+G +TS KV M +P
Sbjct: 321 E-RINRFEKSQGANLYLKNLDDSVDDEKLKEMFSEYGNVTSSKV------------MLNP 367
Query: 363 NGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPV 422
G+SRG GFVA+S PEEA RAL EMNGKMI KPLY+A+AQRKEDRRA LQA FSQ+R
Sbjct: 368 QGMSRGFGFVAYSNPEEALRALSEMNGKMIGRKPLYIALAQRKEDRRAHLQALFSQIRAP 427
Query: 423 AITPSVAPRMPLYPPG--TPGLGQQFMYGQGPPAMMPPQ-AGFGYQQQLVPGMRPGGGPM 479
+PPG PG Q GQ +M+P Q G+G+Q Q +PGMRPG GP
Sbjct: 428 GPMSGFH-----HPPGGPMPGPPQHMYVGQNGASMVPSQPIGYGFQPQFMPGMRPGSGP- 481
Query: 480 PSYFV--PMVQQGQQGQRPGGRRGGPG-QQPPQQVPMMQQQMLPRQRVYRYPPGRSNIQD 536
++ V P+ +Q Q G R G RRG QQ QQ +M + P RY G SN ++
Sbjct: 482 GNFIVPYPLQRQPQTGPRMGFRRGATNVQQHIQQQQLMHRNPSPGM---RYMNGASNGRN 538
Query: 537 APVQNIGGGMMSYDMGGLPL------RDVVPPMPIHALATALANAPPEQQRTMLGEALYP 590
++ G++ + LP+ P +PI L ++LA+A P + MLGE LYP
Sbjct: 539 GMDSSVPQGILP-PIIPLPIDASSISHQKAPLLPISKLTSSLASASPADRTRMLGEQLYP 597
Query: 591 LVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLR 637
LV++ E AKVTGMLLEMDQ E+LHL+ESP+ALK+KV+EA++VLR
Sbjct: 598 LVERHEPLHVAKVTGMLLEMDQAEILHLMESPEALKSKVSEALDVLR 644
>At1g71770 polyadenylate-binding protein 5
Length = 668
Score = 597 bits (1540), Expect = e-171
Identities = 345/673 (51%), Positives = 447/673 (66%), Gaps = 46/673 (6%)
Query: 12 APVPAPSNGVVPNVA---------NQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVR 62
AP P P+ V VA +SLYVGDL+ +VN+S L DLFNQV V ++R
Sbjct: 16 APPPFPAVSQVAAVAAAAAAAEALQTHPNSSLYVGDLDPSVNESHLLDLFNQVAPVHNLR 75
Query: 63 VCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANI 122
VCRDL T RSLGY YVNF NP+DA+RA++ LN+ P+ ++ IR+M S+RDPS+R SG N+
Sbjct: 76 VCRDL-THRSLGYAYVNFANPEDASRAMESLNYAPIRDRPIRIMLSNRDPSTRLSGKGNV 134
Query: 123 FIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNG 182
FIKNLD +ID+KAL++TFSSFG I+SCK+A D G+SKGYGFVQFE E++AQ AIDKLNG
Sbjct: 135 FIKNLDASIDNKALYETFSSFGTILSCKVAMDVVGRSKGYGFVQFEKEETAQAAIDKLNG 194
Query: 183 MLINDKQVFVGHFLRKQDRDNVLSKT--KFNNVYVKNLSESFTEDDLKNEFGAYGTITSA 240
ML+NDKQVFVGHF+R+QDR S F NVYVKNL + T+D+LK FG YG I+SA
Sbjct: 195 MLLNDKQVFVGHFVRRQDRARSESGAVPSFTNVYVKNLPKEITDDELKKTFGKYGDISSA 254
Query: 241 VLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRF 300
V+M+D G S+ FGFVNF + E AA AVE +NG + ++ YVG+AQKKS+RE+EL+ +F
Sbjct: 255 VVMKDQSGNSRSFGFVNFVSPEAAAVAVEKMNGISLGEDVLYVGRAQKKSDREEELRRKF 314
Query: 301 EQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMR 360
EQ + S +K QG NLYLKNLDDS+ DEKLKEMFSE+G +TS KV M
Sbjct: 315 EQE-RISRFEKLQGSNLYLKNLDDSVNDEKLKEMFSEYGNVTSCKV------------MM 361
Query: 361 DPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMR 420
+ G+SRG GFVA+S PEEA A+ EMNGKMI KPLYVA+AQRKE+R+A LQ+ F+Q+R
Sbjct: 362 NSQGLSRGFGFVAYSNPEEALLAMKEMNGKMIGRKPLYVALAQRKEERQAHLQSLFTQIR 421
Query: 421 -PVAITPSVAPRMPL--YPPGTP--GLGQQFMYGQGPPAMMPPQ-AGFGYQQQLVPGMRP 474
P ++P +P +PPG P G G ++P Q G+GYQ Q +PGMRP
Sbjct: 422 SPGTMSPVPSPMSGFHHHPPGGPMSGPHHPMFIGHNGQGLVPSQPMGYGYQVQFMPGMRP 481
Query: 475 GGGPMPSYFV--PMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQQMLPRQRVYRYPPGRS 532
G GP P++ + P+ +Q Q G R G RRG Q QQQ + +Q R+ G
Sbjct: 482 GAGP-PNFMMPFPLQRQTQPGPRVGFRRGANNMQ-----QQFQQQQMLQQNASRFMGGAG 535
Query: 533 NIQDAPVQNIGGGMM------SYDMGGLPLRDVVP-PMPIHALATALANAPPEQQRTMLG 585
N ++ + G++ S + P R P P+ I LA+ LA A P++ MLG
Sbjct: 536 NRRNGMEASAPQGIIPLPLNASANSHNAPQRSHKPTPLTISKLASDLALASPDKHPRMLG 595
Query: 586 EALYPLVDQLEHDSAAKVTGMLLEMDQPEVLHLIESPDALKAKVAEAMEVLRNVSQQQGN 645
+ LYPLV+Q E +AAKVTGMLLEMDQ E+LHL+ESP+ALKAKV+EA++VLR +
Sbjct: 596 DHLYPLVEQQEPANAAKVTGMLLEMDQAEILHLLESPEALKAKVSEALDVLRRSADPAAV 655
Query: 646 SPADQLASLSLND 658
S D +LS ++
Sbjct: 656 SSVDDQFALSSSE 668
>At2g36660 putative poly(A) binding protein
Length = 609
Score = 375 bits (963), Expect = e-104
Identities = 240/628 (38%), Positives = 349/628 (55%), Gaps = 85/628 (13%)
Query: 30 VTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARA 89
VT SLYVGDL +V + LYD F + + SVR+C+D ++ RSL YGY NF + QDA A
Sbjct: 22 VTASLYVGDLHPSVTEGILYDAFAEFKSLTSVRLCKDASSGRSLCYGYANFLSRQDANLA 81
Query: 90 LDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSC 149
++ N + +N K IRVM+S R P +R++G N+F+KNL +++ + L D F FG I+SC
Sbjct: 82 IEKKNNSLLNGKMIRVMWSVRAPDARRNGVGNVFVKNLPESVTNAVLQDMFKKFGNIVSC 141
Query: 150 KIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTK 209
K+AT G+S+GYGFVQFE ED+A AI LN ++ DK+++VG F++K DR V + K
Sbjct: 142 KVATLEDGKSRGYGFVQFEQEDAAHAAIQTLNSTIVADKEIYVGKFMKKTDR--VKPEEK 199
Query: 210 FNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVE 269
+ N+Y+KNL +ED L+ +F +G I S + +D + + + FVNF+N EDA +A E
Sbjct: 200 YTNLYMKNLDADVSEDLLREKFAEFGKIVSLAIAKDENRLCRGYAFVNFDNPEDARRAAE 259
Query: 270 ALNGKKVDDEEWYVGKAQKKSEREQELKGRF-EQTVKESVVDKFQGLNLYLKNLDDSITD 328
+NG K + YVG+AQKK+EREQ L+ +F E+ ++ ++ K N+Y+KN++ ++T+
Sbjct: 260 TVNGTKFGSKCLYVGRAQKKAEREQLLREQFKEKHEEQKMIAKVS--NIYVKNVNVAVTE 317
Query: 329 EKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMN 388
E+L++ FS+ GTITS K +M D G S+G GFV FSTPEEA A+ +
Sbjct: 318 EELRKHFSQCGTITSTK------------LMCDEKGKSKGFGFVCFSTPEEAIDAVKTFH 365
Query: 389 GKMIVSKPLYVAVAQRKEDRRARLQAQFS---QMRPVAITPSVAP--RMPLYPPGT-PGL 442
G+M KPLYVA+AQ+KEDR+ +LQ QF + R + + SV P PLY T PG
Sbjct: 366 GQMFHGKPLYVAIAQKKEDRKMQLQVQFGNRVEARKSSSSASVNPGTYAPLYYTNTHPG- 424
Query: 443 GQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGG 502
M Q P M ++ P + + PMV R R G
Sbjct: 425 ----MVYQSYPLMWK-------SANMIGSSYPNSEAVT--YPPMVANAPSKNRQ-NRIGK 470
Query: 503 PGQQPPQQVPMMQQ--QMLPRQRVY----------RYPPGRSNIQDAPVQNIGGGMMSYD 550
+ VP + Q QMLP R + R + +IQ + +G
Sbjct: 471 LDRNAVSYVPNVYQSTQMLPLSRDFSKQQHSRTYGRGKEMKKSIQQRQSETVG------- 523
Query: 551 MGGLPLRDVVPPMPIHALATALANAPPEQQRTMLGEALYPLVDQLEHDSAAKVTGMLLEM 610
+ +LGE L+PLV++LE A K+TGMLLEM
Sbjct: 524 ----------------------------MEMQLLGELLHPLVEKLEPQLANKITGMLLEM 555
Query: 611 DQPEVLHLIESPDALKAKVAEAMEVLRN 638
D+ E+L L++SP+ L +V EA EVL++
Sbjct: 556 DKSELLLLLKSPEDLAVRVDEAFEVLKS 583
>At1g34140 putative protein
Length = 398
Score = 305 bits (781), Expect = 5e-83
Identities = 173/403 (42%), Positives = 246/403 (60%), Gaps = 48/403 (11%)
Query: 90 LDVLNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSC 149
++VLN+ + K +R+M+S RDPS+R SG N+F+KNLD++ID+K L D FS+FG+++SC
Sbjct: 1 MEVLNYCKLKGKPMRIMFSERDPSNRMSGRGNVFVKNLDESIDNKQLCDMFSAFGKVLSC 60
Query: 150 KIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTK 209
K+A D SG SKGYGFVQF ++ S A + NG LI ++ + V F+ + D
Sbjct: 61 KVARDASGVSKGYGFVQFYSDLSVYTACNFHNGTLIRNQHIHVCPFVSRGQWD---KSRV 117
Query: 210 FNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVE 269
F NVYVKNL E+ T+ DLK FG +G ITSAV+M+D +G+S+ FGFVNFE AE A A+E
Sbjct: 118 FTNVYVKNLVETATDADLKRLFGEFGEITSAVVMKDGEGKSRRFGFVNFEKAEAAVTAIE 177
Query: 270 ALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDE 329
+NG VD++E +VG+AQ+K+ R ++LK +FE + +G+NLY+KNLDDS+ +
Sbjct: 178 KMNGVVVDEKELHVGRAQRKTNRTEDLKAKFELEKIIRDMKTRKGMNLYVKNLDDSVDNT 237
Query: 330 KLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNG 389
KL+E+FSEFGTITS ++G GFV FST EEAS+A+ +MNG
Sbjct: 238 KLEELFSEFGTITS---------------------CNKGVGFVEFSTSEEASKAMLKMNG 276
Query: 390 KMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYG 449
KM+ +KP+YV++AQ KE + LQ QF+ PP +P F
Sbjct: 277 KMVGNKPIYVSLAQCKEQHKLHLQTQFNN-----------------PPPSPHQQPIFSQV 319
Query: 450 QGPPAMM---PPQAGFGYQQQLVPGMR-PGGGP---MPSYFVP 485
P M+ P G+ +Q + G R P P +P++ VP
Sbjct: 320 VAPATMLSQQTPLRGYNFQPYSMCGSRMPNSCPPISIPNFMVP 362
Score = 152 bits (383), Expect = 7e-37
Identities = 96/283 (33%), Positives = 155/283 (53%), Gaps = 26/283 (9%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+++V +L+ ++++ QL D+F+ G+V+S +V RD A+ S GYG+V F + A +
Sbjct: 32 NVFVKNLDESIDNKQLCDMFSAFGKVLSCKVARD-ASGVSKGYGFVQFYSDLSVYTACNF 90
Query: 93 LNFTPMNNKSIRVM-YSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKI 151
N T + N+ I V + R + N+++KNL +T L F FG+I S +
Sbjct: 91 HNGTLIRNQHIHVCPFVSRGQWDKSRVFTNVYVKNLVETATDADLKRLFGEFGEITSAVV 150
Query: 152 ATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFN 211
DG G+S+ +GFV FE ++A AI+K+NG+++++K++ VG RK +R L K KF
Sbjct: 151 MKDGEGKSRRFGFVNFEKAEAAVTAIEKMNGVVVDEKELHVGRAQRKTNRTEDL-KAKFE 209
Query: 212 --------------NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVN 257
N+YVKNL +S L+ F +GTITS +K GFV
Sbjct: 210 LEKIIRDMKTRKGMNLYVKNLDDSVDNTKLEELFSEFGTITSC---------NKGVGFVE 260
Query: 258 FENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRF 300
F +E+A+KA+ +NGK V ++ YV AQ K + + L+ +F
Sbjct: 261 FSTSEEASKAMLKMNGKMVGNKPIYVSLAQCKEQHKLHLQTQF 303
Score = 90.9 bits (224), Expect = 2e-18
Identities = 58/202 (28%), Positives = 102/202 (49%), Gaps = 32/202 (15%)
Query: 30 VTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARA 89
V T++YV +L D+ L LF + G++ S V +D +S +G+VNF + A A
Sbjct: 117 VFTNVYVKNLVETATDADLKRLFGEFGEITSAVVMKD-GEGKSRRFGFVNFEKAEAAVTA 175
Query: 90 LDVLNFTPMNNKSIRVMYSH-------------------RDPSSRKSGTANIFIKNLDKT 130
++ +N ++ K + V + RD +RK N+++KNLD +
Sbjct: 176 IEKMNGVVVDEKELHVGRAQRKTNRTEDLKAKFELEKIIRDMKTRKG--MNLYVKNLDDS 233
Query: 131 IDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQV 190
+D+ L + FS FG I SC +KG GFV+F + A A+ K+NG ++ +K +
Sbjct: 234 VDNTKLEELFSEFGTITSC---------NKGVGFVEFSTSEEASKAMLKMNGKMVGNKPI 284
Query: 191 FVGHFLRKQDRDNVLSKTKFNN 212
+V + +++ + +T+FNN
Sbjct: 285 YVS-LAQCKEQHKLHLQTQFNN 305
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 298 bits (762), Expect = 8e-81
Identities = 176/424 (41%), Positives = 252/424 (58%), Gaps = 22/424 (5%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
SLYVGDL +V + L D F+ VVSV +CR+ T +S+ Y Y+NF +P A+ A+
Sbjct: 22 SLYVGDLSPDVTEKDLIDKFSLNVPVVSVHLCRNSVTGKSMCYAYINFDSPFSASNAMTR 81
Query: 93 LNFTPMNNKSIRVMYSHRDPSSRK---SGTANIFIKNLDKTIDHKALHDTFSSFGQIMSC 149
LN + + K++R+M+S RD + R+ +G AN+++KNLD +I L F FG I+SC
Sbjct: 82 LNHSDLKGKAMRIMWSQRDLAYRRRTRTGFANLYVKNLDSSITSSCLERMFCPFGSILSC 141
Query: 150 KIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTK 209
K+ + +GQSKG+GFVQF+ E SA +A L+G ++ K++FV F+ K +R +
Sbjct: 142 KVVEE-NGQSKGFGFVQFDTEQSAVSARSALHGSMVYGKKLFVAKFINKDERAAMAGNQD 200
Query: 210 FNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVE 269
NVYVKNL E+ T+D L F YGT++S V+MRD GRS+ FGFVNF N E+A KA+E
Sbjct: 201 STNVYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGRSRGFGFVNFCNPENAKKAME 260
Query: 270 ALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDE 329
+L G ++ ++ +VGKA KK ER + LK +F + NLY+KNL +S+ +
Sbjct: 261 SLCGLQLGSKKLFVGKALKKDERREMLKQKFSDNF--IAKPNMRWSNLYVKNLSESMNET 318
Query: 330 KLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNG 389
+L+E+F +G I S KV M NG S+G GFV FS EE+ +A +NG
Sbjct: 319 RLREIFGCYGQIVSAKV------------MCHENGRSKGFGFVCFSNCEESKQAKRYLNG 366
Query: 390 KMIVSKPLYVAVAQRKEDRRARLQAQF-SQMRPVAITPSV-APRMPL--YPPGTPGLGQQ 445
++ KP+ V VA+RKEDR RLQ F +Q R PS +P P+ Y + G Q
Sbjct: 367 FLVDGKPIVVRVAERKEDRIKRLQQYFQAQPRQYTQAPSAPSPAQPVLSYVSSSYGCFQP 426
Query: 446 FMYG 449
F G
Sbjct: 427 FQVG 430
Score = 100 bits (250), Expect = 2e-21
Identities = 62/186 (33%), Positives = 102/186 (54%), Gaps = 16/186 (8%)
Query: 31 TTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARAL 90
+T++YV +L V D L+ LF+Q G V SV V RD RS G+G+VNF NP++A +A+
Sbjct: 201 STNVYVKNLIETVTDDCLHTLFSQYGTVSSVVVMRD-GMGRSRGFGFVNFCNPENAKKAM 259
Query: 91 DVLNFTPMNNKSIRV---------------MYSHRDPSSRKSGTANIFIKNLDKTIDHKA 135
+ L + +K + V +S + +N+++KNL ++++
Sbjct: 260 ESLCGLQLGSKKLFVGKALKKDERREMLKQKFSDNFIAKPNMRWSNLYVKNLSESMNETR 319
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHF 195
L + F +GQI+S K+ +G+SKG+GFV F + ++ A LNG L++ K + V
Sbjct: 320 LREIFGCYGQIVSAKVMCHENGRSKGFGFVCFSNCEESKQAKRYLNGFLVDGKPIVVRVA 379
Query: 196 LRKQDR 201
RK+DR
Sbjct: 380 ERKEDR 385
>At2g18510 putative spliceosome associated protein
Length = 363
Score = 115 bits (289), Expect = 6e-26
Identities = 62/171 (36%), Positives = 106/171 (61%), Gaps = 3/171 (1%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
++YVG L+ +++ L++LF Q G VV+V V +D T YG++ + + +DA A+ V
Sbjct: 26 TVYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKV 85
Query: 93 LNFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMS-CKI 151
LN ++ K IRV + +D S G AN+FI NLD +D K L+DTFS+FG I S KI
Sbjct: 86 LNMIKLHGKPIRVNKASQDKKSLDVG-ANLFIGNLDPDVDEKLLYDTFSAFGVIASNPKI 144
Query: 152 ATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR 201
D +G S+G+GF+ +++ +++ AI+ + G ++++Q+ V + +K +
Sbjct: 145 MRDPDTGNSRGFGFISYDSFEASDAAIESMTGQYLSNRQITVSYAYKKDTK 195
Score = 78.6 bits (192), Expect = 1e-14
Identities = 98/363 (26%), Positives = 144/363 (38%), Gaps = 93/363 (25%)
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEAL 271
VYV L +E+ L F G + + + +D + +GF+ + + EDA A++ L
Sbjct: 27 VYVGGLDAQLSEELLWELFVQAGPVVNVYVPKDRVTNLHQNYGFIEYRSEEDADYAIKVL 86
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
N K+ + V KA S+ ++ L G NL++ NLD + ++ L
Sbjct: 87 NMIKLHGKPIRVNKA---SQDKKSLD---------------VGANLFIGNLDPDVDEKLL 128
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPN-GVSRGSGFVAFSTPEEASRALGEMNGK 390
+ FS FG I S IMRDP+ G SRG GF+++ + E + A+ M G+
Sbjct: 129 YDTFSAFGVIASNPK-----------IMRDPDTGNSRGFGFISYDSFEASDAAIESMTGQ 177
Query: 391 MIVSKPLYVAVAQRKEDRRAR-------------LQAQFSQ------MRPVAITPSV--- 428
+ ++ + V+ A +K+ + R AQ S+ M P + P V
Sbjct: 178 YLSNRQITVSYAYKKDTKGERHGTPAERLLAATNPTAQKSRPHTLFAMGPPSSAPQVNGL 237
Query: 429 ---------------APRMP---------LYPPGTPGLGQ---------QFMYGQGPPAM 455
APR P PP P Q QF QG P
Sbjct: 238 PRPFANGSMQPVPIPAPRQPPPPPPQVYQTQPPSWPSQPQQHSMVPPPMQFRPPQGMPPP 297
Query: 456 MPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQ 515
PPQ F QQ G RP P P + M Q G + ++GGP QQ QQ P
Sbjct: 298 PPPQ--FLNHQQGFGGPRP---PPPPQAMGMHQHGGWPPQHMQQQGGPPQQ--QQPPYQH 350
Query: 516 QQM 518
M
Sbjct: 351 HHM 353
Score = 49.7 bits (117), Expect = 5e-06
Identities = 25/92 (27%), Positives = 55/92 (59%), Gaps = 2/92 (2%)
Query: 30 VTTSLYVGDLEVNVNDSQLYDLFNQVGQVVS-VRVCRDLATRRSLGYGYVNFTNPQDAAR 88
V +L++G+L+ +V++ LYD F+ G + S ++ RD T S G+G++++ + + +
Sbjct: 110 VGANLFIGNLDPDVDEKLLYDTFSAFGVIASNPKIMRDPDTGNSRGFGFISYDSFEASDA 169
Query: 89 ALDVLNFTPMNNKSIRVMYSH-RDPSSRKSGT 119
A++ + ++N+ I V Y++ +D + GT
Sbjct: 170 AIESMTGQYLSNRQITVSYAYKKDTKGERHGT 201
>At5g54900 unknown protein
Length = 387
Score = 103 bits (256), Expect = 4e-22
Identities = 84/327 (25%), Positives = 150/327 (45%), Gaps = 36/327 (11%)
Query: 1 MAQIQVQHQ---TPAPVPAPSNGV-VPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVG 56
M Q Q Q Q + AP+ G+ N + SL++GDL+ ++++ + +F Q G
Sbjct: 25 MQQQQQQQQMQLSAAPLGQHQYGIGSQNPGSASDVKSLWIGDLQQWMDENYIMSVFAQSG 84
Query: 57 QVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPM--NNKSIRVMYSHRDPSS 114
+ S +V R+ T +S GYG++ F + A R L N PM ++ R+ ++
Sbjct: 85 EATSAKVIRNKLTGQSEGYGFIEFVSHSVAERVLQTYNGAPMPSTEQTFRLNWAQAGAGE 144
Query: 115 RKSGTA----NIFIKNLDKTIDHKALHDTFSS-FGQIMSCKIATD-GSGQSKGYGFVQFE 168
++ T IF+ +L + L DTF + +G + K+ D +G+SKGYGFV+F
Sbjct: 145 KRFQTEGPDHTIFVGDLAPEVTDYMLSDTFKNVYGSVKGAKVVLDRTTGRSKGYGFVRFA 204
Query: 169 AEDSAQNAIDKLNGMLINDKQVFVGHFLRKQD---RDNVLSKTKFNN----------VYV 215
E+ A+ ++NG + + + +G K + + T+ N ++V
Sbjct: 205 DENEQMRAMTEMNGQYCSTRPMRIGPAANKNALPMQPAMYQNTQGANAGDNDPNNTTIFV 264
Query: 216 KNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKK 275
L + T+D+LK+ FG +G + + K GFV + N A A+ LNG +
Sbjct: 265 GGLDANVTDDELKSIFGQFGELLHVKI-----PPGKRCGFVQYANKASAEHALSVLNGTQ 319
Query: 276 VDDE----EWYVGKAQKKSEREQELKG 298
+ + W G++ K + + G
Sbjct: 320 LGGQSIRLSW--GRSPNKQSDQAQWNG 344
Score = 86.3 bits (212), Expect = 5e-17
Identities = 66/282 (23%), Positives = 132/282 (46%), Gaps = 24/282 (8%)
Query: 117 SGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQN 175
S +++I +L + +D + F+ G+ S K+ + +GQS+GYGF++F + A+
Sbjct: 57 SDVKSLWIGDLQQWMDENYIMSVFAQSGEATSAKVIRNKLTGQSEGYGFIEFVSHSVAER 116
Query: 176 AIDKLNGM-LINDKQVFVGHFLRK-QDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEF-G 232
+ NG + + +Q F ++ + ++ + ++V +L+ T+ L + F
Sbjct: 117 VLQTYNGAPMPSTEQTFRLNWAQAGAGEKRFQTEGPDHTIFVGDLAPEVTDYMLSDTFKN 176
Query: 233 AYGTITSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSE 291
YG++ A ++ D GRSK +GFV F + + +A+ +NG+ +G A K+
Sbjct: 177 VYGSVKGAKVVLDRTTGRSKGYGFVRFADENEQMRAMTEMNGQYCSTRPMRIGPAANKNA 236
Query: 292 REQELKGRFEQTVKESVVDKF-QGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMI 350
+ ++ T + D +++ LD ++TD++LK +F +FG + K+
Sbjct: 237 LPMQ-PAMYQNTQGANAGDNDPNNTTIFVGGLDANVTDDELKSIFGQFGELLHVKI---- 291
Query: 351 LFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
P G + GFV ++ A AL +NG +
Sbjct: 292 -----------PPG--KRCGFVQYANKASAEHALSVLNGTQL 320
Score = 56.6 bits (135), Expect = 4e-08
Identities = 48/208 (23%), Positives = 92/208 (44%), Gaps = 36/208 (17%)
Query: 206 SKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDA 264
S + ++++ +L + E+ + + F G TSA ++R+ G+S+ +GF+ F + A
Sbjct: 55 SASDVKSLWIGDLQQWMDENYIMSVFAQSGEATSAKVIRNKLTGQSEGYGFIEFVSHSVA 114
Query: 265 AKAVEALNGKKVDDEE------WYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLY 318
+ ++ NG + E W A +K RF+ + + +
Sbjct: 115 ERVLQTYNGAPMPSTEQTFRLNWAQAGAGEK---------RFQTEGPDHTI--------F 157
Query: 319 LKNLDDSITDEKLKEMFSE-FGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTP 377
+ +L +TD L + F +G++ KV ++ G S+G GFV F+
Sbjct: 158 VGDLAPEVTDYMLSDTFKNVYGSVKGAKV-----------VLDRTTGRSKGYGFVRFADE 206
Query: 378 EEASRALGEMNGKMIVSKPLYVAVAQRK 405
E RA+ EMNG+ ++P+ + A K
Sbjct: 207 NEQMRAMTEMNGQYCSTRPMRIGPAANK 234
>At1g47490 unknown protein
Length = 432
Score = 102 bits (253), Expect = 9e-22
Identities = 96/360 (26%), Positives = 164/360 (44%), Gaps = 44/360 (12%)
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQ--IMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAID 178
I++ +L +D L+ +F+S + I+S K+ + +G S+GYGFV+FE+ D A +
Sbjct: 103 IWVGDLHHWMDEAYLNSSFASGDEREIVSVKVIRNKNNGLSEGYGFVEFESHDVADKVLR 162
Query: 179 KLNGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFN-NVYVKNLSESFTEDDLKNEFGA-YG 235
+ NG + N Q F ++ + L + +++V +LS +++ L F Y
Sbjct: 163 EFNGTTMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLSPDVSDNLLHETFSEKYP 222
Query: 236 TITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQ-KKSERE 293
++ +A ++ DA+ GRSK +GFV F + + KA+ +NG K +G A +K+
Sbjct: 223 SVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRIGPATPRKTNGY 282
Query: 294 QELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFT 353
Q+ G +++ LD S+TDE LK+ F+EFG I S K+
Sbjct: 283 QQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSVTDEDLKQPFNEFGEIVSVKI------- 335
Query: 354 DIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQ 413
V +G GFV F A AL ++NG +I + + ++ + +++ R +
Sbjct: 336 ----------PVGKGCGFVQFVNRPNAEEALEKLNGTVIGKQTVRLSWGRNPANKQPRDK 385
Query: 414 AQFSQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMP-----PQAGFGYQQQL 468
+ P Y G G +M Q P M P P G G+QQQ+
Sbjct: 386 YGNQWVDP-------------YYGGQFYNGYGYMVPQPDPRMYPAAPYYPMYG-GHQQQV 431
Score = 92.0 bits (227), Expect = 9e-19
Identities = 72/265 (27%), Positives = 129/265 (48%), Gaps = 29/265 (10%)
Query: 33 SLYVGDLEVNVNDSQLYDLF--NQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARAL 90
+++VGDL ++++ L F ++VSV+V R+ S GYG+V F + A + L
Sbjct: 102 TIWVGDLHHWMDEAYLNSSFASGDEREIVSVKVIRNKNNGLSEGYGFVEFESHDVADKVL 161
Query: 91 DVLNFTPMNN--KSIRVMYSHRDPSSRK----SGTANIFIKNLDKTIDHKALHDTFS-SF 143
N T M N + R+ ++ ++ +IF+ +L + LH+TFS +
Sbjct: 162 REFNGTTMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLSPDVSDNLLHETFSEKY 221
Query: 144 GQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDR- 201
+ + K+ D +G+SKGYGFV+F E+ A+ ++NG+ + + + +G ++
Sbjct: 222 PSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRIGPATPRKTNG 281
Query: 202 ---------DNVLSKTKFN----NVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADG 248
+ L++ + + ++V L S T++DLK F +G I S +
Sbjct: 282 YQQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSVTDEDLKQPFNEFGEIVSVKI-----P 336
Query: 249 RSKCFGFVNFENAEDAAKAVEALNG 273
K GFV F N +A +A+E LNG
Sbjct: 337 VGKGCGFVQFVNRPNAEEALEKLNG 361
Score = 81.3 bits (199), Expect = 2e-15
Identities = 60/205 (29%), Positives = 101/205 (49%), Gaps = 33/205 (16%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQ-VGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V+D+ L++ F++ V + +V D T RS GYG+V F + + +A+
Sbjct: 198 SIFVGDLSPDVSDNLLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMT 257
Query: 92 VLNFTPMNNKSIRV---------------MYSHRDPSSRKSG---TANIFIKNLDKTIDH 133
+N +++++R+ Y +R G IF+ LD ++
Sbjct: 258 EMNGVKCSSRAMRIGPATPRKTNGYQQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSVTD 317
Query: 134 KALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVG 193
+ L F+ FG+I+S KI KG GFVQF +A+ A++KLNG +I + V +
Sbjct: 318 EDLKQPFNEFGEIVSVKIPV-----GKGCGFVQFVNRPNAEEALEKLNGTVIGKQTVRLS 372
Query: 194 ---HFLRKQDRDNVLSKTKFNNVYV 215
+ KQ RD K+ N +V
Sbjct: 373 WGRNPANKQPRD------KYGNQWV 391
Score = 53.9 bits (128), Expect = 3e-07
Identities = 29/98 (29%), Positives = 55/98 (55%), Gaps = 7/98 (7%)
Query: 19 NGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYV 78
NG + + T+++VG L+ +V D L FN+ G++VSV++ G G+V
Sbjct: 291 NGTLTRPEGDIMNTTIFVGGLDSSVTDEDLKQPFNEFGEIVSVKI------PVGKGCGFV 344
Query: 79 NFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRK 116
F N +A AL+ LN T + +++R+ + R+P++++
Sbjct: 345 QFVNRPNAEEALEKLNGTVIGKQTVRLSWG-RNPANKQ 381
>At4g27000 putative DNA binding protein
Length = 415
Score = 101 bits (251), Expect = 2e-21
Identities = 73/289 (25%), Positives = 136/289 (46%), Gaps = 28/289 (9%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
SL++GDL+ ++++ L ++F G+ + +V R+ S GYG++ F N A R L
Sbjct: 81 SLWIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQNGYSEGYGFIEFVNHATAERNLQT 140
Query: 93 LNFTPM--NNKSIRVMYSHRDPSSRKSGTA---NIFIKNLDKTIDHKALHDTFSS-FGQI 146
N PM + ++ R+ ++ R+ +F+ +L + L +TF + + +
Sbjct: 141 YNGAPMPSSEQAFRLNWAQLGAGERRQAEGPEHTVFVGDLAPDVTDHMLTETFKAVYSSV 200
Query: 147 MSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQD---RD 202
K+ D +G+SKGYGFV+F E A+ ++NG + + + G K+ +
Sbjct: 201 KGAKVVNDRTTGRSKGYGFVRFADESEQIRAMTEMNGQYCSSRPMRTGPAANKKPLTMQP 260
Query: 203 NVLSKTKFNN---------VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCF 253
T+ N+ ++V + +S TEDDLK+ FG +G + + K
Sbjct: 261 ASYQNTQGNSGESDPTNTTIFVGAVDQSVTEDDLKSVFGQFGELVHVKI-----PAGKRC 315
Query: 254 GFVNFENAEDAAKAVEALNGKKVDDE----EWYVGKAQKKSEREQELKG 298
GFV + N A +A+ LNG ++ + W + K+++ +Q G
Sbjct: 316 GFVQYANRACAEQALSVLNGTQLGGQSIRLSWGRSPSNKQTQPDQAQYG 364
Score = 80.9 bits (198), Expect = 2e-15
Identities = 67/301 (22%), Positives = 139/301 (45%), Gaps = 23/301 (7%)
Query: 121 NIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDK 179
+++I +L +D L + F G+ + K+ + +G S+GYGF++F +A+ +
Sbjct: 81 SLWIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQNGYSEGYGFIEFVNHATAERNLQT 140
Query: 180 LNGM-LINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGTI 237
NG + + +Q F ++ + + ++ + V+V +L+ T+ L F A Y ++
Sbjct: 141 YNGAPMPSSEQAFRLNWAQLGAGERRQAEGPEHTVFVGDLAPDVTDHMLTETFKAVYSSV 200
Query: 238 TSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQEL 296
A ++ D GRSK +GFV F + + +A+ +NG+ G A K +
Sbjct: 201 KGAKVVNDRTTGRSKGYGFVRFADESEQIRAMTEMNGQYCSSRPMRTGPAANKKPLTMQ- 259
Query: 297 KGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIR 356
++ T S +++ +D S+T++ LK +F +FG + K+
Sbjct: 260 PASYQNTQGNSGESDPTNTTIFVGAVDQSVTEDDLKSVFGQFGELVHVKI---------- 309
Query: 357 VIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARL-QAQ 415
P G + GFV ++ A +AL +NG + + + ++ + +++ + QAQ
Sbjct: 310 -----PAG--KRCGFVQYANRACAEQALSVLNGTQLGGQSIRLSWGRSPSNKQTQPDQAQ 362
Query: 416 F 416
+
Sbjct: 363 Y 363
Score = 60.8 bits (146), Expect = 2e-09
Identities = 54/204 (26%), Positives = 93/204 (45%), Gaps = 27/204 (13%)
Query: 206 SKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDA 264
S + ++++ +L E+ L N FG G T+A ++R+ +G S+ +GF+ F N A
Sbjct: 75 SAGEIRSLWIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQNGYSEGYGFIEFVNHATA 134
Query: 265 AKAVEALNGKKVDDEE--WYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNL 322
+ ++ NG + E + + AQ + ++ +G E TV ++ +L
Sbjct: 135 ERNLQTYNGAPMPSSEQAFRLNWAQLGAGERRQAEGP-EHTV-------------FVGDL 180
Query: 323 DDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASR 382
+TD L E F +S K +V+ G S+G GFV F+ E R
Sbjct: 181 APDVTDHMLTETFK--AVYSSVK--------GAKVVNDRTTGRSKGYGFVRFADESEQIR 230
Query: 383 ALGEMNGKMIVSKPLYVAVAQRKE 406
A+ EMNG+ S+P+ A K+
Sbjct: 231 AMTEMNGQYCSSRPMRTGPAANKK 254
Score = 46.2 bits (108), Expect = 6e-05
Identities = 28/86 (32%), Positives = 52/86 (59%), Gaps = 7/86 (8%)
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+++VG ++ +V + L +F Q G++V V++ A +R G+V + N A +AL
Sbjct: 278 TTIFVGAVDQSVTEDDLKSVFGQFGELVHVKI---PAGKRC---GFVQYANRACAEQALS 331
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKS 117
VLN T + +SIR+ + R PS++++
Sbjct: 332 VLNGTQLGGQSIRLSWG-RSPSNKQT 356
Score = 29.6 bits (65), Expect = 5.6
Identities = 22/58 (37%), Positives = 25/58 (42%), Gaps = 10/58 (17%)
Query: 498 GRRGGPGQQPPQQVPMMQQQ----MLPRQRVYRYPPGRSNIQDAP------VQNIGGG 545
G GPGQ P Q +QQQ M +Q+ PP N Q AP Q GGG
Sbjct: 11 GAATGPGQIPSDQQAYLQQQQSWMMQHQQQQQGQPPAGWNQQSAPSSGQPQQQQYGGG 68
>At1g47500 unknown protein
Length = 434
Score = 100 bits (250), Expect = 2e-21
Identities = 99/395 (25%), Positives = 175/395 (44%), Gaps = 55/395 (13%)
Query: 95 FTPMNNKSIRVMYSHRDPSSRKSGTAN-----IFIKNLDKTIDHKALHDTFSSFGQ--IM 147
F+P + + H+ ++ N I++ +L +D L+ F+S + I+
Sbjct: 73 FSPYHQYPNHHHFHHQSRGNKHQNAFNGENKTIWVGDLQNWMDEAYLNSAFTSAEEREIV 132
Query: 148 SCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKLNGM-LINDKQVFVGHFLRKQDRDNVL 205
S K+ + +G S+GYGFV+FE+ D A + + NG + N Q F ++ + L
Sbjct: 133 SLKVIRNKHNGSSEGYGFVEFESHDVADKVLQEFNGAPMPNTDQPFRLNWASFSTGEKRL 192
Query: 206 SKTKFN-NVYVKNLSESFTEDDLKNEFGA-YGTITSAVLMRDAD-GRSKCFGFVNFENAE 262
+ +++V +L+ ++ L F Y ++ +A ++ DA+ GRSK +GFV F +
Sbjct: 193 ENNGPDLSIFVGDLAPDVSDALLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDEN 252
Query: 263 DAAKAVEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQG----LNLY 318
+ KA+ +NG K +G A + + +G + + + +G ++
Sbjct: 253 ERTKAMTEMNGVKCSSRAMRIGPATPRKTNGYQQQGGY---MPSGAFTRSEGDTINTTIF 309
Query: 319 LKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPE 378
+ LD S+TDE LK+ FSEFG I S K+ V +G GFV F
Sbjct: 310 VGGLDSSVTDEDLKQPFSEFGEIVSVKI-----------------PVGKGCGFVQFVNRP 352
Query: 379 EASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLYPPG 438
A AL ++NG +I + + ++ + +++ R + + P Y G
Sbjct: 353 NAEEALEKLNGTVIGKQTVRLSWGRNPANKQPRDKYGNQWVDP-------------YYGG 399
Query: 439 TPGLGQQFMYGQGPPAMMP-----PQAGFGYQQQL 468
G +M Q P M P P G G+QQQ+
Sbjct: 400 QFYNGYGYMVPQPDPRMYPAAPYYPMYG-GHQQQV 433
Score = 94.7 bits (234), Expect = 1e-19
Identities = 72/265 (27%), Positives = 127/265 (47%), Gaps = 29/265 (10%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQ--VVSVRVCRDLATRRSLGYGYVNFTNPQDAARAL 90
+++VGDL+ ++++ L F + +VS++V R+ S GYG+V F + A + L
Sbjct: 104 TIWVGDLQNWMDEAYLNSAFTSAEEREIVSLKVIRNKHNGSSEGYGFVEFESHDVADKVL 163
Query: 91 DVLNFTPMNN--KSIRVMYSHRDPSSRK----SGTANIFIKNLDKTIDHKALHDTFS-SF 143
N PM N + R+ ++ ++ +IF+ +L + LH+TFS +
Sbjct: 164 QEFNGAPMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLAPDVSDALLHETFSEKY 223
Query: 144 GQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRK---- 198
+ + K+ D +G+SKGYGFV+F E+ A+ ++NG+ + + + +G +
Sbjct: 224 PSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRIGPATPRKTNG 283
Query: 199 -QDRDNVLSKTKF---------NNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADG 248
Q + + F ++V L S T++DLK F +G I S +
Sbjct: 284 YQQQGGYMPSGAFTRSEGDTINTTIFVGGLDSSVTDEDLKQPFSEFGEIVSVKI-----P 338
Query: 249 RSKCFGFVNFENAEDAAKAVEALNG 273
K GFV F N +A +A+E LNG
Sbjct: 339 VGKGCGFVQFVNRPNAEEALEKLNG 363
Score = 82.0 bits (201), Expect = 9e-16
Identities = 61/205 (29%), Positives = 101/205 (48%), Gaps = 33/205 (16%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQ-VGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V+D+ L++ F++ V + +V D T RS GYG+V F + + +A+
Sbjct: 200 SIFVGDLAPDVSDALLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMT 259
Query: 92 VLNFTPMNNKSIRV---------------MYSHRDPSSRKSG---TANIFIKNLDKTIDH 133
+N +++++R+ Y +R G IF+ LD ++
Sbjct: 260 EMNGVKCSSRAMRIGPATPRKTNGYQQQGGYMPSGAFTRSEGDTINTTIFVGGLDSSVTD 319
Query: 134 KALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVG 193
+ L FS FG+I+S KI KG GFVQF +A+ A++KLNG +I + V +
Sbjct: 320 EDLKQPFSEFGEIVSVKIPV-----GKGCGFVQFVNRPNAEEALEKLNGTVIGKQTVRLS 374
Query: 194 ---HFLRKQDRDNVLSKTKFNNVYV 215
+ KQ RD K+ N +V
Sbjct: 375 WGRNPANKQPRD------KYGNQWV 393
Score = 49.3 bits (116), Expect = 7e-06
Identities = 26/87 (29%), Positives = 52/87 (58%), Gaps = 7/87 (8%)
Query: 30 VTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARA 89
+ T+++VG L+ +V D L F++ G++VSV++ G G+V F N +A A
Sbjct: 304 INTTIFVGGLDSSVTDEDLKQPFSEFGEIVSVKI------PVGKGCGFVQFVNRPNAEEA 357
Query: 90 LDVLNFTPMNNKSIRVMYSHRDPSSRK 116
L+ LN T + +++R+ + R+P++++
Sbjct: 358 LEKLNGTVIGKQTVRLSWG-RNPANKQ 383
>At1g11650 putative DNA binding protein
Length = 405
Score = 100 bits (249), Expect = 3e-21
Identities = 80/281 (28%), Positives = 135/281 (47%), Gaps = 27/281 (9%)
Query: 13 PVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRS 72
P A V P A++ T L++GDL+ ++++ LY F G++VS +V R+ T +
Sbjct: 45 PQAAAPPSVQPTTADEIRT--LWIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQV 102
Query: 73 LGYGYVNFTNPQDAARALDVLNFTPM---NNKSIRVMYSHRDPSSRKSGTAN--IFIKNL 127
GYG++ F + A R L N P+ ++ R+ ++ ++ + + IF+ +L
Sbjct: 103 EGYGFIEFASHAAAERVLQTFNNAPIPSFPDQLFRLNWASLSSGDKRDDSPDYTIFVGDL 162
Query: 128 DKTIDHKALHDTF-SSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKLNGMLI 185
+ L +TF +S+ + K+ D +G++KGYGFV+F E A+ ++NG+
Sbjct: 163 AADVTDYILLETFRASYPSVKGAKVVIDRVTGRTKGYGFVRFSDESEQIRAMTEMNGVPC 222
Query: 186 NDKQVFVGHFLRKQ----DRDN-------VLSKTKFNN--VYVKNLSESFTEDDLKNEFG 232
+ + + +G K+ RD+ V + NN V+V L S T+D LKN F
Sbjct: 223 STRPMRIGPAASKKGVTGQRDSYQSSAAGVTTDNDPNNTTVFVGGLDASVTDDHLKNVFS 282
Query: 233 AYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
YG I + K GFV F A +A+ LNG
Sbjct: 283 QYGEIVHVKI-----PAGKRCGFVQFSEKSCAEEALRMLNG 318
Score = 86.7 bits (213), Expect = 4e-17
Identities = 95/400 (23%), Positives = 160/400 (39%), Gaps = 68/400 (17%)
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L+ F+ G+++S K+ + +GQ +GYGF++F + +A+ +
Sbjct: 64 LWIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASHAAAERVLQTF 123
Query: 181 NGMLIND--KQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGTI 237
N I Q+F ++ D + ++V +L+ T+ L F A Y ++
Sbjct: 124 NNAPIPSFPDQLFRLNWASLSSGDKRDDSPDYT-IFVGDLAADVTDYILLETFRASYPSV 182
Query: 238 TSAVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKSEREQEL 296
A ++ D GR+K +GFV F + + +A+ +NG +G A K +
Sbjct: 183 KGAKVVIDRVTGRTKGYGFVRFSDESEQIRAMTEMNGVPCSTRPMRIGPAASKKGVTGQR 242
Query: 297 KGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIR 356
+ + +++ LD S+TD+ LK +FS++G I K+
Sbjct: 243 DSYQSSAAGVTTDNDPNNTTVFVGGLDASVTDDHLKNVFSQYGEIVHVKI---------- 292
Query: 357 VIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQF 416
P G + GFV FS A AL +NG + + ++ + ++++ +QF
Sbjct: 293 -----PAG--KRCGFVQFSEKSCAEEALRMLNGVQLGGTTVRLSWGRSPSNKQSGDPSQF 345
Query: 417 SQMRPVAITPSVAPRMPLYPPGTPGLGQQFMYGQGPPAMMPPQAGFGYQQQLVPGMRPGG 476
Y G YGQG Q +GY P GG
Sbjct: 346 ------------------YYGG---------YGQG-------QEQYGYTMPQDPNAYYGG 371
Query: 477 GPMPSYFVPMVQQGQQGQRPGGRRGGPGQQPPQQVPMMQQ 516
Y G Q P GQQPPQQ P QQ
Sbjct: 372 YSGGGY------SGGYQQTPQA-----GQQPPQQPPQQQQ 400
Score = 48.9 bits (115), Expect = 9e-06
Identities = 29/92 (31%), Positives = 54/92 (58%), Gaps = 7/92 (7%)
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
T+++VG L+ +V D L ++F+Q G++V V++ A +R G+V F+ A AL
Sbjct: 261 TTVFVGGLDASVTDDHLKNVFSQYGEIVHVKIP---AGKRC---GFVQFSEKSCAEEALR 314
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKSGTANIF 123
+LN + ++R+ + R PS+++SG + F
Sbjct: 315 MLNGVQLGGTTVRLSWG-RSPSNKQSGDPSQF 345
>At5g19350 DNA binding protein ACBF - like
Length = 425
Score = 95.9 bits (237), Expect = 6e-20
Identities = 72/299 (24%), Positives = 133/299 (44%), Gaps = 39/299 (13%)
Query: 15 PAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLG 74
P P G + +L++GDL+ V+++ L F+Q G++VSV+V R+ T + G
Sbjct: 7 PQPPQGSYHHPQTLEEVRTLWIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEG 66
Query: 75 YGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA----NIFIKNLDKT 130
YG++ F + A R L N T M + + S + A +IF+ +L
Sbjct: 67 YGFIEFISHAAAERTLQTYNGTQMPGTELTFRLNWASFGSGQKVDAGPDHSIFVGDLAPD 126
Query: 131 IDHKALHDTFS-SFGQIMSCKIATDGS-GQSKGYGFVQFEAEDSAQNAIDKLNGMLINDK 188
+ L +TF + + K+ TD S G+SKGYGFV+F E A+ ++NG+ + +
Sbjct: 127 VTDYLLQETFRVHYSSVRGAKVVTDPSTGRSKGYGFVKFAEESERNRAMAEMNGLYCSTR 186
Query: 189 QVFVGHFLRKQD---RDNVLSKTKF-------------------------NNVYVKNLSE 220
+ + K++ + ++K + + V NL +
Sbjct: 187 PMRISAATPKKNVGVQQQYVTKAVYPVTVPSAVAAPVQAYVAPPESDVTCTTISVANLDQ 246
Query: 221 SFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDE 279
+ TE++LK F G + + +K +G+V F+ A +AV+ + G+ + +
Sbjct: 247 NVTEEELKKAFSQLGEVIYVKI-----PATKGYGYVQFKTRPSAEEAVQRMQGQVIGQQ 300
Score = 82.0 bits (201), Expect = 9e-16
Identities = 66/289 (22%), Positives = 127/289 (43%), Gaps = 35/289 (12%)
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
++I +L +D L FS G+++S K+ + +GQ +GYGF++F + +A+ +
Sbjct: 26 LWIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEGYGFIEFISHAAAERTLQTY 85
Query: 181 NGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGA-YGTITS 239
NG + ++ + ++++V +L+ T+ L+ F Y ++
Sbjct: 86 NGTQMPGTELTFRLNWASFGSGQKVDAGPDHSIFVGDLAPDVTDYLLQETFRVHYSSVRG 145
Query: 240 AVLMRD-ADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKAQKKS----EREQ 294
A ++ D + GRSK +GFV F + +A+ +NG + A K +++
Sbjct: 146 AKVVTDPSTGRSKGYGFVKFAEESERNRAMAEMNGLYCSTRPMRISAATPKKNVGVQQQY 205
Query: 295 ELKGRFEQTVKESVVDKFQGL-----------NLYLKNLDDSITDEKLKEMFSEFGTITS 343
K + TV +V Q + + NLD ++T+E+LK+ FS+ G +
Sbjct: 206 VTKAVYPVTVPSAVAAPVQAYVAPPESDVTCTTISVANLDQNVTEEELKKAFSQLGEVIY 265
Query: 344 YKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
K+ ++G G+V F T A A+ M G++I
Sbjct: 266 VKI-----------------PATKGYGYVQFKTRPSAEEAVQRMQGQVI 297
Score = 71.6 bits (174), Expect = 1e-12
Identities = 51/191 (26%), Positives = 81/191 (41%), Gaps = 38/191 (19%)
Query: 33 SLYVGDLEVNVNDSQLYDLFN-QVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V D L + F V +V D +T RS GYG+V F + RA+
Sbjct: 117 SIFVGDLAPDVTDYLLQETFRVHYSSVRGAKVVTDPSTGRSKGYGFVKFAEESERNRAMA 176
Query: 92 VLN-------------FTPMNNKSIRVMY-------------------SHRDPSSRKSGT 119
+N TP N ++ Y ++ P
Sbjct: 177 EMNGLYCSTRPMRISAATPKKNVGVQQQYVTKAVYPVTVPSAVAAPVQAYVAPPESDVTC 236
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDK 179
I + NLD+ + + L FS G+++ KI +KGYG+VQF+ SA+ A+ +
Sbjct: 237 TTISVANLDQNVTEEELKKAFSQLGEVIYVKIPA-----TKGYGYVQFKTRPSAEEAVQR 291
Query: 180 LNGMLINDKQV 190
+ G +I + V
Sbjct: 292 MQGQVIGQQAV 302
Score = 57.4 bits (137), Expect = 3e-08
Identities = 57/237 (24%), Positives = 104/237 (43%), Gaps = 44/237 (18%)
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEAL 271
+++ +L E+ L + F G + S ++R+ G+ + +GF+ F + A + ++
Sbjct: 26 LWIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEGYGFIEFISHAAAERTLQTY 85
Query: 272 NGKKVDDEE------WY-VGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDD 324
NG ++ E W G QK VD ++++ +L
Sbjct: 86 NGTQMPGTELTFRLNWASFGSGQK--------------------VDAGPDHSIFVGDLAP 125
Query: 325 SITDEKLKEMFS-EFGTITSYKVCTMILFTDIRVIMRDPN-GVSRGSGFVAFSTPEEASR 382
+TD L+E F + ++ KV T DP+ G S+G GFV F+ E +R
Sbjct: 126 DVTDYLLQETFRVHYSSVRGAKVVT------------DPSTGRSKGYGFVKFAEESERNR 173
Query: 383 ALGEMNGKMIVSKPLYVAVAQRKEDRRARLQAQFSQMRPVAITPSVAPRMPLY--PP 437
A+ EMNG ++P+ ++ A K++ + Q + PV + +VA + Y PP
Sbjct: 174 AMAEMNGLYCSTRPMRISAATPKKNVGVQQQYVTKAVYPVTVPSAVAAPVQAYVAPP 230
Score = 47.4 bits (111), Expect = 3e-05
Identities = 28/98 (28%), Positives = 51/98 (51%), Gaps = 6/98 (6%)
Query: 11 PAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATR 70
P+ V AP V + T++ V +L+ NV + +L F+Q+G+V+ V++ AT+
Sbjct: 216 PSAVAAPVQAYVAPPESDVTCTTISVANLDQNVTEEELKKAFSQLGEVIYVKI---PATK 272
Query: 71 RSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYS 108
GYGYV F A A+ + + +++R+ +S
Sbjct: 273 ---GYGYVQFKTRPSAEEAVQRMQGQVIGQQAVRISWS 307
>At1g54080 unknown protein
Length = 426
Score = 94.7 bits (234), Expect = 1e-19
Identities = 74/295 (25%), Positives = 134/295 (45%), Gaps = 39/295 (13%)
Query: 11 PAPVPAPSNGVVPNVANQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATR 70
P P PS + P + S+Y G++ V + L ++F G + S ++ R +
Sbjct: 43 PQMEPLPSGNLPPGF-DPTTCRSVYAGNIHTQVTEILLQEIFASTGPIESCKLIR----K 97
Query: 71 RSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSGTA-NIFIKNLDK 129
YG+V++ + + A+ A+ LN + + ++V +++ + + NIF+ +L
Sbjct: 98 DKSSYGFVHYFDRRCASMAIMTLNGRHIFGQPMKVNWAYATGQREDTSSHFNIFVGDLSP 157
Query: 130 TIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDK 188
+ AL D+FS+F ++ D +G+S+G+GFV F + AQ AI+++NG ++ +
Sbjct: 158 EVTDAALFDSFSAFNSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQTAINEMNGKWVSSR 217
Query: 189 QV---------FVGHFLRKQDRDNVLSKT--------------------KFNNVYVKNLS 219
Q+ G D +V+ T +F VYV NLS
Sbjct: 218 QIRCNWATKGATFGEDKHSSDGKSVVELTNGSSEDGRELSNEDAPENNPQFTTVYVGNLS 277
Query: 220 ESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGK 274
T+ DL F T+ + V+ R K FGFV + ++AA A++ N +
Sbjct: 278 PEVTQLDLHRLF---YTLGAGVIEEVRVQRDKGFGFVRYNTHDEAALAIQMGNAQ 329
Score = 80.5 bits (197), Expect = 3e-15
Identities = 79/292 (27%), Positives = 130/292 (44%), Gaps = 44/292 (15%)
Query: 111 DPSSRKSGTANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAE 170
DP++ +S ++ N+ + L + F+S G I SCK+ YGFV +
Sbjct: 58 DPTTCRS----VYAGNIHTQVTEILLQEIFASTGPIESCKLIRK---DKSSYGFVHYFDR 110
Query: 171 DSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNE 230
A AI LNG I + + V R++ S N++V +LS T+ L +
Sbjct: 111 RCASMAIMTLNGRHIFGQPMKVNWAYATGQREDTSSHF---NIFVGDLSPEVTDAALFDS 167
Query: 231 FGAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKV------------- 276
F A+ + + A +M D GRS+ FGFV+F N +DA A+ +NGK V
Sbjct: 168 FSAFNSCSDARVMWDQKTGRSRGFGFVSFRNQQDAQTAINEMNGKWVSSRQIRCNWATKG 227
Query: 277 ----DDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLK 332
+D+ GK+ + GR E + +++ + Q +Y+ NL +T L
Sbjct: 228 ATFGEDKHSSDGKSVVELTNGSSEDGR-ELSNEDAPENNPQFTTVYVGNLSPEVTQLDLH 286
Query: 333 EMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRAL 384
+F G + ++RV RD +G GFV ++T +EA+ A+
Sbjct: 287 RLFYTLGAG---------VIEEVRV-QRD-----KGFGFVRYNTHDEAALAI 323
Score = 71.6 bits (174), Expect = 1e-12
Identities = 53/190 (27%), Positives = 84/190 (43%), Gaps = 30/190 (15%)
Query: 208 TKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKA 267
T +VY N+ TE L+ F + G I S L+R +GFV++ + A+ A
Sbjct: 60 TTCRSVYAGNIHTQVTEILLQEIFASTGPIESCKLIRKD---KSSYGFVHYFDRRCASMA 116
Query: 268 VEALNGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSIT 327
+ LNG+ + + V A +RE D N+++ +L +T
Sbjct: 117 IMTLNGRHIFGQPMKVNWAYATGQRE----------------DTSSHFNIFVGDLSPEVT 160
Query: 328 DEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEM 387
D L + FS F + + D RV+ G SRG GFV+F ++A A+ EM
Sbjct: 161 DAALFDSFSAFNSCS-----------DARVMWDQKTGRSRGFGFVSFRNQQDAQTAINEM 209
Query: 388 NGKMIVSKPL 397
NGK + S+ +
Sbjct: 210 NGKWVSSRQI 219
Score = 31.6 bits (70), Expect = 1.5
Identities = 26/95 (27%), Positives = 45/95 (47%), Gaps = 19/95 (20%)
Query: 316 NLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFS 375
++Y N+ +T+ L+E+F+ G I S K+ I +D + GFV +
Sbjct: 64 SVYAGNIHTQVTEILLQEIFASTGPIESCKL-----------IRKDKSSY----GFVHYF 108
Query: 376 TPEEASRALGEMNGKMIVSKPLYV----AVAQRKE 406
AS A+ +NG+ I +P+ V A QR++
Sbjct: 109 DRRCASMAIMTLNGRHIFGQPMKVNWAYATGQRED 143
>At3g19130 nuclear acid binding protein, putative
Length = 435
Score = 94.4 bits (233), Expect = 2e-19
Identities = 71/275 (25%), Positives = 132/275 (47%), Gaps = 39/275 (14%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L+VGDL ++++ L+ F+ G+V SV+V R+ T +S GYG+V F + A L
Sbjct: 109 TLWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEEVLQN 168
Query: 93 L--NFTPMNNKSIRVMYSHRDPSSRKS----GTANIFIKNLDKTIDHKALHDTFSS-FGQ 145
+ P +++ R+ ++ +++ ++F+ +L + LH+TFS +
Sbjct: 169 YSGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLLHETFSDRYPS 228
Query: 146 IMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRDN- 203
+ S K+ D +G+SKGYGFV+F E+ A+ ++NG +++Q+ VG K+ N
Sbjct: 229 VKSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRAIANQ 288
Query: 204 --------VLSKTKFNN-----------------VYVKNLSESFTEDDLKNEFGAYGTIT 238
+L+ +N ++V + ++DL+ F +G +
Sbjct: 289 QQHSSQAVILAGGHGSNGSMGYGSQSDGESTNATIFVGGIDPDVIDEDLRQPFSQFGEVV 348
Query: 239 SAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNG 273
S + K GFV F + + A A+E+LNG
Sbjct: 349 SVKI-----PVGKGCGFVQFADRKSAEDAIESLNG 378
Score = 85.9 bits (211), Expect = 7e-17
Identities = 71/289 (24%), Positives = 132/289 (45%), Gaps = 35/289 (12%)
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDG-SGQSKGYGFVQFEAEDSAQNAIDKL 180
+++ +L +D LH FS G++ S K+ + + QS+GYGFV+F + +A+ +
Sbjct: 110 LWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEEVLQNY 169
Query: 181 NGMLI--NDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFG-AYGTI 237
+G ++ +D+ + + +V+V +LS T+ L F Y ++
Sbjct: 170 SGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLLHETFSDRYPSV 229
Query: 238 TSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYVGKA---------Q 287
SA ++ D++ GRSK +GFV F + + ++A+ +NG + + VG A Q
Sbjct: 230 KSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRAIANQQ 289
Query: 288 KKSEREQELKGRFEQTVKESVVDKFQG----LNLYLKNLDDSITDEKLKEMFSEFGTITS 343
+ S + L G + G +++ +D + DE L++ FS+FG + S
Sbjct: 290 QHSSQAVILAGGHGSNGSMGYGSQSDGESTNATIFVGGIDPDVIDEDLRQPFSQFGEVVS 349
Query: 344 YKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
K+ V +G GFV F+ + A A+ +NG +I
Sbjct: 350 VKI-----------------PVGKGCGFVQFADRKSAEDAIESLNGTVI 381
Score = 80.1 bits (196), Expect = 4e-15
Identities = 58/189 (30%), Positives = 87/189 (45%), Gaps = 36/189 (19%)
Query: 33 SLYVGDLEVNVNDSQLYDLFN-QVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V D L++ F+ + V S +V D T RS GYG+V F + + +RAL
Sbjct: 203 SVFVGDLSPDVTDVLLHETFSDRYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALT 262
Query: 92 VLNFTPMNNKSIRV------------------------------MYSHRDPSSRKSGTAN 121
+N +N+ +RV + S +S A
Sbjct: 263 EMNGAYCSNRQMRVGIATPKRAIANQQQHSSQAVILAGGHGSNGSMGYGSQSDGESTNAT 322
Query: 122 IFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLN 181
IF+ +D + + L FS FG+++S KI KG GFVQF SA++AI+ LN
Sbjct: 323 IFVGGIDPDVIDEDLRQPFSQFGEVVSVKIPV-----GKGCGFVQFADRKSAEDAIESLN 377
Query: 182 GMLINDKQV 190
G +I V
Sbjct: 378 GTVIGKNTV 386
Score = 56.2 bits (134), Expect = 6e-08
Identities = 52/195 (26%), Positives = 88/195 (44%), Gaps = 24/195 (12%)
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDA-DGRSKCFGFVNFENAEDAAKAVEAL 271
++V +L E L + F G ++S ++R+ +S+ +GFV F + A A E L
Sbjct: 110 LWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSR---AAAEEVL 166
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
Y G S++ + T ++ V+ L++++ +L +TD L
Sbjct: 167 QN--------YSGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLL 218
Query: 332 KEMFSE-FGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGK 390
E FS+ + ++ S KV ++ G S+G GFV F E SRAL EMNG
Sbjct: 219 HETFSDRYPSVKSAKV-----------VIDSNTGRSKGYGFVRFGDENERSRALTEMNGA 267
Query: 391 MIVSKPLYVAVAQRK 405
++ + V +A K
Sbjct: 268 YCSNRQMRVGIATPK 282
Score = 43.5 bits (101), Expect = 4e-04
Identities = 31/120 (25%), Positives = 59/120 (48%), Gaps = 13/120 (10%)
Query: 5 QVQHQTPAPVPAPSNGVVPNVA------NQFVTTSLYVGDLEVNVNDSQLYDLFNQVGQV 58
Q QH + A + A +G ++ + +++VG ++ +V D L F+Q G+V
Sbjct: 288 QQQHSSQAVILAGGHGSNGSMGYGSQSDGESTNATIFVGGIDPDVIDEDLRQPFSQFGEV 347
Query: 59 VSVRVCRDLATRRSLGYGYVNFTNPQDAARALDVLNFTPMNNKSIRVMYSHRDPSSRKSG 118
VSV++ G G+V F + + A A++ LN T + ++R+ + R P+ + G
Sbjct: 348 VSVKI------PVGKGCGFVQFADRKSAEDAIESLNGTVIGKNTVRLSWG-RSPNKQWRG 400
>At1g49600 DNA binding protein ACBF, putative
Length = 445
Score = 92.4 bits (228), Expect = 7e-19
Identities = 73/270 (27%), Positives = 127/270 (47%), Gaps = 34/270 (12%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L+VGDL ++++ L+ F+ +V SV+V R+ T +S GYG+V F + A AL
Sbjct: 120 TLWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGFVEFLSRSAAEEALQS 179
Query: 93 LNFTPMNN--KSIRVMYSHRDPSSRKSG----TANIFIKNLDKTIDHKALHDTFSS-FGQ 145
+ M N + R+ ++ +++ +IF+ +L + L +TF+ +
Sbjct: 180 FSGVTMPNAEQPFRLNWASFSTGEKRASENGPDLSIFVGDLAPDVSDAVLLETFAGRYPS 239
Query: 146 IMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHFLRKQ----- 199
+ K+ D +G+SKGYGFV+F E+ A+ ++NG + +Q+ VG K+
Sbjct: 240 VKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMTEMNGAFCSSRQMRVGIATPKRAAAYG 299
Query: 200 --------------DRDNVLSKTKFNN--VYVKNLSESFTEDDLKNEFGAYGTITSAVLM 243
+ +S + NN ++V L TE+DL F +G + S +
Sbjct: 300 QQNGSQALTLAGGHGGNGSMSDGESNNSTIFVGGLDADVTEEDLMQPFSDFGEVVSVKI- 358
Query: 244 RDADGRSKCFGFVNFENAEDAAKAVEALNG 273
K GFV F N + A +A+ LNG
Sbjct: 359 ----PVGKGCGFVQFANRQSAEEAIGNLNG 384
Score = 88.2 bits (217), Expect = 1e-17
Identities = 76/300 (25%), Positives = 143/300 (47%), Gaps = 32/300 (10%)
Query: 108 SHRDPSSRKSG--TANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSG-QSKGYGF 164
S + P G +++ +L +D LH FS ++ S K+ + QS+GYGF
Sbjct: 105 SQQQPRGGSGGDDVKTLWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGF 164
Query: 165 VQFEAEDSAQNAIDKLNGMLI-NDKQVFVGHFLRKQDRDNVLSKTKFN-NVYVKNLSESF 222
V+F + +A+ A+ +G+ + N +Q F ++ + S+ + +++V +L+
Sbjct: 165 VEFLSRSAAEEALQSFSGVTMPNAEQPFRLNWASFSTGEKRASENGPDLSIFVGDLAPDV 224
Query: 223 TEDDLKNEF-GAYGTITSAVLMRDAD-GRSKCFGFVNFENAEDAAKAVEALNGKKVDDEE 280
++ L F G Y ++ A ++ D++ GRSK +GFV F + + ++A+ +NG +
Sbjct: 225 SDAVLLETFAGRYPSVKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMTEMNGAFCSSRQ 284
Query: 281 WYVGKAQ-KKSEREQELKGRFEQTV------KESVVD-KFQGLNLYLKNLDDSITDEKLK 332
VG A K++ + G T+ S+ D + +++ LD +T+E L
Sbjct: 285 MRVGIATPKRAAAYGQQNGSQALTLAGGHGGNGSMSDGESNNSTIFVGGLDADVTEEDLM 344
Query: 333 EMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMI 392
+ FS+FG + S K+ V +G GFV F+ + A A+G +NG +I
Sbjct: 345 QPFSDFGEVVSVKI-----------------PVGKGCGFVQFANRQSAEEAIGNLNGTVI 387
Score = 78.6 bits (192), Expect = 1e-14
Identities = 68/237 (28%), Positives = 106/237 (44%), Gaps = 37/237 (15%)
Query: 33 SLYVGDLEVNVNDSQLYDLF-NQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
S++VGDL +V+D+ L + F + V +V D T RS GYG+V F + + +RA+
Sbjct: 214 SIFVGDLAPDVSDAVLLETFAGRYPSVKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMT 273
Query: 92 VLNFTPMNNKSIRVMYS----------------------HRDPSSRKSGTAN---IFIKN 126
+N +++ +RV + H S G +N IF+
Sbjct: 274 EMNGAFCSSRQMRVGIATPKRAAAYGQQNGSQALTLAGGHGGNGSMSDGESNNSTIFVGG 333
Query: 127 LDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLI- 185
LD + + L FS FG+++S KI KG GFVQF SA+ AI LNG +I
Sbjct: 334 LDADVTEEDLMQPFSDFGEVVSVKIPV-----GKGCGFVQFANRQSAEEAIGNLNGTVIG 388
Query: 186 -NDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAV 241
N ++ G KQ R + S ++N Y + + + + Y T +AV
Sbjct: 389 KNTVRLSWGRSPNKQWRSD--SGNQWNGGYSR--GQGYNNGYANQDSNMYATAAAAV 441
Score = 50.1 bits (118), Expect = 4e-06
Identities = 47/194 (24%), Positives = 87/194 (44%), Gaps = 22/194 (11%)
Query: 213 VYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADG-RSKCFGFVNFENAEDAAKAVEAL 271
++V +L E L F ++S ++R+ +S+ +GFV F + A +A+++
Sbjct: 121 LWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGFVEFLSRSAAEEALQSF 180
Query: 272 NGKKVDDEEWYVGKAQKKSEREQELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKL 331
+G + + +E+ L T ++ + L++++ +L ++D L
Sbjct: 181 SGVTMPN-----------AEQPFRLNWASFSTGEKRASENGPDLSIFVGDLAPDVSDAVL 229
Query: 332 KEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKM 391
E F+ G S K +V++ G S+G GFV F E SRA+ EMNG
Sbjct: 230 LETFA--GRYPSVK--------GAKVVIDSNTGRSKGYGFVRFGDENERSRAMTEMNGAF 279
Query: 392 IVSKPLYVAVAQRK 405
S+ + V +A K
Sbjct: 280 CSSRQMRVGIATPK 293
Score = 44.7 bits (104), Expect = 2e-04
Identities = 25/90 (27%), Positives = 45/90 (49%), Gaps = 6/90 (6%)
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
++++VG L+ +V + L F+ G+VVSV++ G G+V F N Q A A+
Sbjct: 327 STIFVGGLDADVTEEDLMQPFSDFGEVVSVKI------PVGKGCGFVQFANRQSAEEAIG 380
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKSGTAN 121
LN T + ++R+ + +S + N
Sbjct: 381 NLNGTVIGKNTVRLSWGRSPNKQWRSDSGN 410
>At5g41695 putative protein
Length = 548
Score = 89.4 bits (220), Expect = 6e-18
Identities = 97/453 (21%), Positives = 187/453 (40%), Gaps = 97/453 (21%)
Query: 33 SLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALDV 92
+L+V +L + D F +VG+VV V++ +L + +G G+V F + +A AL
Sbjct: 56 TLFVANLPYETKIPNIIDFFKKVGEVVRVQLIVNLKGKL-VGCGFVEFASVNEAEEALQK 114
Query: 93 LNFTPMNNKSI---------------------------RVMYSH---RDPSSRKSGTANI 122
N ++N I R + SH D S +
Sbjct: 115 KNGECLDNNKIFLDVANKKATYLPPKYCIDHKVWDKDYRRLESHPIEEDERPPNSVEEVL 174
Query: 123 FIKNLDKTIDHKALHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNG 182
F+ NL + D F+ G+++S ++ + G+ GYGFV+F + D + A++ NG
Sbjct: 175 FVANLSPQTKISDIFDFFNCVGEVVSIRLMVNHEGKHVGYGFVEFASADETKKALENKNG 234
Query: 183 MLINDKQVFVG--------------HFLRKQ----DRDNVLSK------TKFNNVYVKNL 218
++D ++F+ +LR++ D D + + ++V +L
Sbjct: 235 EYLHDHKIFIDVAKTAPYPPGPNYEDYLRRESLPIDEDETPPEFVEAVGVRKKTLFVAHL 294
Query: 219 SESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDA--AKAVEALNGKKV 276
S + N F G + L+ + G+ FV F +A +A +A+E NG+ +
Sbjct: 295 SRKTEITHIINFFKDVGEVVHVRLILNHTGKHVGCAFVEFGSANEAKMVRALETKNGEYL 354
Query: 277 DD----------------------EEWYVGKAQKKS--EREQELKGRFEQTVKESVVDKF 312
+D + WY +++S E E + + T ++V++F
Sbjct: 355 NDCKIFLEVAKMVPYPPPKYCIHHKVWYEDYLRRESLLIEENETEEGLDDT--PALVEEF 412
Query: 313 --QGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSG 370
+ L++ NL + + F + + +R+I+ + G G G
Sbjct: 413 AVRKKTLFVANLSPRTKISHIIKFFKDVAEVVR-----------VRLIV-NHRGEHVGCG 460
Query: 371 FVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQ 403
FV F++ EA +AL +MNG+ + S+ +++ V +
Sbjct: 461 FVEFASVNEAQKALQKMNGENLRSREIFLDVVE 493
Score = 68.9 bits (167), Expect = 8e-12
Identities = 77/367 (20%), Positives = 144/367 (38%), Gaps = 57/367 (15%)
Query: 82 NPQDAARALDVL------NFTPMNNKSIRVMYSHRDPSSRKSGTANIFIKNLDKTIDHKA 135
NPQ A +L N + K+I D +F+ NL
Sbjct: 11 NPQYGLEANPILKRHKKINKKKVTTKAIAFSLQQNDLLEAAVREKTLFVANLPYETKIPN 70
Query: 136 LHDTFSSFGQIMSCKIATDGSGQSKGYGFVQFEAEDSAQNAIDKLNGMLINDKQVFVGHF 195
+ D F G+++ ++ + G+ G GFV+F + + A+ A+ K NG +++ ++F+
Sbjct: 71 IIDFFKKVGEVVRVQLIVNLKGKLVGCGFVEFASVNEAEEALQKKNGECLDNNKIFLDVA 130
Query: 196 LRKQ--------------------------DRDNVLSKTKFNNVYVKNLSESFTEDDLKN 229
+K + D + ++V NLS D+ +
Sbjct: 131 NKKATYLPPKYCIDHKVWDKDYRRLESHPIEEDERPPNSVEEVLFVANLSPQTKISDIFD 190
Query: 230 EFGAYGTITSAVLMRDADGRSKCFGFVNFENAEDAAKAVEALNGKKVDDEEWYV------ 283
F G + S LM + +G+ +GFV F +A++ KA+E NG+ + D + ++
Sbjct: 191 FFNCVGEVVSIRLMVNHEGKHVGYGFVEFASADETKKALENKNGEYLHDHKIFIDVAKTA 250
Query: 284 ----GKAQKKSEREQELKGRFEQTVKESV-VDKFQGLNLYLKNLDDSITDEKLKEMFSEF 338
G + R + L ++T E V + L++ +L + F +
Sbjct: 251 PYPPGPNYEDYLRRESLPIDEDETPPEFVEAVGVRKKTLFVAHLSRKTEITHIINFFKDV 310
Query: 339 GTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVAFSTPEEAS--RALGEMNGKMIVSKP 396
G + +R+I+ + G G FV F + EA RAL NG+ +
Sbjct: 311 GEV-----------VHVRLIL-NHTGKHVGCAFVEFGSANEAKMVRALETKNGEYLNDCK 358
Query: 397 LYVAVAQ 403
+++ VA+
Sbjct: 359 IFLEVAK 365
Score = 32.0 bits (71), Expect = 1.1
Identities = 14/38 (36%), Positives = 24/38 (62%)
Query: 368 GSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRK 405
G GFV F++ EA AL + NG+ + + +++ VA +K
Sbjct: 96 GCGFVEFASVNEAEEALQKKNGECLDNNKIFLDVANKK 133
>At4g00830 unknown protein
Length = 495
Score = 89.0 bits (219), Expect = 8e-18
Identities = 101/410 (24%), Positives = 171/410 (41%), Gaps = 72/410 (17%)
Query: 120 ANIFIKNLDKTIDHKALHDTFSSFGQIMSCKIATD-GSGQSKGYGFVQFEAEDSAQNAID 178
+ +FI L + + + L D G+I ++ D SG SKGY FV F+ +D AQ AI+
Sbjct: 116 SEVFIGGLPRDVGEEDLRDLCEEIGEIFEVRLMKDRDSGDSKGYAFVAFKTKDVAQKAIE 175
Query: 179 KLNGMLINDKQVFVGHFLRKQDRDNVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGT-I 237
+L+ + F G +R LS+TK N +++ N+ +++TED+ + G +
Sbjct: 176 ELHS------KEFKGKTIRCS-----LSETK-NRLFIGNIPKNWTEDEFRKVIEDVGPGV 223
Query: 238 TSAVLMRDADG--RSKCFGFVNFEN--AEDAAKAVEALNGKKVDDEEWYVGKAQKKSERE 293
+ L++D R++ F FV + N D ++ + K++ V A KS E
Sbjct: 224 ENIELIKDPTNTTRNRGFAFVLYYNNACADYSRQKMIDSNFKLEGNAPTVTWADPKSSPE 283
Query: 294 QELKGRFEQTVKESVVDKFQGLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFT 353
Q LY+KN+ ++ + E+LKE+F G +T
Sbjct: 284 HSAAAA-------------QVKALYVKNIPENTSTEQLKELFQRHGEVT----------- 319
Query: 354 DIRVIMRDPNGVSRGSGFVAFSTPEEASRALGEMNGKMIVSKPLYVAVAQRKEDRRARLQ 413
+++ G R GFV ++ A +A+ + + +PL V +A+ + +R+
Sbjct: 320 --KIVTPPGKGGKRDFGFVHYAERSSALKAVKDTERYEVNGQPLEVVLAKPQAERK-HDP 376
Query: 414 AQFSQMRPVAITPSVAPRMPLY---PPGTPGLG--------QQFMYGQGP-------PAM 455
+ +S P V P + P G G G Q +YG+G M
Sbjct: 377 SSYSYGAAPTPAPFVHPTFGGFAAAPYGAMGAGLGIAGSFSQPMIYGRGAMPTGMQMVPM 436
Query: 456 MPPQAGFGYQQQLVPGMRPGGGPMPSYFVPMVQQGQQGQRPGGRRGGPGQ 505
+ P GY Q PGM P Q+ ++ R G GG G+
Sbjct: 437 LLPDGRVGYVLQ-QPGMPMAAAP--------PQRPRRNDRNNGSSGGSGR 477
Score = 75.9 bits (185), Expect = 7e-14
Identities = 69/273 (25%), Positives = 126/273 (45%), Gaps = 22/273 (8%)
Query: 32 TSLYVGDLEVNVNDSQLYDLFNQVGQVVSVRVCRDLATRRSLGYGYVNFTNPQDAARALD 91
+ +++G L +V + L DL ++G++ VR+ +D + S GY +V F A +A++
Sbjct: 116 SEVFIGGLPRDVGEEDLRDLCEEIGEIFEVRLMKDRDSGDSKGYAFVAFKTKDVAQKAIE 175
Query: 92 VLNFTPMNNKSIRVMYSHRDPSSRKSGTAN-IFIKNLDK--TID--HKALHDTFSSFGQI 146
L+ K+IR S T N +FI N+ K T D K + D I
Sbjct: 176 ELHSKEFKGKTIRCSLSE---------TKNRLFIGNIPKNWTEDEFRKVIEDVGPGVENI 226
Query: 147 MSCKIATDGSGQSKGYGFVQFE----AEDSAQNAIDKLNGMLINDKQVFVGHFLRKQDRD 202
K T+ + +++G+ FV + A+ S Q ID + + V K +
Sbjct: 227 ELIKDPTN-TTRNRGFAFVLYYNNACADYSRQKMID--SNFKLEGNAPTVTWADPKSSPE 283
Query: 203 NVLSKTKFNNVYVKNLSESFTEDDLKNEFGAYGTITSAVLMRDADGRSKCFGFVNFENAE 262
+ + + +YVKN+ E+ + + LK F +G +T ++ G + FGFV++
Sbjct: 284 HSAAAAQVKALYVKNIPENTSTEQLKELFQRHGEVTK-IVTPPGKGGKRDFGFVHYAERS 342
Query: 263 DAAKAVEALNGKKVDDEEWYVGKAQKKSEREQE 295
A KAV+ +V+ + V A+ ++ER+ +
Sbjct: 343 SALKAVKDTERYEVNGQPLEVVLAKPQAERKHD 375
Score = 38.9 bits (89), Expect = 0.009
Identities = 22/92 (23%), Positives = 46/92 (49%), Gaps = 11/92 (11%)
Query: 314 GLNLYLKNLDDSITDEKLKEMFSEFGTITSYKVCTMILFTDIRVIMRDPNGVSRGSGFVA 373
G +++ L + +E L+++ E G I ++R++ +G S+G FVA
Sbjct: 115 GSEVFIGGLPRDVGEEDLRDLCEEIGEIF-----------EVRLMKDRDSGDSKGYAFVA 163
Query: 374 FSTPEEASRALGEMNGKMIVSKPLYVAVAQRK 405
F T + A +A+ E++ K K + ++++ K
Sbjct: 164 FKTKDVAQKAIEELHSKEFKGKTIRCSLSETK 195
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,986,196
Number of Sequences: 26719
Number of extensions: 699049
Number of successful extensions: 3813
Number of sequences better than 10.0: 292
Number of HSP's better than 10.0 without gapping: 182
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 2306
Number of HSP's gapped (non-prelim): 974
length of query: 659
length of database: 11,318,596
effective HSP length: 106
effective length of query: 553
effective length of database: 8,486,382
effective search space: 4692969246
effective search space used: 4692969246
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC148486.10


