

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.6 + phase: 0
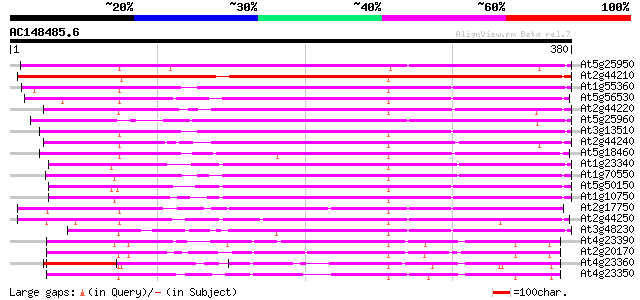
(380 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g25950 putative protein 344 4e-95
At2g44210 unknown protein 333 7e-92
At1g55360 unknown protein 307 7e-84
At5g56530 unknown protein (At5g56530) 304 6e-83
At2g44220 unknown protein 304 6e-83
At5g25960 putative protein 303 8e-83
At3g13510 unknown protein 303 1e-82
At2g44240 unknown protein 295 2e-80
At5g18460 unknown protein 285 2e-77
At1g23340 unknown protein 278 4e-75
At1g70550 unknown protein 275 2e-74
At5g50150 unknown protein 273 1e-73
At1g10750 putative carboxyl-terminal peptidase 272 2e-73
At2g17750 unknown protein 247 7e-66
At2g44250 unknown protein 245 3e-65
At3g48230 putative protein 224 6e-59
At4g23390 unknown protein 205 4e-53
At2g20170 hypothetical protein 201 7e-52
At4g23360 putative protein 194 7e-50
At4g23350 putative protein 184 5e-47
>At5g25950 putative protein
Length = 413
Score = 344 bits (883), Expect = 4e-95
Identities = 184/410 (44%), Positives = 239/410 (57%), Gaps = 39/410 (9%)
Query: 8 IMLLWAFFVAGAISLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLK 67
I ++ F A +I+ KLK N+PA+KTIKSE+GDIIDC+DIYKQ AFDHP LK
Sbjct: 6 IFVILCGFYNEAYGKGSLDIDLKLKALNKPALKTIKSEDGDIIDCIDIYKQHAFDHPALK 65
Query: 68 NHTIQT--------------------------WHKSGRCPKGTVPIRRIQKQDLLRAATL 101
NH IQ W KSG+CP GT+P+RR+ ++D+ RA++
Sbjct: 66 NHKIQMKPSVKFGTKKTTIPNNGSSEHIKSQIWSKSGKCPMGTIPVRRVSREDISRASSP 125
Query: 102 DRFGLK--QSSSFVNSKNTTISNFSKLSGSSNVVSED-HSGVHLATSGSNFIGAEADINV 158
FG K SF+++ NF+ N S + G NF+GA++DIN+
Sbjct: 126 SHFGRKTPHKYSFLDNALQHKGNFNITPAKINEAQPRLRSEAFIVALGFNFVGAQSDINI 185
Query: 159 WNPKVDLPDDSTTAQIWLKAGNGNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTG 218
WNP D +TAQIWL G FES+E GWMVNP ++G+ TRLF WTTD Y TG
Sbjct: 186 WNPPRVEATDYSTAQIWLVGGLSENFESVEGGWMVNPAVFGDSRTRLFISWTTDGYTKTG 245
Query: 219 CFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNID-----EVGHWWLKENHDIPIGY 273
C +L C+GFVQTS ALG + P+SS S TQY + + G+WWL +++ +GY
Sbjct: 246 CINLLCAGFVQTSKKFALGATVEPVSSSSSTQYHITVSIFLDPNSGNWWLTCENNV-LGY 304
Query: 274 WPVELFTSLKHSATLVQWGGQVFS-SQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIK 332
WP LF LKHSAT VQWGG+V S + V K PHT T MGSG A + AC+ N+RIK
Sbjct: 305 WPGTLFNYLKHSATAVQWGGEVHSPNVVLKKPHTTTAMGSGQWASYIWAEACFHTNLRIK 364
Query: 333 DNSLMLKYPESINVASQEPNCYSAF--NDEDVQEPTFYFGGPGQSSPSCP 380
D S+ LKYP+ ++ + E NCYS + EP FYFGGPG++S CP
Sbjct: 365 DYSMQLKYPQFLSEYADEYNCYSTLLHRKTYMSEPHFYFGGPGRNS-RCP 413
>At2g44210 unknown protein
Length = 415
Score = 333 bits (855), Expect = 7e-92
Identities = 166/412 (40%), Positives = 251/412 (60%), Gaps = 46/412 (11%)
Query: 6 VAIMLLWAFFVAGAISLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPV 65
+ +++L V+G S+ +I LK N+PA+K+IKS +GD+IDCV I QPAF HP+
Sbjct: 13 MTVVILAPSVVSGENGFSDLKIRTHLKRLNKPALKSIKSPDGDMIDCVPITDQPAFAHPL 72
Query: 66 LKNHTIQTW------------------------------HKSGRCPKGTVPIRRIQKQDL 95
L NHT+Q W H +G+CPK T+PIRR ++QDL
Sbjct: 73 LINHTVQMWPSLNPESVFSESKVSSKTKNQQSNAIHQLWHVNGKCPKNTIPIRRTRRQDL 132
Query: 96 LRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEAD 155
RA++++ +G+K S K++ N +G + + GV F GA+A
Sbjct: 133 YRASSVENYGMKNQKSIPKPKSSEPPNVLTQNGHQHAIMYVEDGV--------FYGAKAK 184
Query: 156 INVWNPKVDLPDDSTTAQIWLKAGNGN-EFESIEAGWMVNPGLYGNHDTRLFSYWTTDSY 214
INVW P V++P++ + AQIW+ GN N + SIEAGW V+P LYG++ TRLF+YWT+D+Y
Sbjct: 185 INVWKPDVEMPNEFSLAQIWVLGGNFNSDLNSIEAGWQVSPQLYGDNRTRLFTYWTSDAY 244
Query: 215 KSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDI 269
+ TGC++L CSGFVQ + +A+GG I+P+S+ +QY++ I + GHWWL+
Sbjct: 245 QGTGCYNLLCSGFVQINREIAMGGSISPLSNYGNSQYDITILIWKDPKEGHWWLQFGEKY 304
Query: 270 PIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNI 329
IGYWP LF+ L SA++++WGG+V +SQ ++ HT TQMGSG A+E +G A Y +N+
Sbjct: 305 IIGYWPASLFSYLSESASMIEWGGEVVNSQSEEGQHTTTQMGSGRFAEEGWGKASYFKNV 364
Query: 330 RIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQEPT-FYFGGPGQSSPSCP 380
++ D S L+ PE++ V + + NCY+ + + FY+GGPG+ +P+CP
Sbjct: 365 QVVDGSNELRNPENLQVFTDQENCYNVKSGNGGSWGSYFYYGGPGR-NPNCP 415
>At1g55360 unknown protein
Length = 422
Score = 307 bits (786), Expect = 7e-84
Identities = 157/415 (37%), Positives = 241/415 (57%), Gaps = 55/415 (13%)
Query: 9 MLLWAFF-----VAGAISLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDH 63
+ LW FF +S + E++ L N+PAVK+I+S +GD+IDCV I KQPAFDH
Sbjct: 20 LCLWGFFSLSYAARSGVSKQKFEVKKHLNRLNKPAVKSIQSSDGDVIDCVPISKQPAFDH 79
Query: 64 PVLKNHTIQT------------------------------WHKSGRCPKGTVPIRRIQKQ 93
P LK+H IQ WH+ G+C +GT+P+RR ++
Sbjct: 80 PFLKDHKIQMKPNYHPEGLFDDNKVSAPKSNEKEGHIPQLWHRYGKCSEGTIPMRRTKED 139
Query: 94 DLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSED-HSGVHLATSGSNFIGA 152
D+LRA+++ R+G K+ S K S +++++ H G + GA
Sbjct: 140 DVLRASSVKRYGKKKRRSVPLPK----------SAEPDLINQSGHQHAIAYVEGDKYYGA 189
Query: 153 EADINVWNPKVDLPDDSTTAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTT 211
+A INVW PK+ ++ + +QIWL G+ G + SIEAGW V+P LYG+++TRLF+YWT+
Sbjct: 190 KATINVWEPKIQQQNEFSLSQIWLLGGSFGQDLNSIEAGWQVSPDLYGDNNTRLFTYWTS 249
Query: 212 DSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKEN 266
D+Y++TGC++L CSGF+Q ++ +A+G I+P+S +QY+++I + GHWW++
Sbjct: 250 DAYQATGCYNLLCSGFIQINSDIAMGASISPVSGYRNSQYDISILIWKDPKEGHWWMQFG 309
Query: 267 HDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYM 326
+ +GYWP LF+ L SA++++WGG+V +SQ HT TQMGSG +E + A Y
Sbjct: 310 NGYVLGYWPSFLFSYLTESASMIEWGGEVVNSQ-SDGQHTSTQMGSGKFPEEGFSKASYF 368
Query: 327 RNIRIKDNSLMLKYPESINVASQEPNCYSA-FNDEDVQEPTFYFGGPGQSSPSCP 380
RNI++ D S LK P+ + +++ NCY D FY+GGPG++ CP
Sbjct: 369 RNIQVVDGSNNLKAPKGLGTFTEQSNCYDVQTGSNDDWGHYFYYGGPGKNQ-KCP 422
>At5g56530 unknown protein (At5g56530)
Length = 420
Score = 304 bits (778), Expect = 6e-83
Identities = 160/407 (39%), Positives = 242/407 (59%), Gaps = 49/407 (12%)
Query: 11 LWAFFVAGAISLSETEIEAKLKLH--NRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKN 68
L + AG +S+S E L+ N+PAVK+I+S +GDIIDCV I KQPAFDHP LK+
Sbjct: 24 LMSLTCAGRLSVSRQNFEVHKHLNRLNKPAVKSIQSPDGDIIDCVHISKQPAFDHPFLKD 83
Query: 69 HTIQT-----------------------------WHKSGRCPKGTVPIRRIQKQDLLRAA 99
H IQ WH++G C +GT+P+RR +K+D+LRA+
Sbjct: 84 HKIQMGPSYTPESLFGESKVSEKPKESVNPITQLWHQNGVCSEGTIPVRRTKKEDVLRAS 143
Query: 100 TLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVW 159
++ R+G K+ S V + + SG + ++ G F GA+A INVW
Sbjct: 144 SVKRYGKKKHLS-VPLPRSADPDLINQSGHQHAIAY--------VEGGKFYGAKATINVW 194
Query: 160 NPKVDLPDDSTTAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTG 218
PKV ++ + +Q+W+ G+ G + SIEAGW V+P LYG+++TRLF+YWT+D+Y++TG
Sbjct: 195 EPKVQSSNEFSLSQLWILGGSFGQDLNSIEAGWQVSPDLYGDNNTRLFTYWTSDAYQATG 254
Query: 219 CFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGY 273
C++L CSGF+Q ++ +A+G I+P+S QY+++I + GHWW++ +GY
Sbjct: 255 CYNLLCSGFIQINSQIAMGASISPVSGFHNPQYDISITIWKDPKEGHWWMQFGDGYVLGY 314
Query: 274 WPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKD 333
WP LF+ L SA++V+WGG+V + + + HT TQMGSG DE + A Y RNI++ D
Sbjct: 315 WPSFLFSYLADSASIVEWGGEVVNME-EDGHHTTTQMGSGQFPDEGFTKASYFRNIQVVD 373
Query: 334 NSLMLKYPESINVASQEPNCYSA-FNDEDVQEPTFYFGGPGQSSPSC 379
+S LK P+ +N +++ NCY D FY+GGPG+ +P+C
Sbjct: 374 SSNNLKEPKGLNTFTEKSNCYDVEVGKNDDWGHYFYYGGPGR-NPNC 419
>At2g44220 unknown protein
Length = 393
Score = 304 bits (778), Expect = 6e-83
Identities = 160/387 (41%), Positives = 217/387 (55%), Gaps = 45/387 (11%)
Query: 24 ETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ----------- 72
E ++ LK N+PA+K+IKSE+GD+IDCV I QPAFDH +LKNHTIQ
Sbjct: 22 EIKVLRHLKRFNKPALKSIKSEDGDVIDCVPITNQPAFDHHLLKNHTIQMRPSFYPVSDS 81
Query: 73 ------------TWHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTI 120
WHK+G CPK TVPIRR +K+DLLR ++ FG K S +
Sbjct: 82 TYTKREAKAVTQVWHKAGECPKNTVPIRRTKKEDLLRPKSIRSFGRKSHQSIPRT----- 136
Query: 121 SNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN 180
+ F G H + F G E IN+W P V +P + + AQ W+ +GN
Sbjct: 137 TTFDPTLG--------HQYALMGVRNGKFYGTEVAINLWKPYVQIPKEFSLAQTWVVSGN 188
Query: 181 GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGI 240
G+ +IEAGW V P LY +++ R F YWT D Y+ TGC++L CSGFVQTSN +GG I
Sbjct: 189 GSSLNTIEAGWQVYPELYDDNNPRFFVYWTRDGYRKTGCYNLLCSGFVQTSNRYTVGGSI 248
Query: 241 NPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQV 295
+S GTQY+L++ + G+WWL+ N IGYWP LF SL AT V+WGG++
Sbjct: 249 TTMSRYRGTQYDLSVLIWKDQKTGNWWLRVNEKDVIGYWPGSLFNSLGREATRVEWGGEI 308
Query: 296 FSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYS 355
+S+ HT T MGSGH ADE + A Y RN++I D + L+ P+ + + + NCY+
Sbjct: 309 INSKT-GGRHTTTDMGSGHFADEGFKKASYFRNLKIVDGTNTLREPQGLYFFADKHNCYN 367
Query: 356 --AFNDEDVQEPTFYFGGPGQSSPSCP 380
N F++GGPG+ + CP
Sbjct: 368 VKTGNGGTSWGAHFFYGGPGR-NVKCP 393
>At5g25960 putative protein
Length = 352
Score = 303 bits (777), Expect = 8e-83
Identities = 170/368 (46%), Positives = 215/368 (58%), Gaps = 30/368 (8%)
Query: 15 FVAGAISLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQTW 74
F A +I+ KLK N+P++KTIKSE+GDIIDC+DIYKQ AFDHP L+NH IQ
Sbjct: 13 FYNEAYGKGSLDIDMKLKSLNKPSLKTIKSEDGDIIDCIDIYKQHAFDHPALRNHKIQM- 71
Query: 75 HKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVS 134
K +V FG K+++ N + I++ S S N
Sbjct: 72 -------KPSVD-----------------FGTKKTTIPNNGSSEQITS-QIWSKSGNCPK 106
Query: 135 EDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFESIEAGWMVN 194
L G NFIGA++DINVWNP D ++AQIWL G + FESIEAGW VN
Sbjct: 107 GTIPEAVLMALGYNFIGAQSDINVWNPPRVQASDYSSAQIWLLGGLSDTFESIEAGWAVN 166
Query: 195 PGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELN 254
P ++G+ TRLF+YWT D Y TGC +L C+GFVQT+ ALG I P+S+ S Q+ +
Sbjct: 167 PSVFGDSRTRLFTYWTKDGYSKTGCVNLLCAGFVQTTTKFALGAAIEPVSTTSQKQHFIT 226
Query: 255 IDEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGH 314
G+WWL +++ IGYWP LF LKHSAT VQ GG+V S V K PHT+T MGSG
Sbjct: 227 DTNSGNWWLTCANNV-IGYWPGTLFAYLKHSATAVQCGGEVHSPNVGKKPHTRTSMGSGQ 285
Query: 315 LADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSA--FNDEDVQEPTFYFGGP 372
A + ACY NIRIKD SL +KYP+ ++ + E CYS + EP FYFGGP
Sbjct: 286 WASYLWAQACYHSNIRIKDYSLQIKYPKYLSEYADEYECYSTKLHRKTYMSEPYFYFGGP 345
Query: 373 GQSSPSCP 380
GQ+S CP
Sbjct: 346 GQNS-RCP 352
>At3g13510 unknown protein
Length = 419
Score = 303 bits (776), Expect = 1e-82
Identities = 153/397 (38%), Positives = 238/397 (59%), Gaps = 49/397 (12%)
Query: 21 SLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQT------- 73
S + E++ L N+P VKTI+S +GDIIDC+ I KQPAFDHP LK+H IQ
Sbjct: 35 SRQKFEVKKHLNRLNKPPVKTIQSPDGDIIDCIPISKQPAFDHPFLKDHKIQMRPSYHPE 94
Query: 74 ----------------------WHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSS 111
WH+ G+C +GT+P+RR ++ D+LRA+++ R+G K+ S
Sbjct: 95 GLFDDNKVSAEPKGKETHIPQLWHRYGKCTEGTIPMRRTREDDVLRASSVKRYGKKKHRS 154
Query: 112 FVNSKNTTISNFSKLSGSSNVVSED-HSGVHLATSGSNFIGAEADINVWNPKVDLPDDST 170
K S ++++++ H G + GA+A +NVW PK+ ++ +
Sbjct: 155 VPIPK----------SAEPDLINQNGHQHAIAYVEGDKYYGAKATLNVWEPKIQNTNEFS 204
Query: 171 TAQIWLKAGN-GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQ 229
+QIWL G+ G + SIEAGW V+P LYG+++TRLF+YWT+D+Y++TGC++L CSGF+Q
Sbjct: 205 LSQIWLLGGSFGQDLNSIEAGWQVSPDLYGDNNTRLFTYWTSDAYQATGCYNLLCSGFIQ 264
Query: 230 TSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKH 284
++ +A+G I+P+S +QY+++I + GHWW++ + +GYWP LF+ L
Sbjct: 265 INSDIAMGASISPVSGYRNSQYDISILIWKDPKEGHWWMQFGNGYVLGYWPSFLFSYLTE 324
Query: 285 SATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESI 344
SA++++WGG+V +SQ + HT TQMGSGH +E + A Y RNI++ D S LK P+ +
Sbjct: 325 SASMIEWGGEVVNSQ-SEGHHTWTQMGSGHFPEEGFSKASYFRNIQVVDGSNNLKAPKGL 383
Query: 345 NVASQEPNCYSA-FNDEDVQEPTFYFGGPGQSSPSCP 380
+++ NCY D FY+GGPG++ +CP
Sbjct: 384 GTFTEKSNCYDVQTGSNDDWGHYFYYGGPGKNK-NCP 419
>At2g44240 unknown protein
Length = 402
Score = 295 bits (756), Expect = 2e-80
Identities = 158/388 (40%), Positives = 219/388 (55%), Gaps = 48/388 (12%)
Query: 24 ETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQT---------- 73
E +++ LK N+PA+K+IKSE+GDIIDCV I QPAFDHP+LKNHTIQ
Sbjct: 32 EIKVQRFLKQLNKPALKSIKSEDGDIIDCVLITSQPAFDHPLLKNHTIQVKPSFIPEGEG 91
Query: 74 --------------WHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTT 119
W K G CP+ T+PIRR +K+++LRA +L+ FG K++ ++ ++T+
Sbjct: 92 DSTYTKKETKATQVWQKYGECPENTIPIRRTKKEEILRAKSLESFG-KKNHQYI-PEDTS 149
Query: 120 ISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAG 179
N+ H + F G +A INVW P V P + + +Q W+ +G
Sbjct: 150 SPNYH------------HEYAFMGVRNGKFYGTKASINVWKPDVATPSEFSLSQTWIVSG 197
Query: 180 NGNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGG 239
+G +IEAGW V PG+YGN+D RLF YWT+D Y+ TGC++L C GFVQT+N +GG
Sbjct: 198 DGTSRNTIEAGWQVYPGMYGNNDPRLFVYWTSDGYQKTGCYNLVCGGFVQTTNQYTVGGS 257
Query: 240 INPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQ 294
S G Q LN+ + G+WWLK N + IGYWP LF SL A V+WGG+
Sbjct: 258 YVTASQYDGAQLVLNLLIWKDPKTGNWWLKINDNDVIGYWPGSLFNSLGDGAIKVEWGGE 317
Query: 295 VFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCY 354
+F+ + HT T MGSGH A+E A Y++NI I D + L+ P+ + + NCY
Sbjct: 318 IFAPTSDR--HTTTDMGSGHFAEEGIKKASYVKNIMIVDGTNALREPQGLYSYADNRNCY 375
Query: 355 SAF--NDEDVQEPTFYFGGPGQSSPSCP 380
S N F++GGPGQ + CP
Sbjct: 376 SVVPGNAGTSFGTHFFYGGPGQ-NVKCP 402
>At5g18460 unknown protein
Length = 430
Score = 285 bits (730), Expect = 2e-77
Identities = 154/396 (38%), Positives = 228/396 (56%), Gaps = 47/396 (11%)
Query: 21 SLSETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQT------- 73
SL I+ L N+ V TI+S +GD+IDCV KQPA DHP+LK+H IQ
Sbjct: 44 SLRLARIQKHLNKINKSPVFTIQSPDGDVIDCVPKRKQPALDHPLLKHHKIQKAPKKMPK 103
Query: 74 ---------------------WHKSG-RCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSS 111
WH +G RCPKGTVPIRR D+LRA +L FG K+ S
Sbjct: 104 MKGKDDDVKEAENVLEGAWQMWHVNGTRCPKGTVPIRRNTMNDVLRAKSLFDFGKKRRSI 163
Query: 112 FVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTT 171
+++ + ++ + +H+ + S S GA+A INVW+PK++ ++ +
Sbjct: 164 YLDQR-------TEKPDALGTNGHEHA-IAYTESSSEIYGAKATINVWDPKIEEVNEFSL 215
Query: 172 AQIWLKAGN--GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQ 229
+QIW+ +G+ G + SIEAGW V+P LYG++ RLF+YWT+DSY++TGC++L CSGF+Q
Sbjct: 216 SQIWILSGSFVGPDLNSIEAGWQVSPELYGDNRPRLFTYWTSDSYQATGCYNLLCSGFIQ 275
Query: 230 TSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKH 284
T+N +A+G I+P+S+ G Q+++ I ++G+WW+ +GYWP ELFT L
Sbjct: 276 TNNKIAIGAAISPLSTFKGNQFDITILIWKDPKMGNWWMGLGDSTLVGYWPAELFTHLAD 335
Query: 285 SATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESI 344
AT V+WGG+V +++ HT TQMGSGH DE +G A Y RN+ + D+ L +
Sbjct: 336 HATTVEWGGEVVNTRA-SGRHTTTQMGSGHFPDEGFGKASYFRNLEVVDSDNSLVPVHDV 394
Query: 345 NVASQEPNCYSAFNDEDVQEPT-FYFGGPGQSSPSC 379
+ ++ CY + + T FY+GGPG +P C
Sbjct: 395 KILAENTECYDIKSSYSNEWGTYFYYGGPG-FNPRC 429
>At1g23340 unknown protein
Length = 409
Score = 278 bits (711), Expect = 4e-75
Identities = 150/382 (39%), Positives = 225/382 (58%), Gaps = 47/382 (12%)
Query: 27 IEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLK------------------- 67
I +L+ N+PA+KTI S +GD IDCV + QPAFDHP+L+
Sbjct: 47 IRKQLQKINKPAIKTIHSSDGDTIDCVPSHHQPAFDHPLLQGQRPMDPPEMPIGYSQENE 106
Query: 68 -NHTIQTWHKSGR-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
+ Q W G CP+GT+PIRR +QD+LRA ++ RFG K +
Sbjct: 107 SHENFQLWSLYGESCPEGTIPIRRTTEQDMLRANSVRRFGRK---------------IRR 151
Query: 126 LSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN-GNEF 184
+ S+ +H+ ++ SGS + GA+A INVW P+V + + +QIW+ AG+ +
Sbjct: 152 VRRDSSSNGHEHAVGYV--SGSQYYGAKASINVWTPRVISQYEFSLSQIWIIAGSFAGDL 209
Query: 185 ESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPIS 244
+IEAGW ++P LYG+ + R F+YWT+D+Y++TGC++L CSGFVQT+N +A+G I+P+S
Sbjct: 210 NTIEAGWQISPELYGDTNPRFFTYWTSDAYQATGCYNLLCSGFVQTNNRIAIGAAISPVS 269
Query: 245 SDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQ 299
S G Q+++++ + GHWWL+ +GYWPV LFT L+ +VQ+GG++ +++
Sbjct: 270 SYKGGQFDISLLIWKDPKHGHWWLQFGSGTLVGYWPVSLFTHLREHGNMVQFGGEIVNTR 329
Query: 300 VKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFND 359
S HT TQMGSGH A E +G A Y RN+++ D L ++ V + PNCY
Sbjct: 330 PGGS-HTSTQMGSGHFAGEGFGKASYFRNLQMVDWDNTLIPISNLKVLADHPNCYDIRGG 388
Query: 360 ED-VQEPTFYFGGPGQSSPSCP 380
+ V FY+GGPG++S CP
Sbjct: 389 VNRVWGNFFYYGGPGKNS-KCP 409
>At1g70550 unknown protein
Length = 410
Score = 275 bits (704), Expect = 2e-74
Identities = 152/384 (39%), Positives = 224/384 (57%), Gaps = 47/384 (12%)
Query: 25 TEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNH--------------- 69
T I +L N+PAVKTI+S +GD IDCV ++QPAFDHP+L+
Sbjct: 46 TLIRQELDKINKPAVKTIQSSDGDKIDCVSTHQQPAFDHPLLQGQKPLDPPEIPKGYSED 105
Query: 70 -----TIQTWHKSGR-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNF 123
Q W SG CP+GT+PIRR +QD+LRA+++ RFG K +S N
Sbjct: 106 DGSYENSQLWSLSGESCPEGTIPIRRTTEQDMLRASSVQRFGRKIRRVKRDSTN------ 159
Query: 124 SKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN-GN 182
+G + V +G + GA+A INVW+P+V + + +QIW+ AG+ +
Sbjct: 160 ---NGHEHAVGY--------VTGRQYYGAKASINVWSPRVTSQYEFSLSQIWVIAGSFTH 208
Query: 183 EFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINP 242
+ +IEAGW ++P LYG+ R F+YWT+D+Y++TGC++L CSGFVQT+ +A+G I+P
Sbjct: 209 DLNTIEAGWQISPELYGDTYPRFFTYWTSDAYRTTGCYNLLCSGFVQTNRRIAIGAAISP 268
Query: 243 ISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFS 297
SS G Q+++++ + GHWWL+ +GYWP LFT LK ++VQ+GG++ +
Sbjct: 269 RSSYKGGQFDISLLIWKDPKHGHWWLQFGSGALVGYWPAFLFTHLKQHGSMVQFGGEIVN 328
Query: 298 SQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAF 357
++ S HT TQMGSGH A E +G A Y RN++I D L ++ + + PNCY
Sbjct: 329 NRPGGS-HTTTQMGSGHFAGEGFGKASYFRNLQIVDWDNTLIPASNLKILADHPNCYDIR 387
Query: 358 NDED-VQEPTFYFGGPGQSSPSCP 380
+ V FY+GGPG+ +P CP
Sbjct: 388 GGTNRVWGNYFYYGGPGK-NPRCP 410
>At5g50150 unknown protein
Length = 420
Score = 273 bits (698), Expect = 1e-73
Identities = 151/382 (39%), Positives = 228/382 (59%), Gaps = 46/382 (12%)
Query: 27 IEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLK--------------NHTI- 71
+EA L N+P++KTI S +GD+I+CV + QPAFDHP L+ N T
Sbjct: 57 VEAYLSKINKPSIKTIHSPDGDVIECVPSHLQPAFDHPQLQGQKPLDSPYRPSKGNETTY 116
Query: 72 -----QTWHKSGR-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
Q W SG CP G++PIR+ K D+LRA ++ RFG K
Sbjct: 117 EESFNQLWSMSGESCPIGSIPIRKTTKNDVLRANSVRRFGRKLRRP-------------- 162
Query: 126 LSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN-GNEF 184
+ S+ +H+ V + +G + GA+A INVW P+V + + +QIWL +G+ G++
Sbjct: 163 IRRDSSGGGHEHAVVFV--NGEQYYGAKASINVWAPRVTDAYEFSLSQIWLISGSFGHDL 220
Query: 185 ESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPIS 244
+IEAGW V+P LYG++ R F+YWTTD+Y++TGC++L CSGFVQT+N +A+G I+P S
Sbjct: 221 NTIEAGWQVSPELYGDNYPRFFTYWTTDAYQATGCYNLLCSGFVQTNNKIAIGAAISPRS 280
Query: 245 SDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQ 299
S +G Q+++ + + GHWWL+ + + +GYWP LF+ L+ A++VQ+GG+V +S+
Sbjct: 281 SYNGRQFDIGLMIWKDPKHGHWWLELGNGLLVGYWPAFLFSHLRSHASMVQFGGEVVNSR 340
Query: 300 VKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFND 359
HT TQMGSGH ADE + A Y RN+++ D L ++++V + P CY
Sbjct: 341 -SSGAHTGTQMGSGHFADEGFEKAAYFRNLQVVDWDNNLLPLKNLHVLADHPACYDIRQG 399
Query: 360 E-DVQEPTFYFGGPGQSSPSCP 380
+ +V FY+GGPG+ +P CP
Sbjct: 400 KNNVWGTYFYYGGPGR-NPRCP 420
>At1g10750 putative carboxyl-terminal peptidase
Length = 467
Score = 272 bits (696), Expect = 2e-73
Identities = 150/382 (39%), Positives = 223/382 (58%), Gaps = 47/382 (12%)
Query: 27 IEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNH----------------- 69
I L+ N+P++KTI S +GDIIDCV ++ QPAFDHP L+
Sbjct: 105 INQHLRKINKPSIKTIHSPDGDIIDCVLLHHQPAFDHPSLRGQKPLDPPERPRGHNRRGL 164
Query: 70 ---TIQTWHKSGR-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
+ Q W G CP+GTVPIRR +++D+LRA ++ FG K + + T SN
Sbjct: 165 RPKSFQLWGMEGETCPEGTVPIRRTKEEDILRANSVSSFGKKLR----HYRRDTSSN--- 217
Query: 126 LSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN-GNEF 184
+H+ ++ SG + GA+A INVW P+V + + +QIW+ +G+ GN+
Sbjct: 218 --------GHEHAVGYV--SGEKYYGAKASINVWAPQVQNQYEFSLSQIWIISGSFGNDL 267
Query: 185 ESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPIS 244
+IEAGW V+P LYG++ R F+YWT D+Y++TGC++L CSGFVQT++ +A+G I+P S
Sbjct: 268 NTIEAGWQVSPELYGDNYPRFFTYWTNDAYQATGCYNLLCSGFVQTNSEIAIGAAISPSS 327
Query: 245 SDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQ 299
S G Q+++ + + G+WWL+ I +GYWP LFT LK A++VQ+GG++ +S
Sbjct: 328 SYKGGQFDITLLIWKDPKHGNWWLEFGSGILVGYWPSFLFTHLKEHASMVQYGGEIVNSS 387
Query: 300 VKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFND 359
HT TQMGSGH A+E + + Y RNI++ D L ++ V + PNCY
Sbjct: 388 -PFGAHTSTQMGSGHFAEEGFTKSSYFRNIQVVDWDNNLVPSPNLRVLADHPNCYDIQGG 446
Query: 360 ED-VQEPTFYFGGPGQSSPSCP 380
+ FY+GGPG+ +P CP
Sbjct: 447 SNRAWGSYFYYGGPGK-NPKCP 467
>At2g17750 unknown protein
Length = 396
Score = 247 bits (631), Expect = 7e-66
Identities = 145/407 (35%), Positives = 214/407 (51%), Gaps = 54/407 (13%)
Query: 6 VAIMLLWAFFVAGAISLSE------TEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQP 59
+A +L VA +S SE ++E LK N+PA+K+IKS +GDIIDCV + P
Sbjct: 3 IARFVLSILIVAAIVSGSEFSDTRDAKVEKILKKLNKPALKSIKSPDGDIIDCVHMNNHP 62
Query: 60 AFDHPVLKNHTIQT-------------------------WHKSGRCPKGTVPIRRIQKQD 94
+DHP+ KN+TIQ W +G+CPK ++PIRR ++++
Sbjct: 63 IYDHPLFKNYTIQMKPSSYPKGKNNESSDKEKQSVVTQLWTVNGKCPKNSIPIRRTRRKE 122
Query: 95 LLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHLATSGSNFIGAEA 154
+LR + R+ + KN I N K S S+++ H + G F GA A
Sbjct: 123 ILRTEYMQRY---------DKKNPNIINHPKASTSNSI----HEYAQIQAKGK-FHGAHA 168
Query: 155 DINVWNPKVDLPDDSTTAQIWLKAGNGNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSY 214
DINVW P V P + + AQ+W+ AG +E S+EAGW V YG+ + R F +WT D Y
Sbjct: 169 DINVWKPFVQTPKEFSLAQMWVMAGPFSEVNSVEAGWQVYQDRYGDDNPRYFIFWTADGY 228
Query: 215 KSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----DEVGHWWLKENHDI 269
S GC++L C GFV S ALG ++ +S+ G QY ++ G+WWLK +
Sbjct: 229 HS-GCYNLDCQGFVPVSQKFALGAAVSNVSTFDGQQYHISTTIWKDPNSGNWWLKFGDEF 287
Query: 270 PIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNI 329
+GYWP LF LK AT +QWGG++ + + + HT T+MGSGH A+ Y A Y +++
Sbjct: 288 -VGYWPSILFNHLKDGATEIQWGGEIINFK-DGALHTTTRMGSGHFAESGYQKASYFKDV 345
Query: 330 RIKDNSLMLKYPESINVASQEPNCYSAFND-EDVQEPTFYFGGPGQS 375
I D + P+ + +CY+ + V FY+GGPG++
Sbjct: 346 EIIDERDIHSSPKEGYSYMTQESCYNIRSGYAKVWGVYFYYGGPGRN 392
>At2g44250 unknown protein
Length = 427
Score = 245 bits (626), Expect = 3e-65
Identities = 147/426 (34%), Positives = 224/426 (52%), Gaps = 65/426 (15%)
Query: 6 VAIMLLWAFFVAGAISLS---ETEIEAKLKLHNRPAVKTIK------------------S 44
V I L+ +AG I++S E +IE +L N+PAVK+I S
Sbjct: 14 VIIALVVEITLAGKITVSDIHEQKIEQRLNQLNKPAVKSIHVFATTFNYPWFAIPTIFLS 73
Query: 45 ENGDIIDCVDIYKQPAFDHPVLKNHTIQT----------------------WHKSGRCPK 82
+GDIIDCV IY QPAFDHP+LKNHTIQ W G CPK
Sbjct: 74 PDGDIIDCVWIYHQPAFDHPLLKNHTIQMRPKSDSTRDKTGGNKTDIIHQLWRTKGECPK 133
Query: 83 GTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDHSGVHL 142
T+PIRR + DLLR+ ++ G K TIS + +H+ V+L
Sbjct: 134 NTIPIRRRTRDDLLRSDSIKTHGRKNPP--------TISPTTYHLPDDQTEVHEHASVYL 185
Query: 143 ATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNE-FESIEAGWMVNPGLYGNH 201
+ G+++ I++W P V++ + S AQ W+ G+ + ++E+GW + +YG++
Sbjct: 186 --DYGEYHGSKSRISIWRPDVNMTEFSL-AQTWVVGGDWDTVLNTVESGWQILHSMYGDN 242
Query: 202 DTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI-----D 256
+TRLF++WT+DSY C++L C GFVQ + +ALG +N IS+ +G QY+ +
Sbjct: 243 NTRLFAFWTSDSYGDNSCYNLDCPGFVQVNKDIALGAALNTISTYNGNQYDFLLTIEKEQ 302
Query: 257 EVGHWWLKENHDIPIGYWPVELFTSLKHSATLVQWGGQVFSSQVKKSPHTKTQMGSGHLA 316
+ G WWLK + + +GYWP L L SA ++ WGG++ ++ HT TQMGSGH A
Sbjct: 303 DTGLWWLKFDTHL-VGYWPSFLVPKLADSARMIAWGGEIVYDASGQNEHTLTQMGSGHFA 361
Query: 317 DEKYGHACYMRNIRI--KDNSLMLKYPESINVASQEPNCYSAFNDEDVQEPTF-YFGGPG 373
+E + A Y+ NI K N + +P+++ + P CY+ + T+ ++GGPG
Sbjct: 362 EEGFKKAAYINNIEYIDKSNHPIKPFPQNLEASVTRPECYNLKVGSSRRWGTYIFYGGPG 421
Query: 374 QSSPSC 379
+ +P C
Sbjct: 422 R-NPQC 426
>At3g48230 putative protein
Length = 367
Score = 224 bits (571), Expect = 6e-59
Identities = 135/373 (36%), Positives = 200/373 (53%), Gaps = 51/373 (13%)
Query: 40 KTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ---------------------TWHKSG 78
KT++S +GDI+DC D+ QP+ DHP+L+NH IQ W+++G
Sbjct: 14 KTVQSSDGDIVDCFDVRDQPSLDHPLLQNHEIQGAPATGLPYMINKAGKRRVWQVWNQNG 73
Query: 79 R-CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSKLSGSSNVVSEDH 137
CP T+PIRR RF K + ++ T ++ G + E
Sbjct: 74 TSCPDETIPIRR-------SVVGAKRFKKKHWTDVRVNRRTV--PYAAEEGHEYAIGEV- 123
Query: 138 SGVHLATSGSNFIGAEADINVWNPKVDL-PDDSTTAQIWLKAG--NGNEFESIEAGWMVN 194
V+L G A +NVWNP V+ ++ + +QIWL AG N ++ ++EAGW V
Sbjct: 124 --VYLR----GIYGTVATMNVWNPSVEHGTNEFSLSQIWLVAGHYNDSDLNTVEAGWQVF 177
Query: 195 PGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELN 254
P Y + RLF YWT D+Y+ TGC +L C GFVQ ++ A+G +P SS G+QY++
Sbjct: 178 PDHYHDSQPRLFVYWTKDTYQKTGCLNLECPGFVQVTSEFAIGSAFSPTSSYGGSQYDIT 237
Query: 255 I-----DEVGHWWLKENHDIPIGYWPVELFTSLKHS-ATLVQWGGQVFSSQVKKSPHTKT 308
+ + G+WWL + + IGYWP LFT L H ATLVQWGG++ +S+ HT T
Sbjct: 238 MYIWKDTKDGNWWLSIDSSV-IGYWPARLFTHLAHGPATLVQWGGEIVNSR-SYGQHTTT 295
Query: 309 QMGSGHLADEKYGHACYMRNIRIKDNSLMLKYPESINVASQEPNCYSAFNDEDVQEPT-F 367
QMGSGH A+E +G A RN++I D ++ + + ++ P CY+A + + F
Sbjct: 296 QMGSGHFAEEGFGKAGSFRNLKIIDYLSYMQPVQEFILQTKNPTCYTAIKGYSEEWGSHF 355
Query: 368 YFGGPGQSSPSCP 380
Y+GGPG ++ CP
Sbjct: 356 YYGGPGYNA-LCP 367
>At4g23390 unknown protein
Length = 401
Score = 205 bits (521), Expect = 4e-53
Identities = 141/386 (36%), Positives = 191/386 (48%), Gaps = 62/386 (16%)
Query: 26 EIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNH-------TIQTWHKSG 78
E++ L N+PAVK+ ++E+G I DC+DI KQ AFDHP+LKNH TI W K
Sbjct: 36 EMKKLLTYLNKPAVKSFQTEHGYIFDCIDIQKQLAFDHPLLKNHSIKLKPTTIPKWTKDN 95
Query: 79 R----------------CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISN 122
CP GTV ++RI +DL+RA L V++K+ I
Sbjct: 96 NASHKTSSLPFRQDDISCPVGTVIVKRIILEDLIRAQRLQSLRFNYPGQ-VSAKDKKI-- 152
Query: 123 FSKLSGSSNVVSEDHSGVHLATSG--SNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGN 180
D +G H AT + GA+ +INVWNP V PD + A + +
Sbjct: 153 -------------DLTGHHFATISYKDDHYGAKGNINVWNPNVS-PDQFSLAAMAVSGNK 198
Query: 181 GNEFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGI 240
G F+SI AGW+V PGL N+ + LF+YWT D T C++ GFV S A+G
Sbjct: 199 G--FQSISAGWIVYPGLNQNNQSHLFTYWTADGNNKTHCYNTLRPGFVHVSTKYAIGMLA 256
Query: 241 NPISSDSGTQYELNI----DEVGH-WWLKENHDIPIGYWPVELFT--SLKHSATLVQWGG 293
P+S G QY+L + D V WW N++ PIGYWP LFT L A+ V WGG
Sbjct: 257 QPVSIYDGQQYQLEVSIYQDHVTRDWWFVLNNE-PIGYWPKSLFTRQGLADGASAVFWGG 315
Query: 294 QVFSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIR-IKDNSLMLKYP--ESINVASQE 350
+V+SS +KSP MGSGH E + A Y+ ++ I D + + P ++ +
Sbjct: 316 EVYSSVKEKSP----SMGSGHFPQEGFKKAAYVNGLKIITDITKEVSSPLASALKTFASS 371
Query: 351 PNCYSAFNDEDVQE---PTFYFGGPG 373
PNCY+ V E FGGPG
Sbjct: 372 PNCYNVQKILGVGEFWSRAILFGGPG 397
>At2g20170 hypothetical protein
Length = 401
Score = 201 bits (510), Expect = 7e-52
Identities = 128/384 (33%), Positives = 204/384 (52%), Gaps = 58/384 (15%)
Query: 26 EIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ-------TWHKSG 78
E++ L N+PA+K+ ++++G I+DC+DI KQ AFDHP+LKNH+IQ W +
Sbjct: 35 ELKKLLNHINKPAIKSFQTKHGYILDCIDIQKQLAFDHPLLKNHSIQLKPTIIPKWTRDK 94
Query: 79 R----------------CPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISN 122
CP GTV I+R +DL++ L G+K + SK+ +
Sbjct: 95 NTQKSSSLPFRQDEDISCPHGTVIIKRTTLEDLIQIQRLKYLGVK----YTTSKD---KD 147
Query: 123 FSKLSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGN 182
F ++G ++E + + GA +IN+W+P V+ PD + A I+++ G +
Sbjct: 148 FLNMTGRHFAIAEYYRDNY---------GATGNINLWDPPVN-PDQFSLASIYVENGFRD 197
Query: 183 EFESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINP 242
+SI AGW+V+P L N ++ LF+YWT D ++ TGC++ C GFVQ S+ +ALG P
Sbjct: 198 SLQSISAGWIVSPKLNQN-NSGLFTYWTADGHEKTGCYNTVCPGFVQVSSKLALGTLARP 256
Query: 243 ISSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELF--TSLKHSATLVQWGGQV 295
S+ G QY L + G+WW ++ PIGYWP LF L + A+ V WGG+V
Sbjct: 257 TSTYDGEQYYLQAIIYQDNITGNWWFLIKNE-PIGYWPKSLFHVQGLAYGASRVFWGGEV 315
Query: 296 FSSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSL-MLKYP--ESINVASQEPN 352
FS+ +++S T MGSGH E + A ++ +++ D + ++ P + + + + P
Sbjct: 316 FSA-LRQS--TSPLMGSGHFPKEGFKKAAFVNGLKVIDREIEKIRSPPVKDLRLFANSPK 372
Query: 353 CYSAFNDEDVQE---PTFYFGGPG 373
CY V E ++GGPG
Sbjct: 373 CYKVETKTGVGEEWSSAIFYGGPG 396
>At4g23360 putative protein
Length = 760
Score = 194 bits (493), Expect = 7e-50
Identities = 129/380 (33%), Positives = 185/380 (47%), Gaps = 68/380 (17%)
Query: 26 EIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ-------TW---- 74
E++ +LK N+PA+K+ K+E+GDI DC+DI+KQ AFDH +LKNH++Q W
Sbjct: 38 EMKRQLKAINKPAIKSFKTEHGDIFDCIDIHKQLAFDHHLLKNHSVQLKPTTVPEWITGN 97
Query: 75 ---------HKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
+ CP GTV ++R +DL+ A L G F+ ++ K
Sbjct: 98 NISGSFSLLQEGISCPNGTVIVKRTTMEDLMHAQRLKSMGFDGPRPFLTKTTNNTNSNGK 157
Query: 126 LSGSSNVVSEDHSGVHLATSGSNFIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFE 185
L V+ + G L F G ++NVW PK+ L D + A I + G + F
Sbjct: 158 L-----YVARGNYGPDL------FAGVRGNLNVWRPKI-LEDQVSVAYIAVGGGAKDNFA 205
Query: 186 SIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISS 245
SI GW L+G+ +TGC D++C GFVQ S T+ LG I P S
Sbjct: 206 SISVGWK----LHGS---------------NTGCNDMSCPGFVQVSKTIPLGAVIQPTSY 246
Query: 246 DSGTQYELNI----DEV-GHWWLKENHDIPIGYWPVELFTSLKHS--ATLVQWGGQVFSS 298
G QYEL + D + G WW N D +GYWP LF S + S A+ WGGQV+S
Sbjct: 247 YKGPQYELRLTLYQDHIKGDWWFAIN-DEDVGYWPASLFKSWRESNAASYASWGGQVYSP 305
Query: 299 QVKKSPHTKTQMGSGHLADEKYGHACYMRNIR--IKDNSLMLKYPESINVASQEPNCYSA 356
KKSP MGSGH E + + Y+ +++ + D + ++ + NCY A
Sbjct: 306 VTKKSP----PMGSGHWPSEGFQKSAYVSHLQMILGDGRVFNPQTGTVKLYQTNQNCYKA 361
Query: 357 FNDEDVQEP---TFYFGGPG 373
+V +P + Y+GGPG
Sbjct: 362 RLVHEVYKPWLKSIYYGGPG 381
Score = 132 bits (332), Expect = 3e-31
Identities = 85/237 (35%), Positives = 118/237 (48%), Gaps = 37/237 (15%)
Query: 149 FIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNEFESIEAGWMVNPGLYGNHDTRLFSY 208
F G INVW P + L D + A I + G F SI GW L+G++
Sbjct: 545 FAGVRGHINVWKPNI-LQDQVSLAYIAVGGGAKENFASISVGWK----LHGSN------- 592
Query: 209 WTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPISSDSGTQYELNI----DEV-GHWWL 263
TGC D++C GFVQ S T+ALG I P+S G QY+L++ D++ G WW+
Sbjct: 593 --------TGCNDMSCPGFVQVSKTIALGAIIQPLSIYKGPQYQLHLTLYQDQIKGDWWI 644
Query: 264 KENHDIPIGYWPVELFTSLKHS--ATLVQWGGQVFSSQVKKSPHTKTQMGSGHLADEKYG 321
N D +GYWP LF S + S A+ WGGQV+S +KSP MGSGH E Y
Sbjct: 645 SCN-DEDVGYWPASLFKSWRESNAASYASWGGQVYSPVTEKSP----PMGSGHWPSEGYQ 699
Query: 322 HACYMRNIRIK--DNSLMLKYPESINVASQEPNCYSAFNDEDVQEP---TFYFGGPG 373
+ Y R++++ D + ++ + NCY A D +P + Y+GGPG
Sbjct: 700 KSAYFRHVQMSLGDGRVFNPQTGTVKLYETSRNCYKARLVHDHYKPWLKSIYYGGPG 756
Score = 64.3 bits (155), Expect = 1e-10
Identities = 26/49 (53%), Positives = 40/49 (81%)
Query: 24 ETEIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ 72
+ E++ +LK N+PA+K+ K+E+GDI DC+DI+KQ A DH +LKNH++Q
Sbjct: 486 KNEMKRQLKAINKPAIKSFKTEHGDIFDCIDIHKQLALDHHLLKNHSVQ 534
>At4g23350 putative protein
Length = 386
Score = 184 bits (468), Expect = 5e-47
Identities = 126/382 (32%), Positives = 181/382 (46%), Gaps = 71/382 (18%)
Query: 26 EIEAKLKLHNRPAVKTIKSENGDIIDCVDIYKQPAFDHPVLKNHTIQ------------- 72
E++ +LK N+PA+K+ K+E+GDI DC+DI+KQ AFDH +LKNH++Q
Sbjct: 38 EMKRQLKAINKPAIKSFKTEHGDIFDCIDIHKQLAFDHHLLKNHSVQLRPTTVPEYITGN 97
Query: 73 -------TWHKSGRCPKGTVPIRRIQKQDLLRAATLDRFGLKQSSSFVNSKNTTISNFSK 125
+ CP GTV ++R QDL+ A L G + F+ T +N
Sbjct: 98 NISESFSLLQEGISCPDGTVIVKRTTMQDLMHAQRLKSMGFEGPRPFL-----TETNNMN 152
Query: 126 LSGSSNVVSEDHSGVHLATSGSN-FIGAEADINVWNPKVDLPDDSTTAQIWLKAGNGNE- 183
+G D+ G N F G +IN+W PK+ L D + + + G E
Sbjct: 153 FNGKFYDARADY--------GPNPFAGVAGNINIWKPKI-LQDQVSIGYMAVSGGPIEED 203
Query: 184 FESIEAGWMVNPGLYGNHDTRLFSYWTTDSYKSTGCFDLTCSGFVQTSNTVALGGGINPI 243
F SI GW+++ G GC+ ++C GFVQ S T+ +G + P
Sbjct: 204 FASISVGWILHGSSTG------------------GCYGMSCPGFVQVSKTIPVGAVLQPF 245
Query: 244 SSDSGTQYELNI-----DEVGHWWLKENHDIPIGYWPVELFTSL--KHSATLVQWGGQVF 296
S +G QYEL + G+WW +I IGYWP LF S +SA WGGQV+
Sbjct: 246 SIYNGRQYELRLGLFQDSGTGNWWFVFKEEI-IGYWPASLFKSWMESNSANYASWGGQVY 304
Query: 297 SSQVKKSPHTKTQMGSGHLADEKYGHACYMRNIRIKDNSLMLKYP--ESINVASQEPNCY 354
S +KSP MGSGH E + A ++ +++ D + P ++ V P CY
Sbjct: 305 SPIREKSP----PMGSGHWPSEGFHKAAFISGLKLFDGHGKVYNPGRGTVKVHESRPICY 360
Query: 355 SAFNDEDVQEP---TFYFGGPG 373
A DV +P + YFGGPG
Sbjct: 361 KARYVHDVDKPWLKSVYFGGPG 382
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,998,990
Number of Sequences: 26719
Number of extensions: 394379
Number of successful extensions: 1187
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 927
Number of HSP's gapped (non-prelim): 73
length of query: 380
length of database: 11,318,596
effective HSP length: 101
effective length of query: 279
effective length of database: 8,619,977
effective search space: 2404973583
effective search space used: 2404973583
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148485.6


