

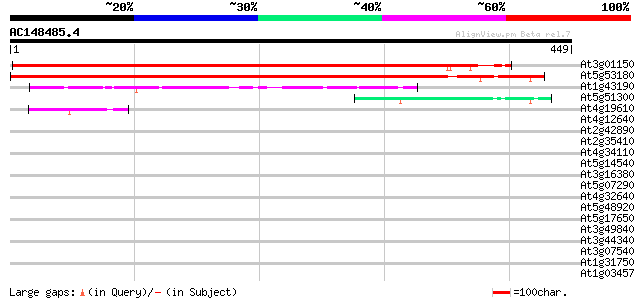
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.4 - phase: 0
(449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01150 polypyrimidine tract-binding protein like 561 e-160
At5g53180 polypyrimidine tract-binding RNA transport protein-like 515 e-146
At1g43190 hypothetical protein 134 1e-31
At5g51300 unknown protein 42 7e-04
At4g19610 putative protein 42 7e-04
At4g12640 putative protein 40 0.003
At2g42890 MEI2-like protein (MEI2) 39 0.004
At2g35410 putative chloroplast RNA binding protein precursor 39 0.008
At4g34110 poly(A)-binding protein 38 0.010
At5g14540 unknown protein 38 0.013
At3g16380 putative poly(A) binding protein 37 0.029
At5g07290 Mei2-like protein 35 0.11
At4g32640 putative protein 35 0.11
At5g48920 unknown protein 34 0.14
At5g17650 glycine/proline-rich protein 34 0.14
At3g49840 putative protein 34 0.14
At3g44340 putative protein 34 0.14
At3g07540 putative protein 34 0.14
At1g31750 unknown protein 34 0.14
At1g03457 ribonucleoprotein like 34 0.14
>At3g01150 polypyrimidine tract-binding protein like
Length = 399
Score = 561 bits (1445), Expect = e-160
Identities = 287/412 (69%), Positives = 328/412 (78%), Gaps = 27/412 (6%)
Query: 3 SSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQF 62
SS Q QFR TQ PSKV+H RNLPWEC +EEL DLC FGK+++TK NVG N+NQAFV+F
Sbjct: 2 SSSGQTQFRYTQTPSKVVHLRNLPWECVEEELIDLCKRFGKIVNTKSNVGANRNQAFVEF 61
Query: 63 AELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGVEA 122
A+LNQAISMVSYYASSSEPAQ+RGKTVYIQYSNRH IVN+ SPGD+PGNVLLVT EGVE+
Sbjct: 62 ADLNQAISMVSYYASSSEPAQIRGKTVYIQYSNRHEIVNNQSPGDVPGNVLLVTFEGVES 121
Query: 123 GDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLP 182
+VSIDVIHLVFSAFGFVHK+ATFEK AGFQAL+Q+TD ETA++A+ +LDGRSIPRYLL
Sbjct: 122 HEVSIDVIHLVFSAFGFVHKIATFEKAAGFQALVQFTDVETASAARSALDGRSIPRYLLS 181
Query: 183 EHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKEHK 242
HVG+C+LR+SYSAH DLNIKFQS+RSRDYTNP LPVNQ A+D ++QPA+G DGK+ E +
Sbjct: 182 AHVGSCSLRMSYSAHTDLNIKFQSHRSRDYTNPYLPVNQTAMDGSMQPALGADGKKVESQ 241
Query: 243 SNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKET 302
SNVLL IENMQYAV +DVLH+VFSA+G VQK+A+F+KNG T ALIQY D+ AA AKE
Sbjct: 242 SNVLLGLIENMQYAVTVDVLHTVFSAYGTVQKIAIFEKNGSTQALIQYSDIPTAAMAKEA 301
Query: 303 LEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQ--------AP--A 352
LEGHCIYDGGYCKL L+YSRHTDLNVKAFSDKSRDYT+ D SL AQ AP
Sbjct: 302 LEGHCIYDGGYCKLRLSYSRHTDLNVKAFSDKSRDYTLPDLSLLVAQKGPAVSGSAPPAG 361
Query: 353 WQTTQAATMYSGSMG---QMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPG 401
WQ QA + YSG G PS DPN G PSGQ PP+ G
Sbjct: 362 WQNPQAQSQYSGYGGSPYMYPSSDPN------------GASPSGQ--PPYYG 399
>At5g53180 polypyrimidine tract-binding RNA transport protein-like
Length = 429
Score = 515 bits (1326), Expect = e-146
Identities = 267/435 (61%), Positives = 321/435 (73%), Gaps = 15/435 (3%)
Query: 1 MSSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFV 60
MSS Q QFR TQ PSKVLH RNLPWEC++EEL +L PFG V++TKCNVG N+NQAF+
Sbjct: 1 MSSVSSQPQFRYTQPPSKVLHLRNLPWECTEEELIELGKPFGTVVNTKCNVGANRNQAFI 60
Query: 61 QFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGV 120
+F +LNQAI M+SYYASSSEPAQVRGKTVY+QYSNR IVN+ + D+ GNVLLVTIEG
Sbjct: 61 EFEDLNQAIQMISYYASSSEPAQVRGKTVYLQYSNRQEIVNNKTTADVVGNVLLVTIEGD 120
Query: 121 EAGDVSIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYL 180
+A VSIDV+HLVFSAFGFVHK+ TFEKTAG+QAL+Q+TDAETA +AK +LDGRSIPRYL
Sbjct: 121 DARMVSIDVLHLVFSAFGFVHKITTFEKTAGYQALVQFTDAETATAAKLALDGRSIPRYL 180
Query: 181 LPEHVGACNLRISYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKE 240
L E VG C+L+I+YSAH DL +KFQS+RSRDYTNP LPV +AIDS Q A+G DGK+ E
Sbjct: 181 LAETVGQCSLKITYSAHTDLTVKFQSHRSRDYTNPYLPVAPSAIDSTGQVAVGVDGKKME 240
Query: 241 HKSNVLLATIENMQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAK 300
+SNVLLA+IENMQYAV LDVLH VF+AFG VQK+AMFDKNG ALIQY D+ A AK
Sbjct: 241 PESNVLLASIENMQYAVTLDVLHMVFAAFGEVQKIAMFDKNGGVQALIQYSDVQTAVVAK 300
Query: 301 ETLEGHCIYDGGYCKLHLTYSRHTDLNVKAFSDKSRDYTVLDPSLHAAQAPAWQTTQAAT 360
E LEGHCIYDGG+CKLH+TYSRHTDL++K +D+SRDYT+ +P + Q P
Sbjct: 301 EALEGHCIYDGGFCKLHITYSRHTDLSIKVNNDRSRDYTMPNPPVPMPQQP------VQN 354
Query: 361 MYSGSMGQMPSWDPN---QQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPH-- 415
Y+G+ Q + + QQ+ Q PG G G++ PP+ +S H
Sbjct: 355 PYAGNPQQYHAAGGSHHQQQQQPQGGWVQPG--GQGSMGMGGGGHNHYMAPPSSSSMHQG 412
Query: 416 --SHMPPSSFAGAFP 428
HMPP + G P
Sbjct: 413 PGGHMPPQHYGGPGP 427
>At1g43190 hypothetical protein
Length = 396
Score = 134 bits (337), Expect = 1e-31
Identities = 99/314 (31%), Positives = 168/314 (52%), Gaps = 36/314 (11%)
Query: 17 SKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVSYYA 76
SKV+H RN+ E S+ +L L PFG + TK + +NQA +Q +++ A+S + ++
Sbjct: 5 SKVVHVRNVGHEISENDLLQLFQPFGVI--TKLVMLRAKNQALLQMQDVSSAVSALQFF- 61
Query: 77 SSSEPAQVRGKTVYIQYSNRHHIV----NSNSPGDIPGNVLLVTIEGVEAGDVSIDVIHL 132
++ +P +RG+ VY+Q+S+ + N + D P +LLVTI + +++DV+H
Sbjct: 62 TNVQPT-IRGRNVYVQFSSHQELTTIEQNIHGREDEPNRILLVTIHHM-LYPITVDVLHQ 119
Query: 133 VFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSIPRYLLPEHVGACNLRI 192
VFS +GFV KL TF+K+AGFQALIQY + AASA+ +L GR+I + G C L I
Sbjct: 120 VFSPYGFVEKLVTFQKSAGFQALIQYQVQQCAASARTALQGRNI-------YDGCCQLDI 172
Query: 193 SYSAHRDLNIKFQSNRSRDYTNPMLPVNQAAIDSALQPAIGPDGKRKEHKSNVLLATIEN 252
+S +L + Q N +AI +A + P + VL++ +
Sbjct: 173 QFS---NLAMVIQE-----------MANTSAIAAAFGGGLPPGITGTNDRCTVLVSNLN- 217
Query: 253 MQYAVPLDVLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDGG 312
++ D L ++FS +G + ++ + +N HAL+Q D A A L+G ++
Sbjct: 218 -ADSIDEDKLFNLFSLYGNIVRIKLL-RNKPDHALVQMGDGFQAELAVHFLKGAMLFGK- 274
Query: 313 YCKLHLTYSRHTDL 326
+L + +S+H ++
Sbjct: 275 --RLEVNFSKHPNI 286
Score = 35.4 bits (80), Expect = 0.064
Identities = 22/72 (30%), Positives = 39/72 (53%), Gaps = 1/72 (1%)
Query: 8 QQFRCTQIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFA-ELN 66
+ +R P+K++H LP + ++EE+ + G V++TK + QA VQF E
Sbjct: 309 KNYRYCCSPTKMIHLSTLPQDVTEEEVMNHVQEHGAVVNTKVFEMNGKKQALVQFENEEE 368
Query: 67 QAISMVSYYASS 78
A ++V +A+S
Sbjct: 369 AAEALVCKHATS 380
>At5g51300 unknown protein
Length = 804
Score = 42.0 bits (97), Expect = 7e-04
Identities = 40/164 (24%), Positives = 62/164 (37%), Gaps = 14/164 (8%)
Query: 277 MFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDG--GYCKLHLTYSRHTDLNVKAFSDK 334
M + +G + + ++ +A K+ + G G Y + + + +N F +
Sbjct: 490 MLEDDGLINLFSSFGEIVMAKVIKDRVTGLSKGYGFVKYADVQMANTAVQAMNGYRFEGR 549
Query: 335 SRDYTVLDPSLHAAQAPAWQTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQ 394
+ + S P Q T Q P P+QQ T Y +AP P G
Sbjct: 550 TLAVRIAGKSPPPIAPPGPPAPQPPTQGYPPSNQPPGAYPSQQYATGGYSTAP--VPWG- 606
Query: 395 AAPPFPGYSPAAVPPAGASPH-----SHMPPSSFAGAFPGSQPH 433
PP P YSP A+PP + HMPP + +P PH
Sbjct: 607 --PPVPSYSPYALPPPPPGSYHPVHGQHMPP--YGMQYPPPPPH 646
>At4g19610 putative protein
Length = 772
Score = 42.0 bits (97), Expect = 7e-04
Identities = 25/83 (30%), Positives = 43/83 (51%), Gaps = 7/83 (8%)
Query: 16 PSKVLHFRNLPWECSDEELADLCSPFGKVMS---TKCNVGVNQNQAFVQFAELNQAISMV 72
P LH +N+ +E + EL L SPFG++ S K N+G AFV+F +A++
Sbjct: 656 PCTKLHVKNIAFEATKRELRQLFSPFGQIKSMRLPKKNIGQYAGYAFVEFVTKQEALNAK 715
Query: 73 SYYASSSEPAQVRGKTVYIQYSN 95
AS+ G+ + ++++N
Sbjct: 716 KALAST----HFYGRHLVLEWAN 734
>At4g12640 putative protein
Length = 695
Score = 40.0 bits (92), Expect = 0.003
Identities = 21/62 (33%), Positives = 39/62 (62%), Gaps = 5/62 (8%)
Query: 261 VLHSVFSAFGFVQKVAMFDKNGHTHALIQYPDLTIAAAAKETLEGHCIYDGGYCKLHLTY 320
+L +VFS+FG + KV +F G ++A +Q+ +L A AKE+L+G G ++H+ +
Sbjct: 39 LLRNVFSSFGEITKVTVFP--GRSYAFVQFRNLMAACKAKESLQGKLF---GNPRVHICF 93
Query: 321 SR 322
++
Sbjct: 94 AK 95
Score = 28.9 bits (63), Expect = 6.0
Identities = 15/42 (35%), Positives = 24/42 (56%), Gaps = 2/42 (4%)
Query: 133 VFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGR 174
VFS+FG + K+ F + A +Q+ + A AK+SL G+
Sbjct: 43 VFSSFGEITKVTVFPGRS--YAFVQFRNLMAACKAKESLQGK 82
>At2g42890 MEI2-like protein (MEI2)
Length = 843
Score = 39.3 bits (90), Expect = 0.004
Identities = 44/171 (25%), Positives = 73/171 (41%), Gaps = 32/171 (18%)
Query: 16 PSKVLHFRNLPWECSDEELADLCSPFGKVMS--TKCNVGVNQNQAFVQFAELNQAISMVS 73
PS+ L RN+ D EL+ L PFG++ S T C +++ FV M+S
Sbjct: 196 PSRTLFVRNINSSVEDSELSALFEPFGEIRSLYTAC-----KSRGFV----------MIS 240
Query: 74 YYASSSEPAQVRG--------KTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGVEAGDV 125
YY + A +R +T+ I +S + +P + N + I V+ V
Sbjct: 241 YYDIRAAHAAMRALQNTLLRKRTLDIHFS-----IPKENPSEKDMNQGTLVIFNVDT-TV 294
Query: 126 SIDVIHLVFSAFGFVHKLATFEKTAGFQALIQYTDAETAASAKDSLDGRSI 176
S D + +F A+G + ++ F I+Y D A +A +L+ I
Sbjct: 295 SNDELLQLFGAYGEIREIRE-TPNRRFHRFIEYYDVRDAETALKALNRSEI 344
>At2g35410 putative chloroplast RNA binding protein precursor
Length = 308
Score = 38.5 bits (88), Expect = 0.008
Identities = 26/111 (23%), Positives = 51/111 (45%), Gaps = 8/111 (7%)
Query: 2 SSSKQQQQFRCTQIPSKVLHFRNLPWECSDEELADL---CSPFGKVMSTKCNVGVNQNQA 58
S ++ ++ + + + K+ F NLPW S ++++L C V + G N+ A
Sbjct: 80 SQEEKTEETQNSNLKRKLFVF-NLPWSMSVNDISELFGQCGTVNNVEIIRQKDGKNRGFA 138
Query: 59 FVQFAELNQAISMVSYYASSSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIP 109
FV A +A + + + + QV G+ + + ++ R SP D+P
Sbjct: 139 FVTMASGEEAQAAIDKF----DTFQVSGRIISVSFARRFKKPTPKSPNDLP 185
>At4g34110 poly(A)-binding protein
Length = 629
Score = 38.1 bits (87), Expect = 0.010
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 6/74 (8%)
Query: 17 SKVLHFRNLPWECSDEELADLCSPFGKVMSTKC---NVGVNQNQAFVQFA---ELNQAIS 70
S L+ +NL SDE+L ++ SPFG V S+K G ++ FV FA E +A+S
Sbjct: 317 SSNLYVKNLDPSISDEKLKEIFSPFGTVTSSKVMRDPNGTSKGSGFVAFATPEEATEAMS 376
Query: 71 MVSYYASSSEPAQV 84
+S S+P V
Sbjct: 377 QLSGKMIESKPLYV 390
Score = 29.3 bits (64), Expect = 4.6
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 5/65 (7%)
Query: 250 IENMQYAVPLDVLHSVFSAFGFVQ--KVAMFDKNGHT--HALIQYPDLTIAAAAKETLEG 305
I+N+ ++ LH FS+FG + KVA+ D +G + + +QY + A A E L G
Sbjct: 128 IKNLDESIDHKALHDTFSSFGNIVSCKVAV-DSSGQSKGYGFVQYANEESAQKAIEKLNG 186
Query: 306 HCIYD 310
+ D
Sbjct: 187 MLLND 191
>At5g14540 unknown protein
Length = 530
Score = 37.7 bits (86), Expect = 0.013
Identities = 26/87 (29%), Positives = 37/87 (41%), Gaps = 9/87 (10%)
Query: 368 QMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSH------MPPS 421
Q P P Q + Y +P Q+ PP P P + PP G++P PPS
Sbjct: 319 QYPQQPPPQLQHPSGYNPEEPPYPQ-QSYPPNPPRQPPSHPPPGSAPSQQYYNAPPTPPS 377
Query: 422 SFAGAFPGSQPHYGWESRFRLNYNPYT 448
+ G PG + + G+ S + PYT
Sbjct: 378 MYDG--PGGRSNSGFPSGYSPESYPYT 402
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 36.6 bits (83), Expect = 0.029
Identities = 38/161 (23%), Positives = 68/161 (41%), Gaps = 17/161 (10%)
Query: 20 LHFRNLPWECSDEELADLCSPFGKVMSTKC--NVGVNQNQAFVQFAELNQAISMVSYYAS 77
L+ +NL + L + PFG ++S K G ++ FVQF A+S S
Sbjct: 114 LYVKNLDSSITSSCLERMFCPFGSILSCKVVEENGQSKGFGFVQFDTEQSAVSA----RS 169
Query: 78 SSEPAQVRGKTVYIQYSNRHHIVNSNSPGDIPGNVLLVTIEGVEAGD-VSIDVIHLVFSA 136
+ + V GK +++ +N + + GN + + V+ D +H +FS
Sbjct: 170 ALHGSMVYGKKLFVA-----KFINKDERAAMAGNQDSTNVYVKNLIETVTDDCLHTLFSQ 224
Query: 137 FGFVHKLATFE----KTAGFQALIQYTDAETAASAKDSLDG 173
+G V + ++ GF + + + E A A +SL G
Sbjct: 225 YGTVSSVVVMRDGMGRSRGF-GFVNFCNPENAKKAMESLCG 264
>At5g07290 Mei2-like protein
Length = 907
Score = 34.7 bits (78), Expect = 0.11
Identities = 42/173 (24%), Positives = 74/173 (42%), Gaps = 33/173 (19%)
Query: 14 QIPSKVLHFRNLPWECSDEELADLCSPFGKVMSTKCNVGVNQNQAFVQFAELNQAISMVS 73
+I S++L RN+ D EL L FG V + + A N+ MVS
Sbjct: 207 EILSRILFVRNVDSSIEDCELGVLFKQFGDVRA-------------LHTAGKNRGFIMVS 253
Query: 74 YY--------ASSSEPAQVRGKTVYIQYS-NRHHIVNSNSPGDIPGNVLLVTIEGVEAGD 124
YY A + +RG+ + I+YS + + ++S G + N L
Sbjct: 254 YYDIRAAQKAARALHGRLLRGRKLDIRYSIPKENPKENSSEGALWVNNL--------DSS 305
Query: 125 VSIDVIHLVFSAFGFVHKL-ATFEKTAGFQALIQYTDAETAASAKDSLDGRSI 176
+S + +H +FS++G + ++ T + + Q I++ D A A L+G +
Sbjct: 306 ISNEELHGIFSSYGEIREVRRTMHENS--QVYIEFFDVRKAKVALQGLNGLEV 356
>At4g32640 putative protein
Length = 1069
Score = 34.7 bits (78), Expect = 0.11
Identities = 34/102 (33%), Positives = 45/102 (43%), Gaps = 13/102 (12%)
Query: 354 QTTQAATMYSGSMGQMPSWDPNQQEVTQSYLSA--PGTFPS-----GQAAPPFPGYSPA- 405
Q + Y GS G + N Q ++ + PG+ P GQ+ PFP SP+
Sbjct: 16 QNSGPPNFYPGSQGNSNALADNMQNLSLNRPPPMMPGSGPRPPPPFGQSPQPFPQQSPSY 75
Query: 406 AVPPAGASPHSHM-PPSSFA---GAFPGSQPHYGWESRFRLN 443
P G SP S PP+ A G P SQP G++S LN
Sbjct: 76 GAPQRGPSPMSRPGPPAGMARPGGPPPVSQP-AGFQSNVPLN 116
Score = 29.3 bits (64), Expect = 4.6
Identities = 27/73 (36%), Positives = 29/73 (38%), Gaps = 7/73 (9%)
Query: 364 GSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAV---PPAGASPHSHMPP 420
GS MP Q + S A G S A PP PG P A PP G+ MPP
Sbjct: 129 GSRPSMPGGPVAQPAASSSGFPAFGPSGSVAAGPP-PGSRPMAFGSPPPVGSG--MSMPP 185
Query: 421 SSFAGAFPGSQPH 433
S G P S H
Sbjct: 186 SGMIGG-PVSNGH 197
>At5g48920 unknown protein
Length = 205
Score = 34.3 bits (77), Expect = 0.14
Identities = 20/52 (38%), Positives = 23/52 (43%), Gaps = 5/52 (9%)
Query: 382 SYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGSQPH 433
S++S P S PP P +SP PP PH H PP S P PH
Sbjct: 29 SHISPPPPPFSPPHHPPPPHFSPPHQPPPSPYPHPHPPPPS-----PYPHPH 75
Score = 29.3 bits (64), Expect = 4.6
Identities = 21/71 (29%), Positives = 27/71 (37%), Gaps = 8/71 (11%)
Query: 363 SGSMGQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSS 422
+ S+ P + P Q L +P P +PP P +SP PP H PP S
Sbjct: 2 AASVEYFPYYSPPSH---QHPLPSPVPPPPSHISPPPPPFSPPHHPPPPHFSPPHQPPPS 58
Query: 423 FAGAFPGSQPH 433
P PH
Sbjct: 59 -----PYPHPH 64
>At5g17650 glycine/proline-rich protein
Length = 173
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/42 (42%), Positives = 21/42 (49%), Gaps = 3/42 (7%)
Query: 397 PPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGSQPHYGWES 438
PP GY P A PP G P + PP AG P P +G+ S
Sbjct: 49 PPPHGYPPVAYPPHGGYPPAGYPP---AGYPPAGYPAHGYPS 87
Score = 29.6 bits (65), Expect = 3.5
Identities = 17/36 (47%), Positives = 17/36 (47%), Gaps = 3/36 (8%)
Query: 382 SYLSAPGTFPSGQ---AAPPFPGYSPAAVPPAGASP 414
SY P P G A PP GY PA PPAG P
Sbjct: 42 SYPYPPPPPPHGYPPVAYPPHGGYPPAGYPPAGYPP 77
>At3g49840 putative protein
Length = 651
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/50 (36%), Positives = 22/50 (44%), Gaps = 1/50 (2%)
Query: 387 PGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGA-FPGSQPHYG 435
P + P Q PP GY PA PP P P + + A +P Q YG
Sbjct: 536 PVSAPPPQGYPPKEGYPPAGYPPPAGYPPPQYPQAGYPPAGYPPPQQGYG 585
>At3g44340 putative protein
Length = 1122
Score = 34.3 bits (77), Expect = 0.14
Identities = 26/68 (38%), Positives = 31/68 (45%), Gaps = 7/68 (10%)
Query: 370 PSWDPNQQEVTQSYLSAPGTFPSGQA--APPFPGYSPAAVPPAGASPHSHMPPSSFAGAF 427
P P + T + AP P GQ A PF G SP + PPA HS PP++F G
Sbjct: 231 PGAFPRGSQFTSGPMMAPPP-PYGQPPNAGPFTGNSPLSSPPA----HSIPPPTNFPGVP 285
Query: 428 PGSQPHYG 435
G P G
Sbjct: 286 YGRPPMPG 293
Score = 30.4 bits (67), Expect = 2.1
Identities = 21/71 (29%), Positives = 25/71 (34%), Gaps = 5/71 (7%)
Query: 367 GQMPSWDPNQQEVTQSYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHS-----HMPPS 421
G P++ P Q S + + PP PG P PP G SP S P
Sbjct: 20 GGPPNFVPGSQGNPNSLAANMQNLNINRPPPPMPGSGPRPSPPFGQSPQSFPQQQQQQPR 79
Query: 422 SFAGAFPGSQP 432
A PG P
Sbjct: 80 PSPMARPGPPP 90
Score = 28.5 bits (62), Expect = 7.9
Identities = 25/75 (33%), Positives = 30/75 (39%), Gaps = 10/75 (13%)
Query: 363 SGSMGQMPSWDPNQQEVTQ--SYLSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPP 420
S GQ P P QQ+ S ++ PG P AA PG P P G PP
Sbjct: 60 SPPFGQSPQSFPQQQQQQPRPSPMARPG--PPPPAAMARPGGPPQVSQPGG------FPP 111
Query: 421 SSFAGAFPGSQPHYG 435
A P +QP +G
Sbjct: 112 VGRPVAPPSNQPPFG 126
>At3g07540 putative protein
Length = 841
Score = 34.3 bits (77), Expect = 0.14
Identities = 18/47 (38%), Positives = 23/47 (48%)
Query: 384 LSAPGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGS 430
L AP T PS + P FP YS + PP + P PP+ FP +
Sbjct: 36 LQAPVTDPSTFSPPFFPLYSSTSPPPPPSPPQPLPPPAPTFATFPAN 82
>At1g31750 unknown protein
Length = 176
Score = 34.3 bits (77), Expect = 0.14
Identities = 19/49 (38%), Positives = 24/49 (48%), Gaps = 5/49 (10%)
Query: 387 PGTFPSGQAAPPFPGYSPAAVPPAGASPHSHMPPSSFAGAFPGSQPHYG 435
PG +P PP GY PAA PP P PP+ + G G +P +G
Sbjct: 40 PGGYPPQGYPPPPHGYPPAAYPP----PPGAYPPAGYPGP-SGPRPGFG 83
>At1g03457 ribonucleoprotein like
Length = 429
Score = 34.3 bits (77), Expect = 0.14
Identities = 27/93 (29%), Positives = 43/93 (46%), Gaps = 17/93 (18%)
Query: 13 TQIPSKV---------LHFRNLPWECSDEELADLCSPFGKVMSTKCNV----GVNQNQAF 59
T IPS + L N+P E D+ELA PFGKV+S K V G+++ F
Sbjct: 316 TSIPSSLQTEGPAGANLFIYNIPREFEDQELAATFQPFGKVLSAKVFVDKATGISKCFGF 375
Query: 60 VQFAELNQAISMVSYYASSSEPAQVRGKTVYIQ 92
+ + A + + ++ Q+ GK + +Q
Sbjct: 376 ISYDSQAAAQNAI----NTMNGCQLSGKKLKVQ 404
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.395
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,355,197
Number of Sequences: 26719
Number of extensions: 450673
Number of successful extensions: 1956
Number of sequences better than 10.0: 94
Number of HSP's better than 10.0 without gapping: 24
Number of HSP's successfully gapped in prelim test: 73
Number of HSP's that attempted gapping in prelim test: 1659
Number of HSP's gapped (non-prelim): 291
length of query: 449
length of database: 11,318,596
effective HSP length: 103
effective length of query: 346
effective length of database: 8,566,539
effective search space: 2964022494
effective search space used: 2964022494
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC148485.4


