

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148485.3 - phase: 0 /pseudo
(138 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
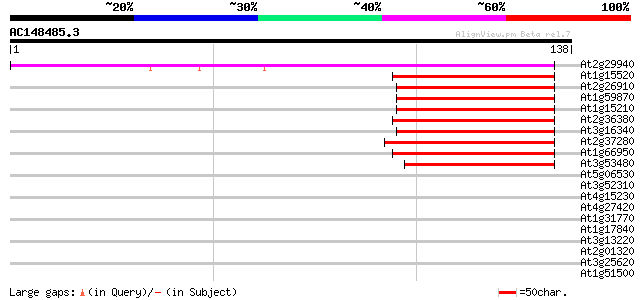
Score E
Sequences producing significant alignments: (bits) Value
At2g29940 putative ABC transporter 59 1e-09
At1g15520 hypothetical protein 56 5e-09
At2g26910 putative ABC transporter 49 6e-07
At1g59870 putative ABC transporter (At1g59870) 49 8e-07
At1g15210 putative ABC transporter 49 8e-07
At2g36380 putative ABC transporter 48 2e-06
At3g16340 putative ABC transporter 47 2e-06
At2g37280 putative ABC transporter 47 3e-06
At1g66950 ABC transporter, putative 46 7e-06
At3g53480 ABC transporter - like protein 41 2e-04
At5g06530 ABC transporter like protein 35 0.012
At3g52310 ABC transporter-like protein 35 0.012
At4g15230 ABC transporter like protein 34 0.028
At4g27420 putative protein 33 0.047
At1g31770 unknown protein 32 0.14
At1g17840 putative ABC transporter (At1g17840) 31 0.18
At3g13220 ABC transporter 30 0.40
At2g01320 putative ABC transporter 30 0.40
At3g25620 membrane transporter, putative 30 0.52
At1g51500 ATP-dependent transmembrane transporter, putative 29 0.89
>At2g29940 putative ABC transporter
Length = 1443
Score = 58.5 bits (140), Expect = 1e-09
Identities = 49/182 (26%), Positives = 77/182 (41%), Gaps = 48/182 (26%)
Query: 1 MSYLHLILSRKKPLAVLHDVSGIIKPKRSSEFW------KDYIVVGIGGKT--------- 45
+S L +I RK L +L D+SGIIKP R + K +++ + GK
Sbjct: 177 LSSLRIIKPRKHKLNILKDISGIIKPGRMTLLLGPPGSGKSTLLLALAGKLDKSLKKTGN 236
Query: 46 --------------RTSAYKSN*YRTVFQL-------------------CGIRRYAGRII 72
RTSAY S + +L G + R+
Sbjct: 237 ITYNGENLNKFHVKRTSAYISQTDNHIAELTVRETLDFAARCQGASEGFAGYMKDLTRLE 296
Query: 73 KRKG*KH*TRF*S*Y*CED*KAKKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKR 132
K +G + + + K +K ++ T Y++K+LGLD C+DTMVG DM+RG G ++
Sbjct: 297 KERGIRPSSEIDAFMKAASVKGEKHSVSTDYVLKVLGLDVCSDTMVGNDMMRGVSGGQRK 356
Query: 133 EL 134
+
Sbjct: 357 RV 358
>At1g15520 hypothetical protein
Length = 1423
Score = 56.2 bits (134), Expect = 5e-09
Identities = 24/40 (60%), Positives = 32/40 (80%)
Query: 95 KKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
+KTN++T YI+KILGL+ CADTMVG DM+RG G K+ +
Sbjct: 295 EKTNVMTDYILKILGLEVCADTMVGDDMLRGISGGQKKRV 334
Score = 36.2 bits (82), Expect = 0.006
Identities = 15/28 (53%), Positives = 22/28 (78%)
Query: 1 MSYLHLILSRKKPLAVLHDVSGIIKPKR 28
++ LHL+ +RKK +L+DVSGI+KP R
Sbjct: 154 LNTLHLVPNRKKKFTILNDVSGIVKPGR 181
>At2g26910 putative ABC transporter
Length = 1420
Score = 49.3 bits (116), Expect = 6e-07
Identities = 21/39 (53%), Positives = 30/39 (76%)
Query: 96 KTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
+T++V Y++KILGLD CADT+VG +MI+G G K+ L
Sbjct: 277 ETSLVVEYVMKILGLDTCADTLVGDEMIKGISGGQKKRL 315
Score = 26.6 bits (57), Expect = 4.4
Identities = 9/25 (36%), Positives = 17/25 (68%)
Query: 4 LHLILSRKKPLAVLHDVSGIIKPKR 28
+H+I ++ L +L +SG+I+P R
Sbjct: 138 IHVIGGKRNKLTILDGISGVIRPSR 162
>At1g59870 putative ABC transporter (At1g59870)
Length = 1469
Score = 48.9 bits (115), Expect = 8e-07
Identities = 21/39 (53%), Positives = 28/39 (70%)
Query: 96 KTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
K ++VT Y +KILGLD C DT+VG DM+RG G K+ +
Sbjct: 313 KNSLVTDYTLKILGLDICKDTIVGDDMMRGISGGQKKRV 351
Score = 26.6 bits (57), Expect = 4.4
Identities = 10/20 (50%), Positives = 15/20 (75%)
Query: 9 SRKKPLAVLHDVSGIIKPKR 28
++K L +L D+SG+IKP R
Sbjct: 179 AKKAQLTILKDISGVIKPGR 198
>At1g15210 putative ABC transporter
Length = 1442
Score = 48.9 bits (115), Expect = 8e-07
Identities = 20/39 (51%), Positives = 29/39 (74%)
Query: 96 KTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
K++++T Y +KILGLD C DT+VG DM+RG G K+ +
Sbjct: 311 KSSLITDYTLKILGLDICKDTIVGDDMMRGISGGQKKRV 349
Score = 29.3 bits (64), Expect = 0.68
Identities = 12/21 (57%), Positives = 16/21 (76%)
Query: 8 LSRKKPLAVLHDVSGIIKPKR 28
L++K L +L DVSGI+KP R
Sbjct: 176 LAKKAQLTILKDVSGIVKPSR 196
>At2g36380 putative ABC transporter
Length = 1450
Score = 47.8 bits (112), Expect = 2e-06
Identities = 20/40 (50%), Positives = 30/40 (75%)
Query: 95 KKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
++T++VT Y++K+LGLD CADT+VG M RG G ++ L
Sbjct: 314 QETSLVTDYVLKLLGLDICADTLVGDVMRRGISGGQRKRL 353
Score = 33.5 bits (75), Expect = 0.036
Identities = 13/28 (46%), Positives = 20/28 (71%)
Query: 1 MSYLHLILSRKKPLAVLHDVSGIIKPKR 28
+ HL+ S+K+ + +L D+SGIIKP R
Sbjct: 173 LGLFHLLPSKKRKIEILKDISGIIKPSR 200
>At3g16340 putative ABC transporter
Length = 1416
Score = 47.4 bits (111), Expect = 2e-06
Identities = 19/39 (48%), Positives = 29/39 (73%)
Query: 96 KTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
K++++T Y ++ILGLD C DT+VG +MIRG G K+ +
Sbjct: 289 KSSLITDYTLRILGLDICKDTVVGDEMIRGISGGQKKRV 327
>At2g37280 putative ABC transporter
Length = 1413
Score = 47.0 bits (110), Expect = 3e-06
Identities = 22/42 (52%), Positives = 28/42 (66%)
Query: 93 KAKKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
K K ++ T YI+KILGLD CA+T+VG M RG G K+ L
Sbjct: 278 KGLKRSLQTDYILKILGLDICAETLVGNAMKRGISGGQKKRL 319
>At1g66950 ABC transporter, putative
Length = 1434
Score = 45.8 bits (107), Expect = 7e-06
Identities = 20/40 (50%), Positives = 28/40 (70%)
Query: 95 KKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
++T++VT Y++KILGLD CAD + G M RG G K+ L
Sbjct: 316 QETSLVTDYVLKILGLDICADILAGDVMRRGISGGQKKRL 355
Score = 34.7 bits (78), Expect = 0.016
Identities = 12/28 (42%), Positives = 21/28 (74%)
Query: 1 MSYLHLILSRKKPLAVLHDVSGIIKPKR 28
+ + HL+ S++K + +L D+SGI+KP R
Sbjct: 175 LGFFHLLPSKRKKIQILKDISGIVKPSR 202
>At3g53480 ABC transporter - like protein
Length = 1450
Score = 40.8 bits (94), Expect = 2e-04
Identities = 18/37 (48%), Positives = 25/37 (66%)
Query: 98 NIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
++ T YI+KILGLD CA+ ++G M RG G K+ L
Sbjct: 318 SLQTDYILKILGLDICAEILIGDVMRRGISGGQKKRL 354
>At5g06530 ABC transporter like protein
Length = 627
Score = 35.0 bits (79), Expect = 0.012
Identities = 13/33 (39%), Positives = 22/33 (66%)
Query: 104 IIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
+I+ LGL+ C DTM+GG +RG G ++ + +
Sbjct: 157 VIQELGLERCQDTMIGGAFVRGVSGGERKRVSI 189
>At3g52310 ABC transporter-like protein
Length = 737
Score = 35.0 bits (79), Expect = 0.012
Identities = 13/31 (41%), Positives = 21/31 (66%)
Query: 104 IIKILGLDFCADTMVGGDMIRGTLSGPKREL 134
+I+ LGL+ C DTM+GG +RG G ++ +
Sbjct: 269 VIQELGLERCQDTMIGGSFVRGVSGGERKRV 299
>At4g15230 ABC transporter like protein
Length = 979
Score = 33.9 bits (76), Expect = 0.028
Identities = 17/30 (56%), Positives = 18/30 (59%)
Query: 105 IKILGLDFCADTMVGGDMIRGTLSGPKREL 134
+KILGLD CADT VG G G KR L
Sbjct: 23 MKILGLDICADTRVGDATRPGISGGEKRRL 52
>At4g27420 putative protein
Length = 635
Score = 33.1 bits (74), Expect = 0.047
Identities = 12/29 (41%), Positives = 19/29 (65%)
Query: 108 LGLDFCADTMVGGDMIRGTLSGPKRELEL 136
LGLD C DT++GG +RG G ++ + +
Sbjct: 174 LGLDRCKDTIIGGPFLRGVSGGERKRVSI 202
>At1g31770 unknown protein
Length = 648
Score = 31.6 bits (70), Expect = 0.14
Identities = 12/33 (36%), Positives = 21/33 (63%)
Query: 104 IIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
+I LGL+ C ++M+GG + RG G K+ + +
Sbjct: 182 VIAELGLNRCTNSMIGGPLFRGISGGEKKRVSI 214
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 31.2 bits (69), Expect = 0.18
Identities = 16/44 (36%), Positives = 27/44 (61%), Gaps = 1/44 (2%)
Query: 93 KAKKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
+++K +V II+ +GL CADT++G +RG G KR + +
Sbjct: 161 RSEKRALVERTIIE-MGLQDCADTVIGNWHLRGISGGEKRRVSI 203
>At3g13220 ABC transporter
Length = 647
Score = 30.0 bits (66), Expect = 0.40
Identities = 13/33 (39%), Positives = 19/33 (57%)
Query: 104 IIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
IIK LGL+ C T VGG ++G G ++ +
Sbjct: 169 IIKELGLERCRRTRVGGGFVKGISGGERKRASI 201
>At2g01320 putative ABC transporter
Length = 725
Score = 30.0 bits (66), Expect = 0.40
Identities = 16/42 (38%), Positives = 23/42 (54%), Gaps = 1/42 (2%)
Query: 95 KKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
++ V ++K LGL CAD+ VG +RG G K+ L L
Sbjct: 186 ERDEYVNNLLLK-LGLVSCADSCVGDAKVRGISGGEKKRLSL 226
>At3g25620 membrane transporter, putative
Length = 646
Score = 29.6 bits (65), Expect = 0.52
Identities = 10/33 (30%), Positives = 21/33 (63%)
Query: 104 IIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
++ LGL C ++++GG +IRG G ++ + +
Sbjct: 199 VVSDLGLTRCCNSVIGGGLIRGISGGERKRVSI 231
>At1g51500 ATP-dependent transmembrane transporter, putative
Length = 687
Score = 28.9 bits (63), Expect = 0.89
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 1/44 (2%)
Query: 93 KAKKTNIVTGYIIKILGLDFCADTMVGGDMIRGTLSGPKRELEL 136
K + +IV G II+ LGL CAD ++G RG G ++ + +
Sbjct: 136 KEEVNDIVEGTIIE-LGLQDCADRVIGNWHSRGVSGGERKRVSV 178
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.160 0.530
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,558,958
Number of Sequences: 26719
Number of extensions: 80802
Number of successful extensions: 332
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 297
Number of HSP's gapped (non-prelim): 37
length of query: 138
length of database: 11,318,596
effective HSP length: 89
effective length of query: 49
effective length of database: 8,940,605
effective search space: 438089645
effective search space used: 438089645
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 54 (25.4 bits)
Medicago: description of AC148485.3


