

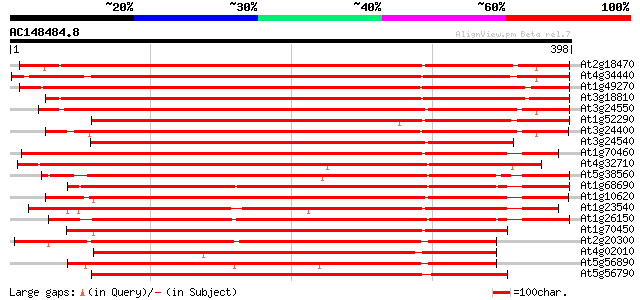
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148484.8 + phase: 0
(398 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g18470 putative protein kinase 521 e-148
At4g34440 serine/threonine protein kinase - like 512 e-145
At1g49270 hypothetical protein 505 e-143
At3g18810 protein kinase, putative 503 e-143
At3g24550 protein kinase, putative 501 e-142
At1g52290 protein kinase, putative 456 e-128
At3g24400 protein kinase, putative 455 e-128
At3g24540 protein kinase, putative 421 e-118
At1g70460 putative protein kinase 415 e-116
At4g32710 putative protein kinase 403 e-113
At5g38560 putative protein 394 e-110
At1g68690 protein kinase, putative 392 e-109
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 390 e-109
At1g23540 putative serine/threonine protein kinase 389 e-108
At1g26150 Pto kinase interactor, putative 388 e-108
At1g70450 putative protein kinase 358 2e-99
At2g20300 protein kinase like protein 301 5e-82
At4g02010 putative NAK-like ser/thr protein kinase 290 1e-78
At5g56890 unknown protein 286 1e-77
At5g56790 putative protein kinase 280 8e-76
>At2g18470 putative protein kinase
Length = 633
Score = 521 bits (1343), Expect = e-148
Identities = 269/395 (68%), Positives = 318/395 (80%), Gaps = 10/395 (2%)
Query: 8 QQNPNGASGVWGAPHP---LMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEEL 64
QQ+ G G +P P + SGE SS YS G P SP L+L TFTY+EL
Sbjct: 219 QQSTGGWGGGGPSPPPPPRMPTSGEDSSMYS-GPSRPVLPPPSPALALGFNKSTFTYQEL 277
Query: 65 ASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHH 124
A+AT GF + N++GQGGFGYVHKG+LP+GKE+AVKSLKAGSGQGEREFQAE+DIISRVHH
Sbjct: 278 AAATGGFTDANLLGQGGFGYVHKGVLPSGKEVAVKSLKAGSGQGEREFQAEVDIISRVHH 337
Query: 125 RHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLH 184
R+LVSLVGYC++ GQRMLVYEFVPNKTLEYHLHGK +P M++ TR+RIALG+A+GLAYLH
Sbjct: 338 RYLVSLVGYCIADGQRMLVYEFVPNKTLEYHLHGKNLPVMEFSTRLRIALGAAKGLAYLH 397
Query: 185 EDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYAS 244
EDC PRIIHRDIK+AN+L+D +F+A VADFGLAKLT+D NTHVSTRVMGTFGY+APEYAS
Sbjct: 398 EDCHPRIIHRDIKSANILLDFNFDAMVADFGLAKLTSDNNTHVSTRVMGTFGYLAPEYAS 457
Query: 245 SGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVD 304
SGKLTEKSDVFS+GVMLLEL+TGKRP+D + MD++LVDWARPL++RAL EDGNF EL D
Sbjct: 458 SGKLTEKSDVFSYGVMLLELITGKRPVDNSITMDDTLVDWARPLMARAL-EDGNFNELAD 516
Query: 305 PFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQ 364
LEGNY+ QEM R+ CAA+SIRHS +KR KMSQIVRALEG+VSL+ L E +
Sbjct: 517 ARLEGNYNPQEMARMVTCAAASIRHSGRKRPKMSQIVRALEGEVSLDALNEGV---KPGH 573
Query: 365 TGVYTTTG--SEYDTMQYNADMAKFRKQIMSSQEF 397
+ VY + G S+Y YNADM KFR+ +SSQEF
Sbjct: 574 SNVYGSLGASSDYSQTSYNADMKKFRQIALSSQEF 608
>At4g34440 serine/threonine protein kinase - like
Length = 670
Score = 512 bits (1318), Expect = e-145
Identities = 256/399 (64%), Positives = 312/399 (78%), Gaps = 15/399 (3%)
Query: 2 DHVVRVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTY 61
DHVV + G WG P+ +SN + P ++ G + + TFTY
Sbjct: 250 DHVVNMAGQ---GGGNWGPQQPVSGPHSDASNLTGRTAIPSPQAATLGHNQS----TFTY 302
Query: 62 EELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISR 121
+EL+ AT+GFA N++GQGGFGYVHKG+LP+GKE+AVKSLK GSGQGEREFQAE+DIISR
Sbjct: 303 DELSIATEGFAQSNLLGQGGFGYVHKGVLPSGKEVAVKSLKLGSGQGEREFQAEVDIISR 362
Query: 122 VHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLA 181
VHHRHLVSLVGYC+SGGQR+LVYEF+PN TLE+HLHGKG P +DWPTR++IALGSARGLA
Sbjct: 363 VHHRHLVSLVGYCISGGQRLLVYEFIPNNTLEFHLHGKGRPVLDWPTRVKIALGSARGLA 422
Query: 182 YLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPE 241
YLHEDC PRIIHRDIKAAN+L+D SFE KVADFGLAKL+ D THVSTRVMGTFGY+APE
Sbjct: 423 YLHEDCHPRIIHRDIKAANILLDFSFETKVADFGLAKLSQDNYTHVSTRVMGTFGYLAPE 482
Query: 242 YASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAE 301
YASSGKL++KSDVFSFGVMLLEL+TG+ PLDLT M++SLVDWARPL +A +DG++ +
Sbjct: 483 YASSGKLSDKSDVFSFGVMLLELITGRPPLDLTGEMEDSLVDWARPLCLKA-AQDGDYNQ 541
Query: 302 LVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSP 361
L DP LE NY HQEM+++A+CAA++IRHSA++R KMSQIVRALEGD+S++DL E
Sbjct: 542 LADPRLELNYSHQEMVQMASCAAAAIRHSARRRPKMSQIVRALEGDMSMDDLSE----GT 597
Query: 362 APQTGVYTTTG---SEYDTMQYNADMAKFRKQIMSSQEF 397
P Y + G SEYD Y ADM KF+K + ++E+
Sbjct: 598 RPGQSTYLSPGSVSSEYDASSYTADMKKFKKLALENKEY 636
>At1g49270 hypothetical protein
Length = 699
Score = 505 bits (1300), Expect = e-143
Identities = 253/391 (64%), Positives = 309/391 (78%), Gaps = 6/391 (1%)
Query: 8 QQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASA 67
Q P+ + V G + ++ SGEMSSN+S G P P ++L TFTYEELASA
Sbjct: 274 QPMPSPPAPVSGGAN-VIQSGEMSSNFSSGPYAPSLPPPHPSVALGFNNSTFTYEELASA 332
Query: 68 TKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHL 127
T+GF+ + ++GQGGFGYVHKGILP GKEIAVKSLKAGSGQGEREFQAE++IISRVHHRHL
Sbjct: 333 TQGFSKDRLLGQGGFGYVHKGILPNGKEIAVKSLKAGSGQGEREFQAEVEIISRVHHRHL 392
Query: 128 VSLVGYCVS-GGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
VSLVGYC + GGQR+LVYEF+PN TLE+HLHGK MDWPTR++IALGSA+GLAYLHED
Sbjct: 393 VSLVGYCSNAGGQRLLVYEFLPNDTLEFHLHGKSGTVMDWPTRLKIALGSAKGLAYLHED 452
Query: 187 CSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSG 246
C P+IIHRDIKA+N+L+D +FEAKVADFGLAKL+ D NTHVSTRVMGTFGY+APEYASSG
Sbjct: 453 CHPKIIHRDIKASNILLDHNFEAKVADFGLAKLSQDNNTHVSTRVMGTFGYLAPEYASSG 512
Query: 247 KLTEKSDVFSFGVMLLELLTGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVDPF 306
KLTEKSDVFSFGVMLLEL+TG+ P+DL+ M++SLVDWARPL R + +DG + ELVDPF
Sbjct: 513 KLTEKSDVFSFGVMLLELITGRGPVDLSGDMEDSLVDWARPLCMR-VAQDGEYGELVDPF 571
Query: 307 LEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTG 366
LE Y+ EM R+ ACAA+++RHS ++R KMSQIVR LEGD SL+DL + + + G
Sbjct: 572 LEHQYEPYEMARMVACAAAAVRHSGRRRPKMSQIVRTLEGDASLDDLDDGVKPKQSSSGG 631
Query: 367 VYTTTGSEYDTMQYNADMAKFRKQIMSSQEF 397
S+Y+ Y A+M KFRK + S+++
Sbjct: 632 ---EGSSDYEMGTYGAEMRKFRKVTLESRDY 659
>At3g18810 protein kinase, putative
Length = 700
Score = 503 bits (1296), Expect = e-143
Identities = 248/372 (66%), Positives = 302/372 (80%), Gaps = 4/372 (1%)
Query: 26 NSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYV 85
NS + SSNYS G P P ++L TFTY+ELA+AT+GF+ ++GQGGFGYV
Sbjct: 293 NSSDFSSNYS-GPHGPSVPPPHPSVALGFNKSTFTYDELAAATQGFSQSRLLGQGGFGYV 351
Query: 86 HKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYE 145
HKGILP GKEIAVKSLKAGSGQGEREFQAE+DIISRVHHR LVSLVGYC++GGQRMLVYE
Sbjct: 352 HKGILPNGKEIAVKSLKAGSGQGEREFQAEVDIISRVHHRFLVSLVGYCIAGGQRMLVYE 411
Query: 146 FVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDD 205
F+PN TLE+HLHGK +DWPTR++IALGSA+GLAYLHEDC PRIIHRDIKA+N+L+D+
Sbjct: 412 FLPNDTLEFHLHGKSGKVLDWPTRLKIALGSAKGLAYLHEDCHPRIIHRDIKASNILLDE 471
Query: 206 SFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELL 265
SFEAKVADFGLAKL+ D THVSTR+MGTFGY+APEYASSGKLT++SDVFSFGVMLLEL+
Sbjct: 472 SFEAKVADFGLAKLSQDNVTHVSTRIMGTFGYLAPEYASSGKLTDRSDVFSFGVMLLELV 531
Query: 266 TGKRPLDLTNAMDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAAS 325
TG+RP+DLT M++SLVDWARP+ A +DG+++ELVDP LE Y+ EM ++ ACAA+
Sbjct: 532 TGRRPVDLTGEMEDSLVDWARPICLNA-AQDGDYSELVDPRLENQYEPHEMAQMVACAAA 590
Query: 326 SIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQYNADMA 385
++RHSA++R KMSQIVRALEGD +L+DL E + G + S+YD+ Y+ADM
Sbjct: 591 AVRHSARRRPKMSQIVRALEGDATLDDLSEGGKAGQSSFLG--RGSSSDYDSSTYSADMK 648
Query: 386 KFRKQIMSSQEF 397
KFRK + S E+
Sbjct: 649 KFRKVALDSHEY 660
>At3g24550 protein kinase, putative
Length = 652
Score = 501 bits (1291), Expect = e-142
Identities = 252/380 (66%), Positives = 304/380 (79%), Gaps = 10/380 (2%)
Query: 21 PHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQG 80
P P S S+YS P SPGL L TFTYEEL+ AT GF+ N++GQG
Sbjct: 233 PPPAFMSSSGGSDYS---DLPVLPPPSPGLVLGFSKSTFTYEELSRATNGFSEANLLGQG 289
Query: 81 GFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQR 140
GFGYVHKGILP+GKE+AVK LKAGSGQGEREFQAE++IISRVHHRHLVSL+GYC++G QR
Sbjct: 290 GFGYVHKGILPSGKEVAVKQLKAGSGQGEREFQAEVEIISRVHHRHLVSLIGYCMAGVQR 349
Query: 141 MLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAAN 200
+LVYEFVPN LE+HLHGKG PTM+W TR++IALGSA+GL+YLHEDC+P+IIHRDIKA+N
Sbjct: 350 LLVYEFVPNNNLEFHLHGKGRPTMEWSTRLKIALGSAKGLSYLHEDCNPKIIHRDIKASN 409
Query: 201 VLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+LID FEAKVADFGLAK+ +DTNTHVSTRVMGTFGY+APEYA+SGKLTEKSDVFSFGV+
Sbjct: 410 ILIDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFGYLAPEYAASGKLTEKSDVFSFGVV 469
Query: 261 LLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
LLEL+TG+RP+D N +D+SLVDWARPLL+RA EE G+F L D + YD +EM R+
Sbjct: 470 LLELITGRRPVDANNVYVDDSLVDWARPLLNRASEE-GDFEGLADSKMGNEYDREEMARM 528
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG--SEYDT 377
ACAA+ +RHSA++R +MSQIVRALEG+VSL DL E M + VY++ G ++YDT
Sbjct: 529 VACAAACVRHSARRRPRMSQIVRALEGNVSLSDLNEGMRPG---HSNVYSSYGGSTDYDT 585
Query: 378 MQYNADMAKFRKQIMSSQEF 397
QYN DM KFRK + +QE+
Sbjct: 586 SQYNDDMIKFRKMALGTQEY 605
>At1g52290 protein kinase, putative
Length = 509
Score = 456 bits (1173), Expect = e-128
Identities = 221/342 (64%), Positives = 280/342 (81%), Gaps = 7/342 (2%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
FTYE+L+ AT F+N N++GQGGFGYVH+G+L G +A+K LK+GSGQGEREFQAEI
Sbjct: 131 FTYEDLSKATSNFSNTNLLGQGGFGYVHRGVLVDGTLVAIKQLKSGSGQGEREFQAEIQT 190
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
ISRVHHRHLVSL+GYC++G QR+LVYEFVPNKTLE+HLH K P M+W RM+IALG+A+
Sbjct: 191 ISRVHHRHLVSLLGYCITGAQRLLVYEFVPNKTLEFHLHEKERPVMEWSKRMKIALGAAK 250
Query: 179 GLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYM 238
GLAYLHEDC+P+ IHRD+KAAN+LIDDS+EAK+ADFGLA+ + DT+THVSTR+MGTFGY+
Sbjct: 251 GLAYLHEDCNPKTIHRDVKAANILIDDSYEAKLADFGLARSSLDTDTHVSTRIMGTFGYL 310
Query: 239 APEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTN--AMDESLVDWARPLLSRALEED 296
APEYASSGKLTEKSDVFS GV+LLEL+TG+RP+D + A D+S+VDWA+PL+ +AL D
Sbjct: 311 APEYASSGKLTEKSDVFSIGVVLLELITGRRPVDKSQPFADDDSIVDWAKPLMIQAL-ND 369
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKES 356
GNF LVDP LE ++D EM R+ ACAA+S+RHSAK+R KMSQIVRA EG++S++DL E
Sbjct: 370 GNFDGLVDPRLENDFDINEMTRMVACAAASVRHSAKRRPKMSQIVRAFEGNISIDDLTEG 429
Query: 357 MIKSPAPQTGVYTTTG-SEYDTMQYNADMAKFRKQIMSSQEF 397
+ Q+ +Y+ G S+Y + QY D+ KF+K S+ F
Sbjct: 430 ---AAPGQSTIYSLDGSSDYSSTQYKEDLKKFKKMAFESKTF 468
>At3g24400 protein kinase, putative
Length = 694
Score = 455 bits (1170), Expect = e-128
Identities = 232/377 (61%), Positives = 293/377 (77%), Gaps = 14/377 (3%)
Query: 26 NSGEMSSNYS-YGMGPPGSMQSSPGLSLTLK--GGTFTYEELASATKGFANENIIGQGGF 82
+SG+ SNYS + PP SPGL+L L GTF YEEL+ AT GF+ N++GQGGF
Sbjct: 287 SSGDYDSNYSDQSVLPP----PSPGLALGLGIYQGTFNYEELSRATNGFSEANLLGQGGF 342
Query: 83 GYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRML 142
GYV KG+L GKE+AVK LK GS QGEREFQAE+ IISRVHHRHLV+LVGYC++ QR+L
Sbjct: 343 GYVFKGMLRNGKEVAVKQLKEGSSQGEREFQAEVGIISRVHHRHLVALVGYCIADAQRLL 402
Query: 143 VYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVL 202
VYEFVPN TLE+HLHGKG PTM+W +R++IA+GSA+GL+YLHE+C+P+IIHRDIKA+N+L
Sbjct: 403 VYEFVPNNTLEFHLHGKGRPTMEWSSRLKIAVGSAKGLSYLHENCNPKIIHRDIKASNIL 462
Query: 203 IDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLL 262
ID FEAKVADFGLAK+ +DTNTHVSTRVMGTFGY+APEYASSGKLTEKSDVFSFGV+LL
Sbjct: 463 IDFKFEAKVADFGLAKIASDTNTHVSTRVMGTFGYLAPEYASSGKLTEKSDVFSFGVVLL 522
Query: 263 ELLTGKRPLDLTNA-MDESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAA 321
EL+TG+RP+D+ N D SLVDWARPLL++ + E GNF +VD L YD +EM R+ A
Sbjct: 523 ELITGRRPIDVNNVHADNSLVDWARPLLNQ-VSELGNFEVVVDKKLNNEYDKEEMARMVA 581
Query: 322 CAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTG--SEYDTMQ 379
CAA+ +R +A +R +M Q+ R LEG++S DL + + + VY ++G ++YD+ Q
Sbjct: 582 CAAACVRSTAPRRPRMDQVARVLEGNISPSDLNQGITPG---HSNVYGSSGGSTDYDSSQ 638
Query: 380 YNADMAKFRKQIMSSQE 396
N M KFRK + +Q+
Sbjct: 639 DNEGMNKFRKVGLETQD 655
>At3g24540 protein kinase, putative
Length = 509
Score = 421 bits (1082), Expect = e-118
Identities = 200/301 (66%), Positives = 252/301 (83%), Gaps = 2/301 (0%)
Query: 58 TFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEID 117
TFTY ELA AT F+ N++G+GGFG+V+KGIL G E+AVK LK GS QGE+EFQAE++
Sbjct: 166 TFTYGELARATNKFSEANLLGEGGFGFVYKGILNNGNEVAVKQLKVGSAQGEKEFQAEVN 225
Query: 118 IISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSA 177
IIS++HHR+LVSLVGYC++G QR+LVYEFVPN TLE+HLHGKG PTM+W R++IA+ S+
Sbjct: 226 IISQIHHRNLVSLVGYCIAGAQRLLVYEFVPNNTLEFHLHGKGRPTMEWSLRLKIAVSSS 285
Query: 178 RGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGY 237
+GL+YLHE+C+P+IIHRDIKAAN+LID FEAKVADFGLAK+ DTNTHVSTRVMGTFGY
Sbjct: 286 KGLSYLHENCNPKIIHRDIKAANILIDFKFEAKVADFGLAKIALDTNTHVSTRVMGTFGY 345
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA-MDESLVDWARPLLSRALEED 296
+APEYA+SGKLTEKSDV+SFGV+LLEL+TG+RP+D N D+SLVDWARPLL +ALEE
Sbjct: 346 LAPEYAASGKLTEKSDVYSFGVVLLELITGRRPVDANNVYADDSLVDWARPLLVQALEE- 404
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKES 356
NF L D L YD +EM R+ ACAA+ +R++A++R +M Q+VR LEG++S DL +
Sbjct: 405 SNFEGLADIKLNNEYDREEMARMVACAAACVRYTARRRPRMDQVVRVLEGNISPSDLNQG 464
Query: 357 M 357
+
Sbjct: 465 I 465
>At1g70460 putative protein kinase
Length = 710
Score = 415 bits (1067), Expect = e-116
Identities = 216/384 (56%), Positives = 276/384 (71%), Gaps = 14/384 (3%)
Query: 9 QNPN-GASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGT-FTYEELAS 66
QNP G SG G ++ S G G S+P ++ G T FTYEEL
Sbjct: 289 QNPTKGYSGPGGYNSQQQSNSGNSFGSQRGGGGYTRSGSAPDSAVMGSGQTHFTYEELTD 348
Query: 67 ATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRH 126
T+GF+ NI+G+GGFG V+KG L GK +AVK LK GSGQG+REF+AE++IISRVHHRH
Sbjct: 349 ITEGFSKHNILGEGGFGCVYKGKLNDGKLVAVKQLKVGSGQGDREFKAEVEIISRVHHRH 408
Query: 127 LVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
LVSLVGYC++ +R+L+YE+VPN+TLE+HLHGKG P ++W R+RIA+GSA+GLAYLHED
Sbjct: 409 LVSLVGYCIADSERLLIYEYVPNQTLEHHLHGKGRPVLEWARRVRIAIGSAKGLAYLHED 468
Query: 187 CSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSG 246
C P+IIHRDIK+AN+L+DD FEA+VADFGLAKL T THVSTRVMGTFGY+APEYA SG
Sbjct: 469 CHPKIIHRDIKSANILLDDEFEAQVADFGLAKLNDSTQTHVSTRVMGTFGYLAPEYAQSG 528
Query: 247 KLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDP 305
KLT++SDVFSFGV+LLEL+TG++P+D + +ESLV+WARPLL +A+ E G+F+ELVD
Sbjct: 529 KLTDRSDVFSFGVVLLELITGRKPVDQYQPLGEESLVEWARPLLHKAI-ETGDFSELVDR 587
Query: 306 FLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQT 365
LE +Y E+ R+ AA+ +RHS KR +M Q+VRAL+ + + D+
Sbjct: 588 RLEKHYVENEVFRMIETAAACVRHSGPKRPRMVQVVRALDSEGDMGDI----------SN 637
Query: 366 GVYTTTGSEYDTMQYNADMAKFRK 389
G S YD+ QYN D KFRK
Sbjct: 638 GNKVGQSSAYDSGQYNNDTMKFRK 661
>At4g32710 putative protein kinase
Length = 731
Score = 403 bits (1036), Expect = e-113
Identities = 212/383 (55%), Positives = 277/383 (71%), Gaps = 13/383 (3%)
Query: 6 RVQQNPNGASGVWGAPHPLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELA 65
+VQ + G +G A H + + + Y G +S ++++ G F+YEEL+
Sbjct: 325 KVQHHRGGNAGTNQA-HVITMPPPIHAKYISSGGCDTKENNSVAKNISMPSGMFSYEELS 383
Query: 66 SATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHR 125
AT GF+ EN++G+GGFGYVHKG+L G E+AVK LK GS QGEREFQAE+D ISRVHH+
Sbjct: 384 KATGGFSEENLLGEGGFGYVHKGVLKNGTEVAVKQLKIGSYQGEREFQAEVDTISRVHHK 443
Query: 126 HLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHE 185
HLVSLVGYCV+G +R+LVYEFVP TLE+HLH ++W R+RIA+G+A+GLAYLHE
Sbjct: 444 HLVSLVGYCVNGDKRLLVYEFVPKDTLEFHLHENRGSVLEWEMRLRIAVGAAKGLAYLHE 503
Query: 186 DCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTN---THVSTRVMGTFGYMAPEY 242
DCSP IIHRDIKAAN+L+D FEAKV+DFGLAK +DTN TH+STRV+GTFGYMAPEY
Sbjct: 504 DCSPTIIHRDIKAANILLDSKFEAKVSDFGLAKFFSDTNSSFTHISTRVVGTFGYMAPEY 563
Query: 243 ASSGKLTEKSDVFSFGVMLLELLTGKRPL-DLTNAMDESLVDWARPLLSRALEEDGNFAE 301
ASSGK+T+KSDV+SFGV+LLEL+TG+ + ++ ++SLVDWARPLL++A+ + +F
Sbjct: 564 ASSGKVTDKSDVYSFGVVLLELITGRPSIFAKDSSTNQSLVDWARPLLTKAISGE-SFDF 622
Query: 302 LVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKE------ 355
LVD LE NYD +M +AACAA+ IR SA R +MSQ+VRALEG+V+L ++E
Sbjct: 623 LVDSRLEKNYDTTQMANMAACAAACIRQSAWLRPRMSQVVRALEGEVALRKVEETGNSVT 682
Query: 356 -SMIKSPAPQTGVYTTTGSEYDT 377
S ++P T Y T +DT
Sbjct: 683 YSSSENPNDITPRYGTNKRRFDT 705
>At5g38560 putative protein
Length = 681
Score = 394 bits (1013), Expect = e-110
Identities = 202/378 (53%), Positives = 270/378 (70%), Gaps = 23/378 (6%)
Query: 23 PLMNSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGF 82
P M S S+Y Y G + + + F+Y+EL+ T GF+ +N++G+GGF
Sbjct: 300 PKMRS-HSGSDYMYASSDSGMVSN--------QRSWFSYDELSQVTSGFSEKNLLGEGGF 350
Query: 83 GYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRML 142
G V+KG+L G+E+AVK LK G QGEREF+AE++IISRVHHRHLV+LVGYC+S R+L
Sbjct: 351 GCVYKGVLSDGREVAVKQLKIGGSQGEREFKAEVEIISRVHHRHLVTLVGYCISEQHRLL 410
Query: 143 VYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVL 202
VY++VPN TL YHLH G P M W TR+R+A G+ARG+AYLHEDC PRIIHRDIK++N+L
Sbjct: 411 VYDYVPNNTLHYHLHAPGRPVMTWETRVRVAAGAARGIAYLHEDCHPRIIHRDIKSSNIL 470
Query: 203 IDDSFEAKVADFGLAKLT--TDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVM 260
+D+SFEA VADFGLAK+ D NTHVSTRVMGTFGYMAPEYA+SGKL+EK+DV+S+GV+
Sbjct: 471 LDNSFEALVADFGLAKIAQELDLNTHVSTRVMGTFGYMAPEYATSGKLSEKADVYSYGVI 530
Query: 261 LLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRL 319
LLEL+TG++P+D + + DESLV+WARPLL +A+E + F ELVDP L N+ EM R+
Sbjct: 531 LLELITGRKPVDTSQPLGDESLVEWARPLLGQAIENE-EFDELVDPRLGKNFIPGEMFRM 589
Query: 320 AACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQ 379
AA+ +RHSA KR KMSQ+VRAL+ +LE+ + G+ +D+ Q
Sbjct: 590 VEAAAACVRHSAAKRPKMSQVVRALD---TLEEATDI-------TNGMRPGQSQVFDSRQ 639
Query: 380 YNADMAKFRKQIMSSQEF 397
+A + F++ SQ++
Sbjct: 640 QSAQIRMFQRMAFGSQDY 657
>At1g68690 protein kinase, putative
Length = 708
Score = 392 bits (1006), Expect = e-109
Identities = 195/357 (54%), Positives = 262/357 (72%), Gaps = 15/357 (4%)
Query: 42 GSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSL 101
GS QS G L F+YEEL AT GF+ EN++G+GGFG V+KGILP G+ +AVK L
Sbjct: 349 GSYQSQSG-GLGNSKALFSYEELVKATNGFSQENLLGEGGFGCVYKGILPDGRVVAVKQL 407
Query: 102 KAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGV 161
K G GQG+REF+AE++ +SR+HHRHLVS+VG+C+SG +R+L+Y++V N L +HLHG+
Sbjct: 408 KIGGGQGDREFKAEVETLSRIHHRHLVSIVGHCISGDRRLLIYDYVSNNDLYFHLHGEK- 466
Query: 162 PTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTT 221
+DW TR++IA G+ARGLAYLHEDC PRIIHRDIK++N+L++D+F+A+V+DFGLA+L
Sbjct: 467 SVLDWATRVKIAAGAARGLAYLHEDCHPRIIHRDIKSSNILLEDNFDARVSDFGLARLAL 526
Query: 222 DTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DES 280
D NTH++TRV+GTFGYMAPEYASSGKLTEKSDVFSFGV+LLEL+TG++P+D + + DES
Sbjct: 527 DCNTHITTRVIGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELITGRKPVDTSQPLGDES 586
Query: 281 LVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQI 340
LV+WARPL+S A+E + F L DP L GNY EM R+ A + +RH A KR +M QI
Sbjct: 587 LVEWARPLISHAIETE-EFDSLADPKLGGNYVESEMFRMIEAAGACVRHLATKRPRMGQI 645
Query: 341 VRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQYNADMAKFRKQIMSSQEF 397
VRA E ++ EDL G+ +++ Q +A++ FR+ SQ +
Sbjct: 646 VRAFE-SLAAEDL----------TNGMRLGESEVFNSAQQSAEIRLFRRMAFGSQNY 691
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 390 bits (1001), Expect = e-109
Identities = 199/376 (52%), Positives = 270/376 (70%), Gaps = 19/376 (5%)
Query: 26 NSGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGT----FTYEELASATKGFANENIIGQGG 81
+S + SS + +G P + +P ++ GT FTYEEL+ T+GF ++G+GG
Sbjct: 324 SSAQNSSPDTNSLGNPKHGRGTPDSAVI---GTSKIHFTYEELSQITEGFCKSFVVGEGG 380
Query: 82 FGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRM 141
FG V+KGIL GK +A+K LK+ S +G REF+AE++IISRVHHRHLVSLVGYC+S R
Sbjct: 381 FGCVYKGILFEGKPVAIKQLKSVSAEGYREFKAEVEIISRVHHRHLVSLVGYCISEQHRF 440
Query: 142 LVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANV 201
L+YEFVPN TL+YHLHGK +P ++W R+RIA+G+A+GLAYLHEDC P+IIHRDIK++N+
Sbjct: 441 LIYEFVPNNTLDYHLHGKNLPVLEWSRRVRIAIGAAKGLAYLHEDCHPKIIHRDIKSSNI 500
Query: 202 LIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVML 261
L+DD FEA+VADFGLA+L +H+STRVMGTFGY+APEYASSGKLT++SDVFSFGV+L
Sbjct: 501 LLDDEFEAQVADFGLARLNDTAQSHISTRVMGTFGYLAPEYASSGKLTDRSDVFSFGVVL 560
Query: 262 LELLTGKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLA 320
LEL+TG++P+D + + +ESLV+WARP L A+E+ G+ +E+VDP LE +Y E+ ++
Sbjct: 561 LELITGRKPVDTSQPLGEESLVEWARPRLIEAIEK-GDISEVVDPRLENDYVESEVYKMI 619
Query: 321 ACAASSIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQY 380
AAS +RHSA KR +M Q+VRAL+ L DL GV YD+ QY
Sbjct: 620 ETAASCVRHSALKRPRMVQVVRALDTRDDLSDL----------TNGVKVGQSRVYDSGQY 669
Query: 381 NADMAKFRKQIMSSQE 396
+ ++ FR+ S +
Sbjct: 670 SNEIRIFRRASEDSSD 685
>At1g23540 putative serine/threonine protein kinase
Length = 731
Score = 389 bits (1000), Expect = e-108
Identities = 213/402 (52%), Positives = 275/402 (67%), Gaps = 44/402 (10%)
Query: 14 ASGVWGAPHPLMNSGEMSSNYSYGMG----PPGSMQSS--PGLSLTLKGGT-FTYEELAS 66
+SG G+ + + S SYG P MQSS P ++ G T F+YEELA
Sbjct: 307 SSGPNGSMYNNSQQQQSSMGNSYGTAGGGYPHHQMQSSGTPDSAILGSGQTHFSYEELAE 366
Query: 67 ATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRH 126
T+GFA +NI+G+GGFG V+KG L GK +AVK LKAGSGQG+REF+AE++IISRVHHRH
Sbjct: 367 ITQGFARKNILGEGGFGCVYKGTLQDGKVVAVKQLKAGSGQGDREFKAEVEIISRVHHRH 426
Query: 127 LVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHED 186
LVSLVGYC+S R+L+YE+V N+TLE+HLH +W R+RIA+GSA+GLAYLHED
Sbjct: 427 LVSLVGYCISDQHRLLIYEYVSNQTLEHHLH-------EWSKRVRIAIGSAKGLAYLHED 479
Query: 187 CSPRIIHRDIKAANVLIDDSFEAK------------------VADFGLAKLTTDTNTHVS 228
C P+IIHRDIK+AN+L+DD +EA+ VADFGLA+L T THVS
Sbjct: 480 CHPKIIHRDIKSANILLDDEYEAQAIMKSSFSLNLSYDCKVLVADFGLARLNDTTQTHVS 539
Query: 229 TRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARP 287
TRVMGTFGY+APEYASSGKLT++SDVFSFGV+LLEL+TG++P+D T + +ESLV+WARP
Sbjct: 540 TRVMGTFGYLAPEYASSGKLTDRSDVFSFGVVLLELVTGRKPVDQTQPLGEESLVEWARP 599
Query: 288 LLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGD 347
LL +A+ E G+ +EL+D LE Y E+ R+ AA+ +RHS KR +M Q+VRAL+ D
Sbjct: 600 LLLKAI-ETGDLSELIDTRLEKRYVEHEVFRMIETAAACVRHSGPKRPRMVQVVRALDCD 658
Query: 348 VSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQYNADMAKFRK 389
D+ G+ + YD+ QYN D+ KFRK
Sbjct: 659 GDSGDI----------SNGIKIGQSTTYDSGQYNEDIMKFRK 690
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 388 bits (996), Expect = e-108
Identities = 203/372 (54%), Positives = 264/372 (70%), Gaps = 24/372 (6%)
Query: 28 GEMSSNYSY-GMGPPGSMQSSPGLSLTLKGGTFTYEELASATKGFANENIIGQGGFGYVH 86
G SSN +Y PG S L F+YEEL AT GF++EN++G+GGFG V+
Sbjct: 394 GNRSSNRTYLSQSEPGGFGQSREL--------FSYEELVIATNGFSDENLLGEGGFGRVY 445
Query: 87 KGILPTGKEIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEF 146
KG+LP + +AVK LK G GQG+REF+AE+D ISRVHHR+L+S+VGYC+S +R+L+Y++
Sbjct: 446 KGVLPDERVVAVKQLKIGGGQGDREFKAEVDTISRVHHRNLLSMVGYCISENRRLLIYDY 505
Query: 147 VPNKTLEYHLHGKGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDS 206
VPN L +HLHG P +DW TR++IA G+ARGLAYLHEDC PRIIHRDIK++N+L++++
Sbjct: 506 VPNNNLYFHLHG--TPGLDWATRVKIAAGAARGLAYLHEDCHPRIIHRDIKSSNILLENN 563
Query: 207 FEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLT 266
F A V+DFGLAKL D NTH++TRVMGTFGYMAPEYASSGKLTEKSDVFSFGV+LLEL+T
Sbjct: 564 FHALVSDFGLAKLALDCNTHITTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVVLLELIT 623
Query: 267 GKRPLDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAAS 325
G++P+D + + DESLV+WARPLLS A E + F L DP L NY EM R+ AA+
Sbjct: 624 GRKPVDASQPLGDESLVEWARPLLSNATETE-EFTALADPKLGRNYVGVEMFRMIEAAAA 682
Query: 326 SIRHSAKKRSKMSQIVRALEGDVSLEDLKESMIKSPAPQTGVYTTTGSEYDTMQYNADMA 385
IRHSA KR +MSQIVRA + ++ EDL G+ ++ Q +A++
Sbjct: 683 CIRHSATKRPRMSQIVRAFD-SLAEEDL----------TNGMRLGESEIINSAQQSAEIR 731
Query: 386 KFRKQIMSSQEF 397
FR+ SQ +
Sbjct: 732 LFRRMAFGSQNY 743
>At1g70450 putative protein kinase
Length = 394
Score = 358 bits (920), Expect = 2e-99
Identities = 178/318 (55%), Positives = 241/318 (74%), Gaps = 6/318 (1%)
Query: 41 PGSMQSSPGLSLTLKGGT---FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIA 97
P S ++P + + G FTYEEL T+GF+ +NI+G+GGFG V+KG L GK +A
Sbjct: 16 PSSSPTAPSVDSAVMGSGQTHFTYEELEDITEGFSKQNILGEGGFGCVYKGKLKDGKLVA 75
Query: 98 VKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLH 157
VK LK GSGQG+REF+AE++IISRVHHRHLVSLVGYC++ +R+L+YE+VPN+TLE+HLH
Sbjct: 76 VKQLKVGSGQGDREFKAEVEIISRVHHRHLVSLVGYCIADSERLLIYEYVPNQTLEHHLH 135
Query: 158 GKGVPTMDWPTRMRIALGSARGLAYLHEDCS-PRIIHRDIKAANVLIDDSFEAKVADFGL 216
GKG P ++W R+RIA+ + + S P+IIHRDIK+AN+L+DD FE +VADFGL
Sbjct: 136 GKGRPVLEWARRVRIAIVLPKVWRICTKTVSHPKIIHRDIKSANILLDDEFEVQVADFGL 195
Query: 217 AKLTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNA 276
AK+ T THVSTRVMGTFGY+APEYA SG+LT++SDVFSFGV+LLEL+TG++P+D
Sbjct: 196 AKVNDTTQTHVSTRVMGTFGYLAPEYAQSGQLTDRSDVFSFGVVLLELITGRKPVDRNQP 255
Query: 277 M-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRS 335
+ +ESLV WARPLL +A+ E G+F+ELVD LE +Y E+ R+ AA+ +R+S KR
Sbjct: 256 LGEESLVGWARPLLKKAI-ETGDFSELVDRRLEKHYVKNEVFRMIETAAACVRYSGPKRP 314
Query: 336 KMSQIVRALEGDVSLEDL 353
+M Q++RAL+ + + D+
Sbjct: 315 RMVQVLRALDSEGDMGDI 332
>At2g20300 protein kinase like protein
Length = 744
Score = 301 bits (770), Expect = 5e-82
Identities = 159/347 (45%), Positives = 232/347 (66%), Gaps = 14/347 (4%)
Query: 4 VVRVQQNPNGASGVWGAPHPLMN----SGEMSSNYSYGMGPPGSMQSSPGLSLTLKGGTF 59
+V+ ++ ++ V A P +N +G M S+ + G M S +L++K TF
Sbjct: 280 IVKWKKIGKSSNAVGPALAPSINKRPGAGSMFSSSARSSGSDSLMSSMATCALSVK--TF 337
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
T EL AT F+ + ++G+GGFG V++G + G E+AVK L + +REF AE++++
Sbjct: 338 TLSELEKATDRFSAKRVLGEGGFGRVYQGSMEDGTEVAVKLLTRDNQNRDREFIAEVEML 397
Query: 120 SRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSARG 179
SR+HHR+LV L+G C+ G R L+YE V N ++E HLH T+DW R++IALG+ARG
Sbjct: 398 SRLHHRNLVKLIGICIEGRTRCLIYELVHNGSVESHLHEG---TLDWDARLKIALGAARG 454
Query: 180 LAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGYMA 239
LAYLHED +PR+IHRD KA+NVL++D F KV+DFGLA+ T+ + H+STRVMGTFGY+A
Sbjct: 455 LAYLHEDSNPRVIHRDFKASNVLLEDDFTPKVSDFGLAREATEGSQHISTRVMGTFGYVA 514
Query: 240 PEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTN-AMDESLVDWARPLLSRALEEDGN 298
PEYA +G L KSDV+S+GV+LLELLTG+RP+D++ + +E+LV WARPLL+
Sbjct: 515 PEYAMTGHLLVKSDVYSYGVVLLELLTGRRPVDMSQPSGEENLVTWARPLLANR----EG 570
Query: 299 FAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
+LVDP L G Y+ +M ++AA A+ + R M ++V+AL+
Sbjct: 571 LEQLVDPALAGTYNFDDMAKVAAIASMCVHQEVSHRPFMGEVVQALK 617
>At4g02010 putative NAK-like ser/thr protein kinase
Length = 725
Score = 290 bits (741), Expect = 1e-78
Identities = 151/292 (51%), Positives = 207/292 (70%), Gaps = 10/292 (3%)
Query: 60 TYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDII 119
+YEEL AT F + +I+G+GGFG V++GIL G +A+K L +G QG++EFQ EID++
Sbjct: 369 SYEELKEATSNFESASILGEGGFGKVYRGILADGTAVAIKKLTSGGPQGDKEFQVEIDML 428
Query: 120 SRVHHRHLVSLVGYCVS--GGQRMLVYEFVPNKTLEYHLHGK-GVP-TMDWPTRMRIALG 175
SR+HHR+LV LVGY S Q +L YE VPN +LE LHG G+ +DW TRM+IAL
Sbjct: 429 SRLHHRNLVKLVGYYSSRDSSQHLLCYELVPNGSLEAWLHGPLGLNCPLDWDTRMKIALD 488
Query: 176 SARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVADFGLAKLTTD-TNTHVSTRVMGT 234
+ARGLAYLHED P +IHRD KA+N+L++++F AKVADFGLAK + H+STRVMGT
Sbjct: 489 AARGLAYLHEDSQPSVIHRDFKASNILLENNFNAKVADFGLAKQAPEGRGNHLSTRVMGT 548
Query: 235 FGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTN-AMDESLVDWARPLLSRAL 293
FGY+APEYA +G L KSDV+S+GV+LLELLTG++P+D++ + E+LV W RP+ L
Sbjct: 549 FGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPSGQENLVTWTRPV----L 604
Query: 294 EEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALE 345
+ ELVD LEG Y ++ IR+ AA+ + A +R M ++V++L+
Sbjct: 605 RDKDRLEELVDSRLEGKYPKEDFIRVCTIAAACVAPEASQRPTMGEVVQSLK 656
>At5g56890 unknown protein
Length = 1113
Score = 286 bits (732), Expect = 1e-77
Identities = 148/316 (46%), Positives = 208/316 (64%), Gaps = 16/316 (5%)
Query: 42 GSMQSSPGLSL-------TLKGGTFTYEELASATKGFANENIIGQGGFGYVHKGILPTGK 94
GS SS LS TL TFT E+ AT F ++G+GGFG V++G+ G
Sbjct: 687 GSRFSSTSLSFESSIAPFTLSAKTFTASEIMKATNNFDESRVLGEGGFGRVYEGVFDDGT 746
Query: 95 EIAVKSLKAGSGQGEREFQAEIDIISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEY 154
++AVK LK QG REF AE++++SR+HHR+LV+L+G C+ R LVYE +PN ++E
Sbjct: 747 KVAVKVLKRDDQQGSREFLAEVEMLSRLHHRNLVNLIGICIEDRNRSLVYELIPNGSVES 806
Query: 155 HLHG--KGVPTMDWPTRMRIALGSARGLAYLHEDCSPRIIHRDIKAANVLIDDSFEAKVA 212
HLHG K +DW R++IALG+ARGLAYLHED SPR+IHRD K++N+L+++ F KV+
Sbjct: 807 HLHGIDKASSPLDWDARLKIALGAARGLAYLHEDSSPRVIHRDFKSSNILLENDFTPKVS 866
Query: 213 DFGLAK--LTTDTNTHVSTRVMGTFGYMAPEYASSGKLTEKSDVFSFGVMLLELLTGKRP 270
DFGLA+ L + N H+STRVMGTFGY+APEYA +G L KSDV+S+GV+LLELLTG++P
Sbjct: 867 DFGLARNALDDEDNRHISTRVMGTFGYVAPEYAMTGHLLVKSDVYSYGVVLLELLTGRKP 926
Query: 271 LDLTNAM-DESLVDWARPLLSRALEEDGNFAELVDPFLEGNYDHQEMIRLAACAASSIRH 329
+D++ E+LV W RP L+ A A ++D L + ++AA A+ ++
Sbjct: 927 VDMSQPPGQENLVSWTRPFLTSA----EGLAAIIDQSLGPEISFDSIAKVAAIASMCVQP 982
Query: 330 SAKKRSKMSQIVRALE 345
R M ++V+AL+
Sbjct: 983 EVSHRPFMGEVVQALK 998
>At5g56790 putative protein kinase
Length = 669
Score = 280 bits (717), Expect = 8e-76
Identities = 141/297 (47%), Positives = 199/297 (66%), Gaps = 7/297 (2%)
Query: 59 FTYEELASATKGFANENIIGQGGFGYVHKGILPTGKEIAVKSLKAGSGQGEREFQAEIDI 118
FTY EL +ATKGF+ + + +GGFG VH G LP G+ IAVK K S QG+REF +E+++
Sbjct: 378 FTYSELETATKGFSKGSFLAEGGFGSVHLGTLPDGQIIAVKQYKIASTQGDREFCSEVEV 437
Query: 119 ISRVHHRHLVSLVGYCVSGGQRMLVYEFVPNKTLEYHLHGKGVPTMDWPTRMRIALGSAR 178
+S HR++V L+G CV G+R+LVYE++ N +L HL+G G + W R +IA+G+AR
Sbjct: 438 LSCAQHRNVVMLIGLCVEDGKRLLVYEYICNGSLHSHLYGMGREPLGWSARQKIAVGAAR 497
Query: 179 GLAYLHEDCSPR-IIHRDIKAANVLIDDSFEAKVADFGLAKLTTDTNTHVSTRVMGTFGY 237
GL YLHE+C I+HRD++ N+L+ FE V DFGLA+ + + V TRV+GTFGY
Sbjct: 498 GLRYLHEECRVGCIVHRDMRPNNILLTHDFEPLVGDFGLARWQPEGDKGVETRVIGTFGY 557
Query: 238 MAPEYASSGKLTEKSDVFSFGVMLLELLTGKRPLDLTNAM-DESLVDWARPLLSRALEED 296
+APEYA SG++TEK+DV+SFGV+L+EL+TG++ +D+ + L +WARPLL +
Sbjct: 558 LAPEYAQSGQITEKADVYSFGVVLVELITGRKAMDIKRPKGQQCLTEWARPLLQKQA--- 614
Query: 297 GNFAELVDPFLEGNYDHQEMIRLAACAASSIRHSAKKRSKMSQIVRALEGDVSLEDL 353
EL+DP L Y QE+ +A CA IR R +MSQ++R LEGDV + +
Sbjct: 615 --INELLDPRLMNCYCEQEVYCMALCAYLCIRRDPNSRPRMSQVLRMLEGDVVMNPI 669
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,958,929
Number of Sequences: 26719
Number of extensions: 397896
Number of successful extensions: 4079
Number of sequences better than 10.0: 997
Number of HSP's better than 10.0 without gapping: 875
Number of HSP's successfully gapped in prelim test: 122
Number of HSP's that attempted gapping in prelim test: 1107
Number of HSP's gapped (non-prelim): 1100
length of query: 398
length of database: 11,318,596
effective HSP length: 101
effective length of query: 297
effective length of database: 8,619,977
effective search space: 2560133169
effective search space used: 2560133169
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 61 (28.1 bits)
Medicago: description of AC148484.8


